

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0333b.9
(488 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
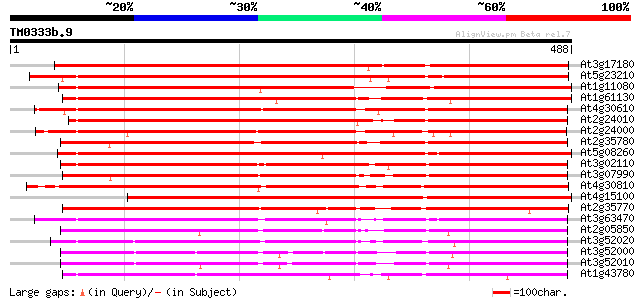
Score E
Sequences producing significant alignments: (bits) Value
At3g17180 putative serine carboxypeptidase II 611 e-175
At5g23210 serine carboxypeptidase II-like protein 485 e-137
At1g11080 Serine carboxypeptidase isolog 476 e-135
At1g61130 452 e-127
At4g30610 SERINE CARBOXYPEPTIDASE II-like protein 445 e-125
At2g24010 putative serine carboxypeptidase II 426 e-119
At2g24000 putative serine carboxypeptidase II 426 e-119
At2g35780 serine carboxypeptidase II like protein 419 e-117
At5g08260 serine-type carboxypeptidase II-like protein 417 e-117
At3g02110 serine carboxypeptidase II like protein 417 e-117
At3g07990 putative serine carboxypeptidase II 416 e-116
At4g30810 serine carboxypeptidase II like protein 408 e-114
At4g15100 hydroxynitrile lyase like protein 400 e-112
At2g35770 serine carboxypeptidase II like protein 387 e-108
At3g63470 serin carboxypeptidase - like protein 373 e-103
At2g05850 putative serine carboxypeptidase II 338 5e-93
At3g52020 serine-type carboxypeptidase-like protein 337 9e-93
At3g52000 serine-type carboxypeptidase like protein 325 5e-89
At3g52010 serine-type carboxypeptidase-like protein 318 4e-87
At1g43780 serine carboxypeptidase II, putative 313 1e-85
>At3g17180 putative serine carboxypeptidase II
Length = 478
Score = 611 bits (1576), Expect = e-175
Identities = 299/452 (66%), Positives = 355/452 (78%), Gaps = 10/452 (2%)
Query: 40 NVGANESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSE-PSKKPLLLWL 98
++ A SD++V+LP P +P +SHFSGY+ VN+++ R+LF+WFFEA SE PS +PL+LWL
Sbjct: 30 HIEAQNSDKVVNLPEQPLNPKISHFSGYVNVNQENTRSLFFWFFEALSESPSTRPLVLWL 89
Query: 99 NGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSS 158
NGGPGCSS+GYGAA E+GP V +NG L+FN YSW QEAN+LF+ESPVGVGFSYTN+SS
Sbjct: 90 NGGPGCSSIGYGAASELGPFRVVENGTSLSFNQYSWVQEANMLFLESPVGVGFSYTNSSS 149
Query: 159 DLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKN 218
DL L D FVAEDAYNF+V W R+PQ+KSR FFI+GESY GHY PQLAELI+DRNK +
Sbjct: 150 DLENLNDAFVAEDAYNFMVAWFARYPQYKSRDFFIAGESYAGHYSPQLAELIYDRNKVQP 209
Query: 219 KYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQWSNECN 278
K FINLKGFIVGNP T+D YD KG+LEYAWSHAVISD YD AK CDFK WS CN
Sbjct: 210 KDSFINLKGFIVGNPLTDDEYDNKGILEYAWSHAVISDHLYDSAKHNCDFKSSNWSEPCN 269
Query: 279 KAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIA--SEGLEQLTKGRNDYRMKRKRIFGGYD 336
AMN VF Y EIDIYNIYAPKC NS+S + G+ + D+ KR R F GYD
Sbjct: 270 VAMNTVFTKYKEIDIYNIYAPKCISNSSSGASYLGFGVNDKSPAVKDW-FKRVRWFEGYD 328
Query: 337 PCYSTYAEKYFNRIDVQSSFHVNTERGNTNIT-WEVCNNSILQTYNFSVFSILPIYTKLI 395
PCYS YAE+YFNR+DV+ S H T N+ W+VCN+SILQTY+F+V S+LP Y+KLI
Sbjct: 329 PCYSNYAEEYFNRVDVRLSLHATTR----NVARWKVCNDSILQTYHFTVSSMLPTYSKLI 384
Query: 396 KGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYE-GLTYV 454
K GLKIW+YSGDADGRVPVIG+RYCVEALG+ +KS WRSW+ ++QVGGRI EYE GLT+V
Sbjct: 385 KAGLKIWVYSGDADGRVPVIGSRYCVEALGISVKSEWRSWFHNHQVGGRITEYEGGLTFV 444
Query: 455 TVRGAGHLVPLNKPKEALSLIHSFLAGDRLPT 486
TVRGAGHLVPLNKP+EAL+L SFL G LP+
Sbjct: 445 TVRGAGHLVPLNKPEEALALFRSFLNGQELPS 476
>At5g23210 serine carboxypeptidase II-like protein
Length = 499
Score = 485 bits (1248), Expect = e-137
Identities = 247/491 (50%), Positives = 317/491 (64%), Gaps = 26/491 (5%)
Query: 18 ILSVHFLCFFLLTTSFIKTYGINVGAN-----------ESDRIVDLPGVPSSPSVSHFSG 66
+L + + F L +TS K N G N +DR+ +LPG P ++G
Sbjct: 10 VLVLFLVSFLLGSTSAEKLCSDNDGDNGCFRSRVLAAQRADRVKELPGQPPV-KFRQYAG 68
Query: 67 YITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSSVGYGAAIEIGPLLV-NKNGE 125
Y+TVNE HGRALFYWFFEA PSKKP+LLWLNGGPGCSS+G+GAA E+GP N +
Sbjct: 69 YVTVNETHGRALFYWFFEATQNPSKKPVLLWLNGGPGCSSIGFGAAEELGPFFPQNSSQP 128
Query: 126 GLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQ 185
L NPYSWN+ ANLLF+ESPVGVGFSYTNTS D+ L D A D+YNFLVNW +RFPQ
Sbjct: 129 KLKLNPYSWNKAANLLFLESPVGVGFSYTNTSRDIKQLGDTVTARDSYNFLVNWFKRFPQ 188
Query: 186 FKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFINLKGFIVGNPETEDYYDYKGLL 245
+KS F+I+GESY GHY+PQL+ELI+ NK +K FINLKG ++GN +D D KG++
Sbjct: 189 YKSHDFYIAGESYAGHYVPQLSELIYKENKIASKKDFINLKGLMIGNALLDDETDQKGMI 248
Query: 246 EYAWSHAVISDQQYDKAKQVCDFKQFQWSNECNKAMNEVFQDYSEIDIYNIYAPKCRLNS 305
EYAW HAVISD Y+K + CDFKQ + ECN A++E F Y +D+Y++YAPKC S
Sbjct: 249 EYAWDHAVISDALYEKVNKNCDFKQKLVTKECNDALDEYFDVYKILDMYSLYAPKCVPTS 308
Query: 306 TSAIASE---GLEQLTKGRNDYRMKR-------KRIFGGYDPCYSTYAEKYFNRIDVQSS 355
T++ S G L R+ R + +R+ GYDPC S Y EKY NR DVQ +
Sbjct: 309 TNSSTSHSVAGNRPLPAFRSILRPRLISHNEGWRRMAAGYDPCASEYTEKYMNRKDVQEA 368
Query: 356 FHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVI 415
H N N + W C++++ ++ + S+LP L+ GL++W++SGD DGR+PV
Sbjct: 369 LHANVT--NISYPWTHCSDTV-SFWSDAPASMLPTLRTLVSAGLRVWVFSGDTDGRIPVT 425
Query: 416 GTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLI 475
TRY ++ LGL + W WY QVGG VEY+GL +VTVRGAGH VP KP+EAL LI
Sbjct: 426 ATRYSLKKLGLKIVQDWTPWYTKLQVGGWTVEYDGLMFVTVRGAGHQVPTFKPREALQLI 485
Query: 476 HSFLAGDRLPT 486
H FL +LPT
Sbjct: 486 HHFLGNKKLPT 496
>At1g11080 Serine carboxypeptidase isolog
Length = 465
Score = 476 bits (1226), Expect = e-135
Identities = 236/451 (52%), Positives = 298/451 (65%), Gaps = 35/451 (7%)
Query: 43 ANESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGP 102
ANE D + LPG P S H++GY+ V+E +GRA+FYWFFEA P +KPL+LWLNGGP
Sbjct: 45 ANEQDLVTGLPGQPDV-SFRHYAGYVPVDESNGRAMFYWFFEAMDLPKEKPLVLWLNGGP 103
Query: 103 GCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTI 162
GCSSVGYGA EIGP LV+ NG GLNFNPY+WN+EAN+LF+ESPVGVGFSY+NTSSD
Sbjct: 104 GCSSVGYGATQEIGPFLVDTNGNGLNFNPYAWNKEANMLFLESPVGVGFSYSNTSSDYQK 163
Query: 163 LEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKD--KNKY 220
L D+F A DAY FL NW ++FP+ K F+I+GESY G Y+P+LAE+++D N + KN
Sbjct: 164 LGDDFTARDAYTFLCNWFEKFPEHKENTFYIAGESYAGKYVPELAEVVYDNNNNNKKNGS 223
Query: 221 PF-INLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQ-WSN-EC 277
F INLKG ++GNPET D D++G ++YAWSHAVISD+ + + C+F WSN EC
Sbjct: 224 SFHINLKGILLGNPETSDAEDWRGWVDYAWSHAVISDETHRIITRTCNFSSDNTWSNDEC 283
Query: 278 NKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDP 337
N+A+ EV + Y EIDIY+IY R+ GGYDP
Sbjct: 284 NEAVAEVLKQYHEIDIYSIYTSM---------------------------PPRLMGGYDP 316
Query: 338 CYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKG 397
C YA ++NR DVQ S H + N W +CN I + S S+LPIY KLI G
Sbjct: 317 CLDDYARVFYNRADVQKSLHASDGVNLKN--WSICNMEIFNNWTGSNPSVLPIYEKLIAG 374
Query: 398 GLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVR 457
GL+IW+YSGD DGRVPV+ TRY + AL LP+K++WR WY + QV G + EYEGLT+ T R
Sbjct: 375 GLRIWVYSGDTDGRVPVLATRYSLNALELPIKTAWRPWYHEKQVSGWLQEYEGLTFATFR 434
Query: 458 GAGHLVPLNKPKEALSLIHSFLAGDRLPTRR 488
GAGH VP KP +L+ +FL+G P R
Sbjct: 435 GAGHAVPCFKPSSSLAFFSAFLSGVPPPPSR 465
>At1g61130
Length = 476
Score = 452 bits (1163), Expect = e-127
Identities = 226/460 (49%), Positives = 298/460 (64%), Gaps = 31/460 (6%)
Query: 47 DRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSS 106
D + + PG P S H++GY+TVN GRALFYWFFEA + P+ KPL+LWLNGGPGCSS
Sbjct: 30 DLVTNFPGQPKV-SFRHYAGYVTVNIISGRALFYWFFEAMTHPNVKPLVLWLNGGPGCSS 88
Query: 107 VGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDN 166
VGYGA EIGP LV+ G L FNPY+WN+EAN+LF+ESP GVGFSY+NTSSD L D+
Sbjct: 89 VGYGATQEIGPFLVDNKGNSLKFNPYAWNKEANILFLESPAGVGFSYSNTSSDYRKLGDD 148
Query: 167 FVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKD-KNKYPFINL 225
F A D+Y FL W RFP +K + FFI+GESY G Y+P+LAE+I+D+NKD +N INL
Sbjct: 149 FTARDSYTFLQKWFLRFPAYKEKDFFIAGESYAGKYVPELAEVIYDKNKDNENLSLHINL 208
Query: 226 KGFIV-------------GNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDF-KQF 271
KG +V GNP T D+ G ++YAW+HAV+SD+ Y KQ C+F
Sbjct: 209 KGILVLNTFDLNFKDIFLGNPLTSYAEDWTGWVDYAWNHAVVSDETYRVIKQSCNFSSDT 268
Query: 272 QWS-NECNKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKR 330
W +C + ++E+ + Y EID +++Y P C ++ +S + S +Y+ R
Sbjct: 269 TWDVKDCKEGVDEILKQYKEIDQFSLYTPIC-MHHSSKVDSYA---------NYKTTIPR 318
Query: 331 IFGGYDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNF--SVFSIL 388
+F G+DPC YA+ ++NR DVQ + H G W +CN+ IL +N+ S S+L
Sbjct: 319 LFDGFDPCLDDYAKVFYNRADVQKALHATD--GVHLKNWTICNDDILNHWNWTDSKRSVL 376
Query: 389 PIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEY 448
PIY KLI GG ++W+YSGD DGRVPV+ TRYC+ L LP+K++WR WY + QV G EY
Sbjct: 377 PIYKKLIAGGFRVWVYSGDTDGRVPVLSTRYCINKLELPIKTAWRPWYHETQVSGWFQEY 436
Query: 449 EGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLPTRR 488
EGLT+ T RGAGH VP KP E+L+ +FL G P R
Sbjct: 437 EGLTFATFRGAGHDVPSFKPSESLAFFSAFLNGVPPPLSR 476
>At4g30610 SERINE CARBOXYPEPTIDASE II-like protein
Length = 465
Score = 445 bits (1144), Expect = e-125
Identities = 231/470 (49%), Positives = 302/470 (64%), Gaps = 18/470 (3%)
Query: 22 HFLCFFLLTTSFIKTYGINVGANES--DRIVDLPGVPSSPSVSHFSGYITVNEDHGRALF 79
HF+ F LL T+ + + E DRI LPG P + S +SGY+ VN+ HGRALF
Sbjct: 5 HFI-FLLLVALLSTTFPSSSSSREQEKDRIKALPGQPKV-AFSQYSGYVNVNQSHGRALF 62
Query: 80 YWFFEAQS-EPSKKPLLLWLNGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEA 138
YW E+ S P KPLLLWLNGGPGCSS+ YGA+ EIGP +NK G L N ++WN++A
Sbjct: 63 YWLTESSSPSPHTKPLLLWLNGGPGCSSIAYGASEEIGPFRINKTGSNLYLNKFAWNKDA 122
Query: 139 NLLFVESPVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESY 198
NLLF+ESP GVG+SYTNTSSDL D A+D FL+ WL RFPQ+K R F+I+GESY
Sbjct: 123 NLLFLESPAGVGYSYTNTSSDLKDSGDERTAQDNLIFLIKWLSRFPQYKYRDFYIAGESY 182
Query: 199 GGHYIPQLAELIFDRNKDKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQ 258
GHY+PQLA+ I D NK +K P INLKGF+VGN T++ YD G + Y W+HA+ISD+
Sbjct: 183 AGHYVPQLAKKINDYNKAFSK-PIINLKGFLVGNAVTDNQYDSIGTVTYWWTHAIISDKS 241
Query: 259 YDKAKQVCDFKQFQWSNECNKAMNEVF-QDYSEIDIYNIYAPKCRLNSTSAIASEGLEQL 317
Y + C+F + S++C+ A+N ++ +ID Y+IY P C +A++ +
Sbjct: 242 YKSILKYCNFTVERVSDDCDNAVNYAMNHEFGDIDQYSIYTPTC-------VAAQQKKNT 294
Query: 318 TK--GRNDYRMKRKRIFGGYDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNS 375
T R + R+R+ GYDPC +YAEKYFNR DVQ + H N W C++
Sbjct: 295 TGFFVRMKNTLLRRRLVSGYDPCTESYAEKYFNRPDVQRAMHANVT--GIRYKWTACSDV 352
Query: 376 ILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSW 435
+++T+ S ++LPIY +L GL+IWI+SGD D VPV TR+ + L LP+K+ W W
Sbjct: 353 LIKTWKDSDKTMLPIYKELAASGLRIWIFSGDTDSVVPVTATRFSLSHLNLPVKTRWYPW 412
Query: 436 YLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
Y DNQVGG Y+GLT+ TVRGAGH VPL +PK AL L SFLAG LP
Sbjct: 413 YTDNQVGGWTEVYKGLTFATVRGAGHEVPLFEPKRALILFRSFLAGKELP 462
>At2g24010 putative serine carboxypeptidase II
Length = 425
Score = 426 bits (1096), Expect = e-119
Identities = 213/436 (48%), Positives = 279/436 (63%), Gaps = 20/436 (4%)
Query: 52 LPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSSVGYGA 111
LPG P S FSGY+TVNE HGR+LFYW E+ S KPLLLWLNGGPGCSS+GYGA
Sbjct: 5 LPGQPQV-GFSQFSGYVTVNESHGRSLFYWLTESPSSSHTKPLLLWLNGGPGCSSIGYGA 63
Query: 112 AIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDNFVAED 171
+ EIGP +NK G L N ++WN EAN+LF+ESP GVGFSYTNTSSDL D A++
Sbjct: 64 SEEIGPFRINKTGSNLYLNKFTWNTEANILFLESPAGVGFSYTNTSSDLKDSGDERTAQE 123
Query: 172 AYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFINLKGFIVG 231
FL+ W+ RFPQ++ R F+I GESY GHY+PQLA+ I NK N P INLKGF+VG
Sbjct: 124 NLIFLIKWMSRFPQYQYRDFYIVGESYAGHYVPQLAKKIHLYNKAFNNTPIINLKGFMVG 183
Query: 232 NPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQWSNECNKAMNEVFQDYSEI 291
N + + +YD G YAWSHA+ISD+ Y + C F + S++CN A+ ++++ ++
Sbjct: 184 NGDMDKHYDRLGAAMYAWSHAMISDKTYKSILKHCSFTADKTSDKCNWALYFAYREFGKV 243
Query: 292 DIYNIYAPKC--RLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPCYSTYAEKYFNR 349
+ Y+IY+P C + N T + L + +Y YDPC +YAE Y+NR
Sbjct: 244 NGYSIYSPSCVHQTNQTKFLHGRLLVE------EYE---------YDPCTESYAEIYYNR 288
Query: 350 IDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDAD 409
DVQ + H N + W +CN + + S FS+LPIY +L GL+IW++SGD D
Sbjct: 289 PDVQRAMHANLT--SIPYKWTLCNMVVNNNWKDSEFSMLPIYKELTAAGLRIWVFSGDTD 346
Query: 410 GRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPK 469
VPV GTR + L LP+K+ W WY + QVGG YEGLT+ T+RGAGH VP+ +P+
Sbjct: 347 AVVPVTGTRLALSKLNLPVKTPWYPWYSEKQVGGWTEVYEGLTFATIRGAGHEVPVLQPE 406
Query: 470 EALSLIHSFLAGDRLP 485
AL+L+ SFLAG LP
Sbjct: 407 RALTLLRSFLAGKELP 422
>At2g24000 putative serine carboxypeptidase II
Length = 474
Score = 426 bits (1095), Expect = e-119
Identities = 232/479 (48%), Positives = 296/479 (61%), Gaps = 33/479 (6%)
Query: 23 FLCFFLLTTSFIKTYGINVGANESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWF 82
FL F LL+ + T + E DRI LPG P S FSGY+TVNE HGR+LFYW
Sbjct: 8 FLLFVLLS---LATSSTSTKEQEEDRIKALPGQPKV-GFSQFSGYVTVNESHGRSLFYWL 63
Query: 83 FEAQSE-PSKKPLLLWLNGG----------PGCSSVGYGAAIEIGPLLVNKNGEGLNFNP 131
E+ S P KPLLLWLNGG PGCSS+ YGA+ EIGP ++K G L N
Sbjct: 64 TESSSHSPHTKPLLLWLNGGWIFFLPTFPRPGCSSIAYGASEEIGPFRISKTGCNLYLNN 123
Query: 132 YSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGF 191
+SWN EANLLF+ESPVGVGFSYTNTSSD D A++ FL++W+ RFPQ++ R F
Sbjct: 124 FSWNTEANLLFLESPVGVGFSYTNTSSDFEESGDERTAQENLIFLISWMSRFPQYRYRDF 183
Query: 192 FISGESYGGHYIPQLAELIFDRNKDKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSH 251
+I GESY GHY+PQLA+ I + N + K P INLKGF+VGNPE + D G + Y WSH
Sbjct: 184 YIVGESYAGHYVPQLAQKIHEYN-NAYKNPVINLKGFMVGNPEMDKNNDRLGTITYWWSH 242
Query: 252 AVISDQQYDKAKQVCDFKQFQWSNECNKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIAS 311
A+ISD Y++ + CDF ++S EC+ A+ D+ +ID Y+IY PKC +
Sbjct: 243 AMISDASYNRILKNCDFTADRFSKECDSAIYVAAADFGDIDQYSIYTPKC-------VPP 295
Query: 312 EGLEQLTKGRNDYRMKRKRIF--GGYDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNI-- 367
+ TK +M + F YDPC YAE Y+NR +VQ + H N +T I
Sbjct: 296 QDQTNQTKFEQMMQMHTTKRFLEDQYDPCTENYAEIYYNRPEVQRAMHAN----HTAIPY 351
Query: 368 TWEVCNNSILQTYNF--SVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALG 425
W C++S+ +N+ S S+LPIY +LI GL+IW+YSGD D +PV TRY + L
Sbjct: 352 KWTACSDSVFNNWNWRDSDNSMLPIYKELIAAGLRIWVYSGDTDSVIPVTATRYSLGKLN 411
Query: 426 LPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRL 484
L +K+ W WY NQVGGR YEGLT+VTVRGAGH VP +P+ AL L+ SFLAG+ L
Sbjct: 412 LRVKTRWYPWYSGNQVGGRTEVYEGLTFVTVRGAGHEVPFFQPQSALILLRSFLAGNEL 470
>At2g35780 serine carboxypeptidase II like protein
Length = 452
Score = 419 bits (1078), Expect = e-117
Identities = 216/444 (48%), Positives = 277/444 (61%), Gaps = 21/444 (4%)
Query: 45 ESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEA--QSEPSKKPLLLWLNGGP 102
E DRI LPG P+ S SHFSGYITVNE GRALFYW E+ P KPL+LWLNGGP
Sbjct: 26 EKDRIFHLPGEPNDVSFSHFSGYITVNESAGRALFYWLTESPPSENPESKPLVLWLNGGP 85
Query: 103 GCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTI 162
GCSSV YGAA EIGP +N +G+ L NPYSWN+ ANLLF+ESP GVGFSY+NT+SDL
Sbjct: 86 GCSSVAYGAAEEIGPFRINPDGKTLYHNPYSWNKLANLLFLESPAGVGFSYSNTTSDLYT 145
Query: 163 LEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPF 222
D AEDAY FLV W +RFPQ+K R F+I+GESY GHY+PQL+++++++ + P
Sbjct: 146 AGDQRTAEDAYVFLVKWFERFPQYKHREFYIAGESYAGHYVPQLSQIVYEK-----RNPA 200
Query: 223 INLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQW-SNECNKAM 281
IN KGFIVGN +DY+DY GL EY W+H +ISD Y + C+F + S++C KAM
Sbjct: 201 INFKGFIVGNAVIDDYHDYVGLFEYWWAHGLISDLTYHNLRITCEFGSSEHPSSKCTKAM 260
Query: 282 NEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPCYST 341
+ ID Y+IY C+ +A+ S R++ ++ YDPC
Sbjct: 261 EAADLEQGNIDPYSIYTVTCK-KEAAALRS----------RFSRVRHPWMWRAYDPCTEK 309
Query: 342 YAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGGLKI 401
Y+ YFN +VQ + H N W+ C++ + + + S S+LPIY +LI GL+I
Sbjct: 310 YSGMYFNSPEVQKAMHANIT--GLAYPWKGCSDIVGEKWADSPLSMLPIYKELIAAGLRI 367
Query: 402 WIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGH 461
W++SGD D VP+ GTRY + AL L S W W D QVGG Y+GLT VT+ GAGH
Sbjct: 368 WVFSGDTDSVVPITGTRYSIRALKLQPLSKWYPWNDDGQVGGWSQVYKGLTLVTIHGAGH 427
Query: 462 LVPLNKPKEALSLIHSFLAGDRLP 485
VPL +P+ A L SFL LP
Sbjct: 428 EVPLFRPRRAFLLFQSFLDNKPLP 451
>At5g08260 serine-type carboxypeptidase II-like protein
Length = 480
Score = 417 bits (1072), Expect = e-117
Identities = 208/451 (46%), Positives = 289/451 (63%), Gaps = 8/451 (1%)
Query: 42 GANESDRIVDLPGVPSSPSVSHFSGYITVN-EDHGRALFYWFFEAQSEPSKKPLLLWLNG 100
G E D + LPG P + H++GY+ + E +ALFYWFFEAQ S++PL+LWLNG
Sbjct: 33 GRKEDDLVTGLPGQPPV-NFKHYAGYVNLGPEQKQKALFYWFFEAQQNSSRRPLVLWLNG 91
Query: 101 GPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDL 160
GPGCSS+ YGAA E+GP LV+ NG L +N +SWN+EAN+LF+E+PVGVGFSYTN S DL
Sbjct: 92 GPGCSSIAYGAAQELGPFLVHDNGGKLTYNHFSWNKEANMLFLEAPVGVGFSYTNNSMDL 151
Query: 161 TILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKY 220
L D A D+ FL+NW +FP+F+S F+ISGESY GHY+PQLAE+I+DRNK K
Sbjct: 152 QKLGDEVTASDSLAFLINWFMKFPEFRSSEFYISGESYAGHYVPQLAEVIYDRNKKVTKD 211
Query: 221 PFINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQ--FQWSNECN 278
INLKGF++GN + D GL++YAWSHA+ISD+ + C F++ + +C
Sbjct: 212 SSINLKGFMIGNAVINEATDMAGLVDYAWSHAIISDEVHTSIHGSCSFEEDTTNKTEQCY 271
Query: 279 KAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPC 338
Y++IDIY+IY P C + S+ + ++ + + GYDPC
Sbjct: 272 NNFKGFMDAYNDIDIYSIYTPVCLSSLLSSSPRKPKIVVSPRLLTFDDLWDKFPAGYDPC 331
Query: 339 YSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGG 398
+YAE YFNR DVQ + H N N + C + +++ ++ + +++PI KL+ GG
Sbjct: 332 TESYAENYFNRKDVQVALHANVT--NLPYPYSPC-SGVIKRWSDAPSTMIPIIQKLLTGG 388
Query: 399 LKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYE-GLTYVTVR 457
L+IWIYSGD DGRVPV TRY ++ +GL ++S WRSW+ +QV G + Y GL +VTVR
Sbjct: 389 LRIWIYSGDTDGRVPVTSTRYSIKKMGLKVESPWRSWFHKSQVAGWVETYAGGLNFVTVR 448
Query: 458 GAGHLVPLNKPKEALSLIHSFLAGDRLPTRR 488
GAGH VP P ++L+L F++ LP++R
Sbjct: 449 GAGHQVPALAPAQSLTLFSHFISSVPLPSKR 479
>At3g02110 serine carboxypeptidase II like protein
Length = 473
Score = 417 bits (1071), Expect = e-117
Identities = 212/447 (47%), Positives = 281/447 (62%), Gaps = 15/447 (3%)
Query: 45 ESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGC 104
E+DRI LPG P+ + FSGY+TV++ GR+LFYW EA P KPL++WLNGGPGC
Sbjct: 34 EADRITSLPGQPNV-TFEQFSGYVTVDKLSGRSLFYWLTEASDLPLSKPLVIWLNGGPGC 92
Query: 105 SSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILE 164
SSV YGA+ EIGP ++K G GL N ++WN +NLLF+E+P GVGFSYTN SSDL
Sbjct: 93 SSVAYGASEEIGPFRISKGGSGLYLNKFAWNSISNLLFLEAPAGVGFSYTNRSSDLFNTG 152
Query: 165 DNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFIN 224
D A+D+ FL+ WL RFP++ R +I+GESY GHY+PQLA+ I + NK ++K P +N
Sbjct: 153 DRRTAKDSLQFLIQWLHRFPRYNHREIYITGESYAGHYVPQLAKEIMNYNK-RSKNP-LN 210
Query: 225 LKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQWSNECNKAMNEV 284
LKG +VGN T+++YD G + Y WSHA+ISD+ Y + CDF + + S+EC +
Sbjct: 211 LKGIMVGNAVTDNHYDNLGTVSYWWSHAMISDRTYHQLISTCDFSRQKESDECETLYSYA 270
Query: 285 F-QDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKR-----KRIFGGYDPC 338
Q++ ID YNIYAP C +S + G + GR R+ R GYDPC
Sbjct: 271 MEQEFGNIDQYNIYAPPCNKSSDGGGSYNG----SSGRRSMRLPHLPHSVLRKISGYDPC 326
Query: 339 YSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGG 398
YAE Y+NR DVQ + H NT + W C+ + + +N + ++LPIY ++I GG
Sbjct: 327 TERYAEIYYNRPDVQKALHANTTK--IPYKWTACSEVLNRNWNDTDSTVLPIYREMIAGG 384
Query: 399 LKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRG 458
+++W++SGD D VPV TRY + L L K W WY+ QVGG YEGLT+VTVRG
Sbjct: 385 IRVWVFSGDVDSVVPVTATRYSLARLSLSTKLPWYPWYVKKQVGGWTEVYEGLTFVTVRG 444
Query: 459 AGHLVPLNKPKEALSLIHSFLAGDRLP 485
AGH VPL KP+ A L FL G LP
Sbjct: 445 AGHEVPLFKPRAAFELFKYFLRGKPLP 471
>At3g07990 putative serine carboxypeptidase II
Length = 459
Score = 416 bits (1069), Expect = e-116
Identities = 215/442 (48%), Positives = 283/442 (63%), Gaps = 19/442 (4%)
Query: 47 DRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQ--SEPSKKPLLLWLNGGPGC 104
DRI +LPG PS+ +SGY+TV+E+ GRALFYW E+ +P +PL+LWLNGGPGC
Sbjct: 32 DRISNLPGQPSNVDFRQYSGYVTVHEERGRALFYWLVESPLARDPKSRPLVLWLNGGPGC 91
Query: 105 SSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILE 164
SSV YGAA EIGP V +G+ L+ Y+WN+ ANLLF+ESP GVGFSY+NT+SDL
Sbjct: 92 SSVAYGAAEEIGPFRVGSDGKTLHSKLYAWNKLANLLFLESPAGVGFSYSNTTSDLYTTG 151
Query: 165 DNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFIN 224
D AED+Y FLVNW +RFPQ+K R F+I GESY GH++PQL++L+ +RNK K P IN
Sbjct: 152 DQRTAEDSYIFLVNWFERFPQYKHREFYIVGESYAGHFVPQLSKLVHERNKG-FKNPAIN 210
Query: 225 LKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQW-SNECNKAMNE 283
LKGF+VGN T+DY+DY G EY W+H +ISD Y + K C Q S +C A+
Sbjct: 211 LKGFMVGNAVTDDYHDYIGTFEYWWNHGLISDSTYHQLKTACYSVSSQHPSMQCMVALRN 270
Query: 284 VFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPCYSTYA 343
+ ID Y+I+ C NST A L++ KGR + + YDPC Y+
Sbjct: 271 AELEQGNIDPYSIFTKPC--NSTVA-----LKRFLKGRYPWMSR------AYDPCTERYS 317
Query: 344 EKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGGLKIWI 403
YFNR+DVQ + H N R + W+ C++ + ++ S S+LPIY +LI GLKIW+
Sbjct: 318 NVYFNRLDVQKALHANVTR--LSYPWKACSDIVGSYWDDSPLSMLPIYKELITAGLKIWV 375
Query: 404 YSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLV 463
+SGD D VP+ TRY V+AL L ++W WY +VGG Y+GLT VTV GAGH V
Sbjct: 376 FSGDTDAVVPITATRYSVDALKLATITNWYPWYDHGKVGGWSQVYKGLTLVTVAGAGHEV 435
Query: 464 PLNKPKEALSLIHSFLAGDRLP 485
PL++P++A L SFL +P
Sbjct: 436 PLHRPRQAFILFRSFLESKPMP 457
>At4g30810 serine carboxypeptidase II like protein
Length = 479
Score = 408 bits (1048), Expect = e-114
Identities = 215/475 (45%), Positives = 295/475 (61%), Gaps = 25/475 (5%)
Query: 15 MNVILSVHFLCFFLLTTSFIKTYGINVGANESDRIVDLPGVPSSPSVSHFSGYITVNEDH 74
+N ++++ FL T+ + G++ E D++ LPG + S +H+SG++ NE
Sbjct: 11 VNALIAIAFLA-----TAHLCEAGLS--QKEQDKVSKLPGQNFNVSFAHYSGFVATNEQL 63
Query: 75 GRALFYWFFEAQSEPSKKPLLLWLNGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSW 134
GRALFYW FEA + KPL+LWLNGGPGCSSV YG A EIGP + +G+ L N YSW
Sbjct: 64 GRALFYWLFEAVEDAKSKPLVLWLNGGPGCSSVAYGEAEEIGPFHIKADGKTLYLNQYSW 123
Query: 135 NQEANLLFVESPVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFIS 194
NQ AN+LF+++PVGVG+SY+NTSSDL D AED+ FL+ W++RFP++K R F+I
Sbjct: 124 NQAANILFLDAPVGVGYSYSNTSSDLKSNGDKRTAEDSLKFLLKWVERFPEYKGRDFYIV 183
Query: 195 GESYGGHYIPQLAELIFDRNK--DKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHA 252
GESY GHYIPQL+E I N+ DKN INLKG++VGN +D++D GL +Y WS
Sbjct: 184 GESYAGHYIPQLSEAIVKHNQGSDKNS---INLKGYMVGNGLMDDFHDRLGLFQYIWSLG 240
Query: 253 VISDQQYDKAKQVCDFKQF-QWSNECNKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIAS 311
ISDQ Y + C F+ F S +CNK + ++ ID Y+++ P C N AS
Sbjct: 241 FISDQTYSLLQLQCGFESFIHSSKQCNKILEIADKEIGNIDQYSVFTPACVAN-----AS 295
Query: 312 EGLEQLTKGRNDYRMKRKRIFGGYDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEV 371
+ L K R R+ YDPC + YFN +VQ + HV G W+
Sbjct: 296 QSNMLLKK-----RPMTSRVSEQYDPCTEKHTTVYFNLPEVQKALHV--PPGLAPSKWDT 348
Query: 372 CNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSS 431
C++ + + +N S S+L IY +LI GL+IW++SGDAD VPV TRY ++AL L S+
Sbjct: 349 CSDVVSEHWNDSPSSVLNIYHELIAAGLRIWVFSGDADAVVPVTSTRYSIDALNLRPLSA 408
Query: 432 WRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLPT 486
+ WYLD QVGG +Y GL +VTVRGAGH VPL++PK+AL+L +F++G L T
Sbjct: 409 YGPWYLDGQVGGWSQQYAGLNFVTVRGAGHEVPLHRPKQALALFKAFISGTPLST 463
>At4g15100 hydroxynitrile lyase like protein
Length = 407
Score = 400 bits (1029), Expect = e-112
Identities = 190/389 (48%), Positives = 261/389 (66%), Gaps = 5/389 (1%)
Query: 103 GCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTI 162
GCSSVGYGA EIGP L + N +GL FNPY+WN+E N+LF+ESPVGVGFSY+NTSSD
Sbjct: 17 GCSSVGYGATQEIGPFLADTNEKGLIFNPYAWNKEVNMLFLESPVGVGFSYSNTSSDYLN 76
Query: 163 LEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPF 222
L+D+F +DAY FL NW ++FP+ K F+I+GESY G Y+P+LAEL++D N+ N
Sbjct: 77 LDDHFAKKDAYTFLCNWFEKFPEHKGNEFYIAGESYAGIYVPELAELVYDNNEKNNDLSL 136
Query: 223 -INLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQ-WSNE-CNK 279
INLKGF++GNP+ + D++G ++YAWSHAVISD+ + ++C+F W+N+ CN+
Sbjct: 137 HINLKGFLLGNPDISNPDDWRGWVDYAWSHAVISDETHRNINRLCNFSSDDVWNNDKCNE 196
Query: 280 AMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPCY 339
A+ EV + Y+EIDIY++Y C+ +S + + T + R GYDPC
Sbjct: 197 AIAEVDKQYNEIDIYSLYTSACKGDSAKSSYFASAQFKTNYHISSKRMPPRRLAGYDPCL 256
Query: 340 STYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGGL 399
Y + Y+NR DVQ + H + G W +CN I + + V S+LPIY KLI GGL
Sbjct: 257 DDYVKVYYNRADVQKALHASD--GVNLKNWSICNMEIFHNWTYVVQSVLPIYQKLIAGGL 314
Query: 400 KIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGA 459
+IW+YSGD DG +PV+GTRY + ALGLP+K++WR WY + QV G + EY+GLT+ T RGA
Sbjct: 315 RIWVYSGDTDGCIPVLGTRYSLNALGLPIKTAWRPWYHEKQVSGWVQEYDGLTFATFRGA 374
Query: 460 GHLVPLNKPKEALSLIHSFLAGDRLPTRR 488
GH VP KP +L+ I +F+ G L + R
Sbjct: 375 GHTVPSFKPSSSLAFISAFVKGVPLSSSR 403
>At2g35770 serine carboxypeptidase II like protein
Length = 462
Score = 387 bits (994), Expect = e-108
Identities = 206/445 (46%), Positives = 275/445 (61%), Gaps = 26/445 (5%)
Query: 47 DRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSS 106
D+I+ LPG P + + S FSGY+TV+ GRALFYW EA KPL+LWLNGGPGCSS
Sbjct: 36 DKIISLPGQPPNLNFSQFSGYVTVDPAAGRALFYWLTEAPRPSGTKPLVLWLNGGPGCSS 95
Query: 107 VGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDN 166
+ YGA+ E+GP VN +G+ L N Y+WN+ AN+LF++SP GVGFSYTNTSSD + D
Sbjct: 96 IAYGASEEVGPFRVNPDGKTLRLNLYAWNKVANVLFLDSPAGVGFSYTNTSSDELTVGDK 155
Query: 167 FVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFINLK 226
EDAY FLV WL+RFP++K R F+I+GESY GHYIP+LA+LI +RNK K P INLK
Sbjct: 156 RTGEDAYRFLVRWLERFPEYKERAFYIAGESYAGHYIPELAQLIVNRNKGA-KNPTINLK 214
Query: 227 GFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVC--DFKQFQWSNECNKAMNEV 284
G ++GNP +DY D KG+ +Y W+H +ISD+ Y+ + C D F N CN A+N+
Sbjct: 215 GILMGNPLVDDYNDNKGMRDYWWNHGLISDESYNDLTKWCLNDSILFPKLN-CNAALNQA 273
Query: 285 FQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPCYSTYAE 344
++ +ID YNI +P C +T A ++E ++ + G D C Y
Sbjct: 274 LSEFGDIDPYNINSPAC---TTHASSNEWMQAWR-------------YRGNDECVVGYTR 317
Query: 345 KYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGGLKIWIY 404
KY N +V SFH R N + W C+ I + + S S+LPI L++ L+IWI+
Sbjct: 318 KYMNDPNVHKSFHA---RLNGSTPWTPCSRVIRKNWKDSPKSMLPIIKNLLQAHLRIWIF 374
Query: 405 SGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDN-QVGGRIVEYEG--LTYVTVRGAGH 461
SGD+D +P+ GTR+ + A+ L W WY + VGG YE LTY TVR AGH
Sbjct: 375 SGDSDAVLPLSGTRHSINAMKLKSSKRWYPWYHSHGLVGGWSQVYEDGLLTYTTVRAAGH 434
Query: 462 LVPLNKPKEALSLIHSFLAGDRLPT 486
VPL++P+ AL L FLA LP+
Sbjct: 435 EVPLSQPRLALFLFTHFLANHSLPS 459
>At3g63470 serin carboxypeptidase - like protein
Length = 502
Score = 373 bits (957), Expect = e-103
Identities = 202/468 (43%), Positives = 275/468 (58%), Gaps = 29/468 (6%)
Query: 22 HFLCFFLLTTSFIKTYGINVGANESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYW 81
HF L S +++ G + D I LPG P S + GY+TVNE GR+ FY+
Sbjct: 55 HFKAVKDLKPSSLRSAANQEGLRKRDLIRRLPGQPPV-SFDQYGGYVTVNESAGRSFFYY 113
Query: 82 FFEAQSEPSKKPLLLWLNGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLL 141
F EA PLLLWLNGGPGCSS+ YGA E+GP V+ +G+ L N Y+WN AN+L
Sbjct: 114 FVEASKSKDSSPLLLWLNGGPGCSSLAYGALQELGPFRVHSDGKTLFRNRYAWNNAANVL 173
Query: 142 FVESPVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGH 201
F+ESP GVGFSYTNT+SDL D A D Y FLVNWL+RFP++K R +I+GESY GH
Sbjct: 174 FLESPAGVGFSYTNTTSDLEKHGDRNTAADNYIFLVNWLERFPEYKGRDLYIAGESYAGH 233
Query: 202 YIPQLAELIFDRNKDKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDK 261
Y+PQLA I ++ F NLKG ++GN D D G+ ++ SHA+IS+ +
Sbjct: 234 YVPQLAHTILLHHRS-----FFNLKGILIGNAVINDETDLMGMYDFFESHALISEDSLAR 288
Query: 262 AKQVCDFKQFQWS---NECNKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLT 318
K CD K S EC +++ D +DIYNIYAP C LNST LT
Sbjct: 289 LKSNCDLKTESASVMTEECAVVSDQIDMDTYYLDIYNIYAPLC-LNST----------LT 337
Query: 319 KGRNDYRMKRKRIFGGYDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQ 378
+ R KR +DPC Y + Y NR +VQ++ H N + W+ C +S+++
Sbjct: 338 R-----RPKRGTTIREFDPCSDHYVQAYLNRPEVQAALHANATK--LPYEWQPC-SSVIK 389
Query: 379 TYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLD 438
+N S +++P+ +L+ G+++W++SGD DGR+PV T+Y ++ + L K++W WYL
Sbjct: 390 KWNDSPTTVIPLIKELMGQGVRVWVFSGDTDGRIPVTSTKYSLKKMNLTAKTAWHPWYLG 449
Query: 439 NQVGGRIVEYEG-LTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
+VGG EY+G LT+ TVRGAGH VP +PK +LSL FL LP
Sbjct: 450 GEVGGYTEEYKGKLTFATVRGAGHQVPSFQPKRSLSLFIHFLNDTPLP 497
>At2g05850 putative serine carboxypeptidase II
Length = 487
Score = 338 bits (866), Expect = 5e-93
Identities = 184/448 (41%), Positives = 260/448 (57%), Gaps = 30/448 (6%)
Query: 45 ESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGC 104
E D I LPG PS S + GY+ VNE R L+Y+F EA PL+LW NGGPGC
Sbjct: 60 EKDLIEKLPGQPSGISFRQYGGYVAVNEPATRFLYYYFVEAIKPSKSTPLVLWFNGGPGC 119
Query: 105 SSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTIL- 163
SSVG+GA E+GP V+ +G+ L NPYSWN EAN+LF E P+ VGFSY++T D I
Sbjct: 120 SSVGFGAFEELGPFRVHSDGKTLYRNPYSWNNEANMLFFEGPISVGFSYSSTPFDWEIFG 179
Query: 164 --EDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYP 221
D AED Y FLVNWL+RFP++K R +ISG+SY GHYIPQLA++I RN
Sbjct: 180 EQADKLTAEDNYMFLVNWLERFPEYKGRDVYISGQSYAGHYIPQLAQIILHRNNQ----T 235
Query: 222 FINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQWSNECNKAM 281
FINL+G +GNP + + ++ SH ++S + +++ +VCDF + +EC K M
Sbjct: 236 FINLRGISIGNPGLDLLIEADNENKFILSHGLVSQKDFEEYSKVCDFANYD-MDECPKIM 294
Query: 282 NEVFQDYSE-IDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPCYS 340
+ ++++ +D+YNIYAP C LNST ++SE K+ DPC S
Sbjct: 295 PKFSIEHNKHLDVYNIYAPVC-LNST--LSSE-------------PKKCTTIMEVDPCRS 338
Query: 341 TYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTY--NFSVFSILPIYTKLIKGG 398
Y + Y N +VQ + H NT + W+ CN+ + + S++PI L+ G
Sbjct: 339 NYVKAYLNSENVQEAMHANTTK--LPYEWKACNHYLNSVWIDADKDASMVPILHDLMGEG 396
Query: 399 LKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYE-GLTYVTVR 457
+++ +YSGD D +P T ++ + L + + WR W+ Q+GG +YE LTY TV+
Sbjct: 397 VRVLVYSGDVDAAIPFTATMAVLKTMNLTVVNEWRPWFTGGQLGGFTEDYERNLTYATVK 456
Query: 458 GAGHLVPLNKPKEALSLIHSFLAGDRLP 485
G+GH VPL++P AL+L SF+ LP
Sbjct: 457 GSGHSVPLDQPVHALNLFTSFIRNTPLP 484
>At3g52020 serine-type carboxypeptidase-like protein
Length = 501
Score = 337 bits (864), Expect = 9e-93
Identities = 193/455 (42%), Positives = 261/455 (56%), Gaps = 27/455 (5%)
Query: 36 TYGINVGAN-ESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPL 94
T G+N E D I +LPG PS S + GY+TVNE GR+L+Y+F EA PL
Sbjct: 66 TSGVNQQEQKERDLIENLPGQPSV-SFRQYGGYVTVNESAGRSLYYYFVEATKTKKSLPL 124
Query: 95 LLWLNGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYT 154
+LWLNGGPGCSS+ YGA E+GP + +G+ L NPYSWN AN+LF+ESPVG GFSYT
Sbjct: 125 VLWLNGGPGCSSL-YGAFQELGPFRIYGDGKTLYTNPYSWNNVANILFLESPVGTGFSYT 183
Query: 155 NTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRN 214
NT SDL D A D Y FLV WL+RFP++K R F+I+GESY GHY+PQLA+ I N
Sbjct: 184 NTESDLENPGDMKAAADKYIFLVKWLERFPEYKGREFYIAGESYAGHYVPQLAQTILVHN 243
Query: 215 KDKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQWS 274
K++N FINL+G ++GNP D + G +Y SHA++S K+ C +
Sbjct: 244 KNQN---FINLRGILIGNPTLNDIVETTGSFDYLVSHALLSQDSLLSYKENCATDTPKME 300
Query: 275 NECNKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGG 334
+C ++ D ++++YNI P C +N+T L LT + K
Sbjct: 301 VDCIALSMKIDDDIKKMNLYNILTPTC-INAT-------LTPLTN-----QSKECTTVLQ 347
Query: 335 YDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVF--SILPIYT 392
Y+PC Y Y NR DVQ S HV TW +CN + +N + + S+LPI
Sbjct: 348 YEPCGMQYIAAYLNREDVQRSMHVT----KLPHTWMLCNEATGFNWNQTDYSASMLPILK 403
Query: 393 KLIK-GGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEG- 450
+L+K L++W+Y+GD D +P+ T + ++ + L + W W+ + QVGG EY+G
Sbjct: 404 ELMKHDQLRVWVYTGDTDTVIPLTVTMHALKMMNLTAVTDWLPWFSEGQVGGFTEEYKGN 463
Query: 451 LTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
Y TV GAGH VPL KPK AL+L F+ LP
Sbjct: 464 FRYATVIGAGHEVPLYKPKAALTLFKHFIRNSPLP 498
>At3g52000 serine-type carboxypeptidase like protein
Length = 482
Score = 325 bits (832), Expect = 5e-89
Identities = 179/449 (39%), Positives = 257/449 (56%), Gaps = 37/449 (8%)
Query: 45 ESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGC 104
+ D I LPG PS + + GY+ VN+ GR L+Y+F E + PL++W NGGPGC
Sbjct: 60 DKDLIQQLPGQPSDVTFKQYGGYVAVNKPAGRFLYYYFVETIKPGNTTPLVIWFNGGPGC 119
Query: 105 SSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILE 164
SS+G GA E+GP V+ +G+ L NPYSWN EAN+LF+E+PVG GFSY+N+ +
Sbjct: 120 SSLG-GAFKELGPFRVHSDGKTLFRNPYSWNNEANVLFLETPVGTGFSYSNSPINGK-QG 177
Query: 165 DNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFIN 224
D AED Y FLVNWL+RFP++K R +I+G+SY GHY+PQLA++I RN IN
Sbjct: 178 DKATAEDNYMFLVNWLERFPEYKGRDIYIAGQSYAGHYVPQLAQIILHRNNQ----TLIN 233
Query: 225 LKGFIVGNP----ETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVC-DFKQFQWSNECNK 279
L+G ++GNP E +D + YK + +SH +IS QQ D + C D + W ++C+
Sbjct: 234 LRGILIGNPSLNREIQDDFGYK----FMFSHGLISQQQMDNYNKFCTDSDLYDW-DKCHL 288
Query: 280 AMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDPCY 339
A ++ + +DIYNIYAP C LNST + + + K DPC
Sbjct: 289 ASQKIEAQKTHLDIYNIYAPLC-LNSTLSSEPKKCTTIMKA---------------DPCS 332
Query: 340 STYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFS--VFSILPIYTKLIKG 397
Y + Y N +VQ + H NT + W CN +L +N S+ PI +L+
Sbjct: 333 GNYLKAYLNIKEVQEAIHANTTK--IPYEWTSCNTKLLWEWNEKDRYVSLTPILQELMGK 390
Query: 398 GLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEG-LTYVTV 456
G+++ +Y+GD D +P T V+ + L + WR W+ VGG +Y+G LT+VTV
Sbjct: 391 GVRVMLYNGDVDLVIPFTSTLAVVKTMNLTVVKEWRPWFTGGHVGGFTEDYKGNLTFVTV 450
Query: 457 RGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
+GAGH VP ++P AL++ SF+ LP
Sbjct: 451 KGAGHSVPTDQPIHALNIFTSFIRNTPLP 479
>At3g52010 serine-type carboxypeptidase-like protein
Length = 487
Score = 318 bits (815), Expect = 4e-87
Identities = 178/451 (39%), Positives = 252/451 (55%), Gaps = 37/451 (8%)
Query: 45 ESDRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGC 104
E D I LPG PS S + GY+ VNE R L+Y+F EA + PL++W NGGP C
Sbjct: 61 EKDLIKKLPGQPSGVSFRQYGGYVPVNEPSSRFLYYYFVEAIKPNTSTPLVIWFNGGPAC 120
Query: 105 SSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILE 164
SS+G GA +E+GP V+ G L NPYSWN EAN+LF+ESPV GFSY++ DL L
Sbjct: 121 SSLG-GAFLELGPFRVHSGGRKLFRNPYSWNNEANVLFLESPVTTGFSYSSNPIDLEELG 179
Query: 165 ---DNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYP 221
D AED Y FL+NWL+RFP++K R +I+G+SY GHY+PQLA++I RNK
Sbjct: 180 EKGDKATAEDNYIFLMNWLERFPEYKGRDIYIAGQSYAGHYVPQLAQIIIHRNKK----T 235
Query: 222 FINLKGFIVGNP----ETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQWSNEC 277
+NL+G ++GNP +D Y Y E+ SH ++S QQ D Q C +++C
Sbjct: 236 LVNLRGILIGNPSLLTSIQDPYGY----EFMLSHGLMSQQQMDNYNQFCLRDDLYDNDKC 291
Query: 278 NKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKRKRIFGGYDP 337
++ + +D YNIYAP C LNST + S+ + + DP
Sbjct: 292 ALSVKTIDDAKKHLDTYNIYAPVC-LNSTLSRISKKCTTVLE---------------VDP 335
Query: 338 CYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTY--NFSVFSILPIYTKLI 395
C Y + Y NR VQ + H NT + W CNN + + + N ++PI +L+
Sbjct: 336 CSKDYLKAYLNRKKVQKAIHANTTK--LPYEWTSCNNELTENWSENDRDTPMIPILHELM 393
Query: 396 KGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEG-LTYV 454
G+++ IY+GD D +P T V+ + L + +R W+ Q+GG +Y+G LT+V
Sbjct: 394 GEGVRVMIYNGDVDLEIPFASTLAVVKEMNLTVVKEFRPWFTGGQLGGFTEDYKGNLTFV 453
Query: 455 TVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
TV+GAGH VP ++P AL++ SF+ LP
Sbjct: 454 TVKGAGHSVPTDQPIHALNIFTSFIRNTPLP 484
>At1g43780 serine carboxypeptidase II, putative
Length = 479
Score = 313 bits (803), Expect = 1e-85
Identities = 172/453 (37%), Positives = 260/453 (56%), Gaps = 32/453 (7%)
Query: 47 DRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSS 106
D + LPG P + F+GY+ ++ GR+LFY+F EA+ +P KPL LWLNGGPGCSS
Sbjct: 36 DLVTKLPGQPEV-AFRQFAGYVDIDVKAGRSLFYYFVEAEKQPHSKPLTLWLNGGPGCSS 94
Query: 107 VGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDN 166
+G GA E+GP + GL NP SWN+ +NLLFV+SP GVG+SY+NT+SD T D
Sbjct: 95 IGGGAFTELGPFYPTGDARGLRRNPKSWNKASNLLFVDSPAGVGWSYSNTTSDYT-TGDE 153
Query: 167 FVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFINLK 226
A+D F++ WL++FPQFK+R F++GESY GHY+PQLA++I + N ++ NLK
Sbjct: 154 STAKDMLVFMLRWLEKFPQFKTRNLFLAGESYAGHYVPQLADVILEYNAQRSNRFKFNLK 213
Query: 227 GFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQVCDFKQFQWSNE------CNKA 280
G +GNP + D + E+ WSH +ISD+ CDF+ + +++ C A
Sbjct: 214 GIAIGNPLLKLDRDVPAIYEFFWSHGMISDELGLTIMNQCDFEDYTFTDSHNISKLCEAA 273
Query: 281 MNEVFQDYSE-IDIYNIYAPKCRLNSTSAIASEGLEQLTKGRNDYRMKR--KRIFGGYDP 337
+N+ ++ ++ Y+I C + EQ + R+K+ R+ G D
Sbjct: 274 VNQAGTIITQYVNYYDILLDVCYPSL--------FEQ------ELRLKKMGTRMSFGVDV 319
Query: 338 CYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKG 397
C S + Y N +VQ + H N R W +C++ + Y ++LPI +++K
Sbjct: 320 CMSFEEQLYLNLPEVQKALHAN--RTKLPYEWSMCSSLLNYKYTDGNANMLPILKRIVKS 377
Query: 398 GLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSS----WRSWYLDNQVGGRIVEYEG-LT 452
+ +W++SGD D +P++G+R V+ L L + + +W+ QVGG +VEY LT
Sbjct: 378 KVPVWVFSGDEDSVIPLLGSRTLVKELADDLNFNTTVPYGAWFDKGQVGGWVVEYGNLLT 437
Query: 453 YVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
+ TVRGA H+VP ++P AL L SF+ G +LP
Sbjct: 438 FATVRGAAHMVPYSQPSRALHLFTSFVLGRKLP 470
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,258,669
Number of Sequences: 26719
Number of extensions: 581223
Number of successful extensions: 1497
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1185
Number of HSP's gapped (non-prelim): 86
length of query: 488
length of database: 11,318,596
effective HSP length: 103
effective length of query: 385
effective length of database: 8,566,539
effective search space: 3298117515
effective search space used: 3298117515
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0333b.9


