

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0333b.4
(162 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
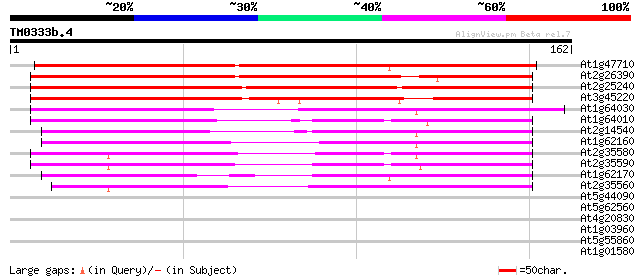
At1g47710 serpin, putative 132 1e-31
At2g26390 putative serpin 127 2e-30
At2g25240 putative serpin 127 2e-30
At3g45220 serpin-like protein 123 4e-29
At1g64030 hypothetical protein 111 2e-25
At1g64010 hypothetical protein 104 2e-23
At2g14540 putative serpin 103 4e-23
At1g62160 hypothetical protein 101 2e-22
At2g35580 putative serpin 99 1e-21
At2g35590 putative serpin 95 1e-20
At1g62170 hypothetical protein 95 2e-20
At2g35560 putative serpin 89 1e-18
At5g44090 protein phosphatase 2A 62 kDa B'' regulatory subunit (... 28 2.9
At5g62560 unknown protein 27 4.9
At4g20830 reticuline oxidase -like protein 27 4.9
At1g03960 protein phosphatase 2A like protein 27 4.9
At5g55860 myosin heavy chain-like 26 8.3
At1g01580 hypothetical protein 26 8.3
>At1g47710 serpin, putative
Length = 391
Score = 132 bits (331), Expect = 1e-31
Identities = 71/147 (48%), Positives = 100/147 (67%), Gaps = 3/147 (2%)
Query: 8 DGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQG 67
+GLS L+ K+ S PGFL+ +P+ +VK+ FKIP+F SF F+AS+VLK LG+ PFS G
Sbjct: 245 NGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVLKGLGLTSPFS-G 303
Query: 68 DADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAAT--IRALRGGGGPPQGLKFV 125
+ T+MV+ L V +IF K I+V+EEGTEAAAA+ + LRG + FV
Sbjct: 304 EEGLTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAAASAGVIKLRGLLMEEDEIDFV 363
Query: 126 ADHPFLFLIREDFSGTILFVGQVLNPL 152
ADHPFL ++ E+ +G +LF+GQV++PL
Sbjct: 364 ADHPFLLVVTENITGVVLFIGQVVDPL 390
>At2g26390 putative serpin
Length = 389
Score = 127 bits (320), Expect = 2e-30
Identities = 68/149 (45%), Positives = 101/149 (67%), Gaps = 10/149 (6%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL+ L+ K+++EPGFL+ +P H+ + + +IP+ + SF F+AS+VLK++G+ PF+
Sbjct: 244 KDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKASEVLKDMGLTSPFT- 302
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGL---- 122
+ T+MV + ++L+V SI K I+V EEGTEAAA ++ + PQ L
Sbjct: 303 SKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIMM-----PQCLMRNP 357
Query: 123 KFVADHPFLFLIREDFSGTILFVGQVLNP 151
FVADHPFLF +RED SG ILF+GQVL+P
Sbjct: 358 DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>At2g25240 putative serpin
Length = 385
Score = 127 bits (320), Expect = 2e-30
Identities = 67/145 (46%), Positives = 96/145 (66%), Gaps = 2/145 (1%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
++GL+PL+ K+ SEP F + +P H + + +F+IP+F SF F AS+VLK++G+ PF+
Sbjct: 240 KEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMGLTSPFNN 299
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVA 126
G T+MV + ++LYV SI K I+V EEGTEAAA ++ + + FVA
Sbjct: 300 G-GGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEAAAVSVGVV-SCTSFRRNPDFVA 357
Query: 127 DHPFLFLIREDFSGTILFVGQVLNP 151
D PFLF +RED SG ILF+GQVL+P
Sbjct: 358 DRPFLFTVREDKSGVILFMGQVLDP 382
>At3g45220 serpin-like protein
Length = 393
Score = 123 bits (309), Expect = 4e-29
Identities = 71/157 (45%), Positives = 102/157 (64%), Gaps = 22/157 (14%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL L+ +++S+P FL+ +P+ ++ +FKIP+F SF F+ASDVLKE+G+ LPF+
Sbjct: 244 RDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVLKEMGLTLPFTH 303
Query: 67 GDADFTKMVK-VNTPFN-----ELYVESIFQKVFIKVHEEGTEAAAATIRA------LRG 114
G T+MV+ + P N L+V ++F K I+V EEGTEAAA ++ + L G
Sbjct: 304 G--SLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVSVASMTKDMLLMG 361
Query: 115 GGGPPQGLKFVADHPFLFLIREDFSGTILFVGQVLNP 151
FVADHPFLF +RE+ SG ILF+GQVL+P
Sbjct: 362 --------DFVADHPFLFTVREEKSGVILFMGQVLDP 390
>At1g64030 hypothetical protein
Length = 385
Score = 111 bits (277), Expect = 2e-25
Identities = 63/158 (39%), Positives = 88/158 (54%), Gaps = 28/158 (17%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL L+ KMAS PGFL+ +P ++ +L F+IP+F I F F + VL LG
Sbjct: 247 KDGLDDLLEKMASTPGFLDSHIPTYRDELEKFRIPKFKIEFGFSVTSVLDRLG------- 299
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGG----GPPQGL 122
L S++ K +++ EEG EAAAAT G PP+ +
Sbjct: 300 -----------------LRSMSMYHKACVEIDEEGAEAAAATADGDCGCSLDFVEPPKKI 342
Query: 123 KFVADHPFLFLIREDFSGTILFVGQVLNPLGGANGTTT 160
FVADHPFLFLIRE+ +GT+LFVGQ+ +P G +G+ +
Sbjct: 343 DFVADHPFLFLIREEKTGTVLFVGQIFDPSGPCSGSNS 380
>At1g64010 hypothetical protein
Length = 185
Score = 104 bits (260), Expect = 2e-23
Identities = 62/147 (42%), Positives = 85/147 (57%), Gaps = 28/147 (19%)
Query: 7 QDGLSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQ 66
+DGL L+ KMAS GFL+ +P KVK+ F IP+F I F F AS LG+
Sbjct: 61 KDGLDNLVEKMASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRLGL------ 114
Query: 67 GDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPP--QGLKF 124
+E+ +++QK +++ EEG EA AAT A+ GG G + + F
Sbjct: 115 ---------------DEM---ALYQKACVEIDEEGAEAIAAT--AVVGGFGCAFVKRIDF 154
Query: 125 VADHPFLFLIREDFSGTILFVGQVLNP 151
VADHPFLF+IRED +GT+LFVGQ+ +P
Sbjct: 155 VADHPFLFMIREDKTGTVLFVGQIFDP 181
>At2g14540 putative serpin
Length = 407
Score = 103 bits (257), Expect = 4e-23
Identities = 60/145 (41%), Positives = 82/145 (56%), Gaps = 28/145 (19%)
Query: 10 LSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGDA 69
L L+ ++ S PGFL+ +P+++V + F+IP+F I F FEAS V +
Sbjct: 278 LDNLLERITSNPGFLDSHIPEYRVDVGDFRIPKFKIEFGFEASSVFNDF----------- 326
Query: 70 DFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGG---GPPQGLKFVA 126
EL V S+ QK I++ EEGTEAAAAT + G P + + FVA
Sbjct: 327 -------------ELNV-SLHQKALIEIDEEGTEAAAATTVVVVTGSCLWEPKKKIDFVA 372
Query: 127 DHPFLFLIREDFSGTILFVGQVLNP 151
DHPFLFLIRED +GT+LF GQ+ +P
Sbjct: 373 DHPFLFLIREDKTGTLLFAGQIFDP 397
>At1g62160 hypothetical protein
Length = 265
Score = 101 bits (252), Expect = 2e-22
Identities = 58/143 (40%), Positives = 79/143 (54%), Gaps = 26/143 (18%)
Query: 10 LSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGDA 69
L L+ +M S PGFL+ P+ +V++ F+IP+F I F FEAS V + + + F
Sbjct: 146 LDDLLKRMTSTPGFLDSHTPRERVEVDEFRIPKFKIEFGFEASSVFSDFEIDVSF----- 200
Query: 70 DFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGG-GPPQGLKFVADH 128
+QK I++ EEGTEAAAAT G G + L FVADH
Sbjct: 201 --------------------YQKALIEIDEEGTEAAAATAFVDNEDGCGFVETLDFVADH 240
Query: 129 PFLFLIREDFSGTILFVGQVLNP 151
PFLFLIRE+ +GT+LF GQ+ +P
Sbjct: 241 PFLFLIREEQTGTVLFAGQIFDP 263
>At2g35580 putative serpin
Length = 374
Score = 98.6 bits (244), Expect = 1e-21
Identities = 62/150 (41%), Positives = 83/150 (55%), Gaps = 29/150 (19%)
Query: 7 QDGLSPLIGKMASEPGFLEGR--LPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPF 64
+DGL ++ ++AS GFL+ LP H + KIPRF FAFEAS+ LK G+V+P
Sbjct: 246 KDGLPSMLERLASTRGFLKDNEVLPSHSAVIKELKIPRFKFDFAFEASEALKGFGLVVPL 305
Query: 65 SQGDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGG---GPPQG 121
S I K I+V E G++AAAA A RG G PP+
Sbjct: 306 SM----------------------IMHKSCIEVDEVGSKAAAAA--AFRGIGCRRPPPEK 341
Query: 122 LKFVADHPFLFLIREDFSGTILFVGQVLNP 151
FVADHPFLF+++E SG +LF+GQV++P
Sbjct: 342 HDFVADHPFLFIVKEYRSGLVLFLGQVMDP 371
>At2g35590 putative serpin
Length = 331
Score = 95.1 bits (235), Expect = 1e-20
Identities = 61/150 (40%), Positives = 82/150 (54%), Gaps = 29/150 (19%)
Query: 7 QDGLSPLIGKMASEPGFLEGR--LPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPF 64
+DGL ++ +AS GFL+ LP K + KIPRF +F FEAS LK LG+ +P
Sbjct: 203 KDGLPSMLESLASTRGFLKDNKVLPSQKAGVKELKIPRFKFAFDFEASKALKGLGLKVPL 262
Query: 65 SQGDADFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGG---PPQG 121
S +I K I+V E G++AAAA ALR GG PP+
Sbjct: 263 S----------------------TIIHKSCIEVDEVGSKAAAAA--ALRSCGGCYFPPKK 298
Query: 122 LKFVADHPFLFLIREDFSGTILFVGQVLNP 151
FVADHPFLF+++E SG +LF+G V++P
Sbjct: 299 YDFVADHPFLFIVKEYISGLVLFLGHVMDP 328
>At1g62170 hypothetical protein
Length = 433
Score = 94.7 bits (234), Expect = 2e-20
Identities = 56/143 (39%), Positives = 77/143 (53%), Gaps = 26/143 (18%)
Query: 10 LSPLIGKMASEPGFLEGRLPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGDA 69
L L+ +M S PGFL+ P+ +VK+ F+IP+F I F FEAS FS +
Sbjct: 314 LDDLLERMTSTPGFLDSHNPERRVKVGKFRIPKFKIEFGFEASSA---------FSDFEL 364
Query: 70 DFTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAAT-IRALRGGGGPPQGLKFVADH 128
D S +QK I++ E+GTEA T R+ G + + FVADH
Sbjct: 365 DV----------------SFYQKTLIEIDEKGTEAVTFTAFRSAYLGCALVKPIDFVADH 408
Query: 129 PFLFLIREDFSGTILFVGQVLNP 151
PFLFLIRE+ +GT+LF GQ+ +P
Sbjct: 409 PFLFLIREEQTGTVLFAGQIFDP 431
>At2g35560 putative serpin
Length = 121
Score = 88.6 bits (218), Expect = 1e-18
Identities = 54/141 (38%), Positives = 76/141 (53%), Gaps = 25/141 (17%)
Query: 13 LIGKMASEPGFLEGR--LPQHKVKLASFKIPRFDISFAFEASDVLKELGVVLPFSQGDAD 70
++ ++AS GFL G+ +P H + KIPRF F FEAS+ LK LG+ +P
Sbjct: 1 MLERLASTRGFLNGKEDIPSHWADVGELKIPRFKFDFGFEASEALKGLGLEVP------- 53
Query: 71 FTKMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGGGGPPQGLKFVADHPF 130
+I K I+V E G++AAAA + + G + FVADHPF
Sbjct: 54 ----------------STIIHKSCIEVDEVGSKAAAAAVIFVGCCGPAEKRYDFVADHPF 97
Query: 131 LFLIREDFSGTILFVGQVLNP 151
LFL++E SG ILF+GQV++P
Sbjct: 98 LFLVKEYRSGLILFLGQVMDP 118
>At5g44090 protein phosphatase 2A 62 kDa B'' regulatory subunit
(gb|AAD45158.1)
Length = 538
Score = 27.7 bits (60), Expect = 2.9
Identities = 14/39 (35%), Positives = 22/39 (55%), Gaps = 3/39 (7%)
Query: 78 NTP-FNELYVESIFQKVFIKVHEEGTEAAAATIRALRGG 115
NTP F E Y E++ ++F ++ GT T+R L+ G
Sbjct: 256 NTPEFQERYAETVIYRIFYYINRSGT--GCITLRELKRG 292
>At5g62560 unknown protein
Length = 559
Score = 26.9 bits (58), Expect = 4.9
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 4/44 (9%)
Query: 78 NTPFNELYVESIFQKVFIKVHEEGTEA----AAATIRALRGGGG 117
N F L E+ ++V ++V E G E A+ + A+RGGGG
Sbjct: 474 NLRFRGLASEAGAEEVLMEVEENGNERVKEKASKILLAMRGGGG 517
>At4g20830 reticuline oxidase -like protein
Length = 570
Score = 26.9 bits (58), Expect = 4.9
Identities = 21/80 (26%), Positives = 35/80 (43%), Gaps = 12/80 (15%)
Query: 64 FSQGDADFTKMVKV---NTPFNELYVESIFQKVFIKVHEEGTEAAAAT-------IRALR 113
FSQ + F+ +++ N FN ++ + I E +AA T + +R
Sbjct: 59 FSQTNPAFSSVLRAYIRNARFNTS--STLKPTIIITPRSESHVSAAVTCSKTLNFLLKIR 116
Query: 114 GGGGPPQGLKFVADHPFLFL 133
GG GL +++D PF L
Sbjct: 117 SGGHDYDGLSYISDKPFFIL 136
>At1g03960 protein phosphatase 2A like protein
Length = 529
Score = 26.9 bits (58), Expect = 4.9
Identities = 12/43 (27%), Positives = 23/43 (52%), Gaps = 2/43 (4%)
Query: 73 KMVKVNTPFNELYVESIFQKVFIKVHEEGTEAAAATIRALRGG 115
+ ++ + F E Y E++ ++F ++ GT T+R LR G
Sbjct: 243 EFLRTISEFQERYAETVIYRIFYYINRSGT--GCLTLRELRRG 283
>At5g55860 myosin heavy chain-like
Length = 649
Score = 26.2 bits (56), Expect = 8.3
Identities = 20/66 (30%), Positives = 34/66 (51%), Gaps = 2/66 (3%)
Query: 50 EASDVLKELGVVLPFSQGDADFTKMVKVNTPFNELYVESIFQKV-FIKVHEEGTEAAAAT 108
+ASD+ GV L ++ F K+V+ EL VES+ ++ +K+ + EA A
Sbjct: 294 KASDIDSVNGVSLELNEAKGLFEKLVEEEKSLQEL-VESLKAELKNVKMEHDEVEAKEAE 352
Query: 109 IRALRG 114
I ++ G
Sbjct: 353 IESVAG 358
>At1g01580 hypothetical protein
Length = 725
Score = 26.2 bits (56), Expect = 8.3
Identities = 17/74 (22%), Positives = 32/74 (42%), Gaps = 11/74 (14%)
Query: 39 KIPRFDISFAFEASDVLKELGVVLPFSQGDADFTKMVKVNTPFNELYVESIFQKVFIKVH 98
KIP+ + AF+ S + L +VLP S + + + + + + + +
Sbjct: 473 KIPKITLICAFKKSSEISMLDLVLPLSGLETELSSDINIK-----------IEAFITRDN 521
Query: 99 EEGTEAAAATIRAL 112
+ G EA A I+ L
Sbjct: 522 DAGDEAKAGKIKTL 535
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.143 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,706,463
Number of Sequences: 26719
Number of extensions: 147500
Number of successful extensions: 346
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 310
Number of HSP's gapped (non-prelim): 18
length of query: 162
length of database: 11,318,596
effective HSP length: 91
effective length of query: 71
effective length of database: 8,887,167
effective search space: 630988857
effective search space used: 630988857
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0333b.4


