

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0333b.12
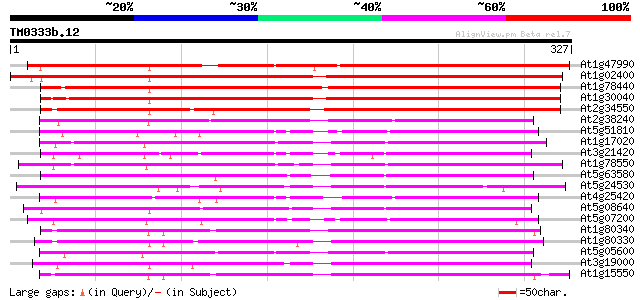
(327 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g47990 dioxygenase, putative 319 1e-87
At1g02400 unknown protein 311 4e-85
At1g78440 putative dioxygenase 273 7e-74
At1g30040 unknown protein 272 2e-73
At2g34550 putative gibberellin 2-oxidase 252 2e-67
At2g38240 putative anthocyanidin synthase 147 1e-35
At5g51810 gibberellin 20-oxidase (emb|CAA58294.1) 140 9e-34
At1g17020 SRG1-like protein 140 9e-34
At3g21420 unknown protein 139 2e-33
At1g78550 unknown protein 139 3e-33
At5g63580 flavonol synthase 138 5e-33
At5g24530 flavanone 3-hydroxylase-like protein 137 6e-33
At4g25420 gibberellin 20-oxidase - Arabidopsis thaliana 135 3e-32
At5g08640 flavonol synthase (FLS) (sp|Q96330) 134 7e-32
At5g07200 gibberellin 20-oxidase 134 9e-32
At1g80340 gibberellin 3 beta-hydroxylase 132 3e-31
At1g80330 putative gibberellin 3 beta-hydroxylase 130 1e-30
At5g05600 leucoanthocyanidin dioxygenase-like protein 129 2e-30
At3g19000 unknown protein 129 2e-30
At1g15550 Gibberillin 3 beta-hydroxylase (AtGA3ox1) 128 5e-30
>At1g47990 dioxygenase, putative
Length = 321
Score = 319 bits (818), Expect = 1e-87
Identities = 174/328 (53%), Positives = 223/328 (67%), Gaps = 23/328 (7%)
Query: 11 SEKIVP--GDLPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFF 68
S+KIV D+PI+D+ E+S+V+ IVKA E GFFKVINHG+ TI++ME+ FF
Sbjct: 5 SQKIVAVDQDIPIIDMSQERSQVSMQIVKACESLGFFKVINHGVDQTTISRMEQESINFF 64
Query: 69 AKPMPQKKQAAP-----GYGCKNIGFNGDMGEVEYLLLNANTPSI-AQISKSVSIDPSNF 122
AKP +KK P YG ++IG NGD GEVEYLL + N P+ +Q+S F
Sbjct: 65 AKPAHEKKSVRPVNQPFRYGFRDIGLNGDSGEVEYLLFHTNDPAFRSQLS---------F 115
Query: 123 RSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPP---ILN 179
S V+ Y EAV++LA EIL++ AEGL V FSRLI ++SDSVLR+NHYPP
Sbjct: 116 SSAVNCYIEAVKQLAREILDLTAEGLHVPPHS-FSRLISSVDSDSVLRVNHYPPSDQFFG 174
Query: 180 SNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNV 239
N D+S S +VGFGEH+DPQILT+LRSN V GLQ+S DG+W+ V PDP AFCVNV
Sbjct: 175 EANLSDQSVSLT-RVGFGEHTDPQILTVLRSNGVGGLQVSNSDGMWVSVSPDPSAFCVNV 233
Query: 240 GDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACI-VAPPVLVTPERPSLFRP 298
GD+L+VMTNGRF+SVRHRA+T +SR+S AYF PPL A I +++T +P L++
Sbjct: 234 GDLLQVMTNGRFISVRHRALTYGEESRLSTAYFAGPPLQAKIGPLSAMVMTMNQPRLYQT 293
Query: 299 FTWADYKKATYSLRLGDSRMELFRNNND 326
FTW +YKK YSLRL DSR+++FR D
Sbjct: 294 FTWGEYKKRAYSLRLEDSRLDMFRTCKD 321
>At1g02400 unknown protein
Length = 329
Score = 311 bits (796), Expect = 4e-85
Identities = 169/334 (50%), Positives = 218/334 (64%), Gaps = 19/334 (5%)
Query: 1 MVLASRNPIRS--EKIVPG---DLPIVDLKA-EKSEVTRLIVKASEEYGFFKVINHGISH 54
MVL S P+++ +K + + P++D ++S+++ IVKA E GFFKVINHG+
Sbjct: 1 MVLPSSTPLQTTGKKTISSPEYNFPVIDFSLNDRSKLSEKIVKACEVNGFFKVINHGVKP 60
Query: 55 ETIAKMEEAGFGFFAKPMPQKKQAAP----GYGCKNIGFNGDMGEVEYLLLNANTPSIAQ 110
E I + E G FF KP K +A P GYGCKNIGFNGD+GE+EYLLL+AN ++A
Sbjct: 61 EIIKRFEHEGEEFFNKPESDKLRAGPASPFGYGCKNIGFNGDLGELEYLLLHANPTAVAD 120
Query: 111 ISKSVSID-PSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVL 169
S+++S D P F S + Y VR+LACEI+++ E L Q + S LIRD+ SDS+L
Sbjct: 121 KSETISHDDPFKFSSATNDYIRTVRDLACEIIDLTIENLWGQKSSEVSELIRDVRSDSIL 180
Query: 170 RLNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVK 229
RLNHYPP + S ++GFGEHSDPQILT+LRSNDV GL+I +DG+WIP+
Sbjct: 181 RLNHYPPA-------PYALSGVGQIGFGEHSDPQILTVLRSNDVDGLEICSRDGLWIPIP 233
Query: 230 PDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKS-RMSMAYFGAPPLHACIVAPPVLV 288
DP F V VGD L+ +TNGRF SVRHR + N+ K RMS YF APPL A I P +V
Sbjct: 234 SDPTCFFVLVGDCLQALTNGRFTSVRHRVLANTAKKPRMSAMYFAAPPLEAKISPLPKMV 293
Query: 289 TPERPSLFRPFTWADYKKATYSLRLGDSRMELFR 322
+PE P + FTW DYKKATYSLRL R+E F+
Sbjct: 294 SPENPRRYNSFTWGDYKKATYSLRLDVPRLEFFK 327
>At1g78440 putative dioxygenase
Length = 329
Score = 273 bits (699), Expect = 7e-74
Identities = 143/307 (46%), Positives = 198/307 (63%), Gaps = 9/307 (2%)
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+P++D+ +S+ +VKA E++GFFKVINHG+S E ++ +E FF+ P +K Q
Sbjct: 18 IPVIDMSDPESK--HALVKACEDFGFFKVINHGVSAELVSVLEHETVDFFSLPKSEKTQV 75
Query: 79 AP---GYGCKNIGFNGDMGEVEYLLLNANTPS-IAQISKSVSIDPSNFRSTVSAYTEAVR 134
A GYG IG NGD+G VEYLL+NAN S + S+ P FR+ + YT +VR
Sbjct: 76 AGYPFGYGNSKIGRNGDVGWVEYLLMNANHDSGSGPLFPSLLKSPGTFRNALEEYTTSVR 135
Query: 135 ELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYNNKV 194
++ ++LE + +GLG++ S+L+ D +DS+LRLNHYPP SN K+ N +
Sbjct: 136 KMTFDVLEKITDGLGIKPRNTLSKLVSDQNTDSILRLNHYPPCPLSNK---KTNGGKNVI 192
Query: 195 GFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRFVSV 254
GFGEH+DPQI+++LRSN+ SGLQI+L DG WI V PD +F NVGD L+VMTNGRF SV
Sbjct: 193 GFGEHTDPQIISVLRSNNTSGLQINLNDGSWISVPPDHTSFFFNVGDSLQVMTNGRFKSV 252
Query: 255 RHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYKKATYSLRLG 314
RHR + N KSR+SM YF P L I L+ E L+ FTW++YK +TY+ RL
Sbjct: 253 RHRVLANCKKSRVSMIYFAGPSLTQRIAPLTCLIDNEDERLYEEFTWSEYKNSTYNSRLS 312
Query: 315 DSRMELF 321
D+R++ F
Sbjct: 313 DNRLQQF 319
>At1g30040 unknown protein
Length = 341
Score = 272 bits (695), Expect = 2e-73
Identities = 143/308 (46%), Positives = 201/308 (64%), Gaps = 14/308 (4%)
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+P+V+L A+ TR IVKA EE+GFFKV+NHG+ E + ++E+ GFF P K +A
Sbjct: 31 IPVVNL-ADPEAKTR-IVKACEEFGFFKVVNHGVRPELMTRLEQEAIGFFGLPQSLKNRA 88
Query: 79 AP----GYGCKNIGFNGDMGEVEYLLLNANTP-SIAQISKSVSIDPSNFRSTVSAYTEAV 133
P GYG K IG NGD+G +EYLLLNAN S + S P FR +V Y + +
Sbjct: 89 GPPEPYGYGNKRIGPNGDVGWIEYLLLNANPQLSSPKTSAVFRQTPQIFRESVEEYMKEI 148
Query: 134 RELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYNNK 193
+E++ ++LE++AE LG++ S+++RD +SDS LRLNHYP ++ K
Sbjct: 149 KEVSYKVLEMVAEELGIEPRDTLSKMLRDEKSDSCLRLNHYPAA-------EEEAEKMVK 201
Query: 194 VGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRFVS 253
VGFGEH+DPQI+++LRSN+ +GLQI ++DG W+ V PD +F +NVGD L+VMTNGRF S
Sbjct: 202 VGFGEHTDPQIISVLRSNNTAGLQICVKDGSWVAVPPDHSSFFINVGDALQVMTNGRFKS 261
Query: 254 VRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYKKATYSLRL 313
V+HR + ++ +SR+SM YFG PPL I P LV + L++ FTW+ YK + Y +L
Sbjct: 262 VKHRVLADTRRSRISMIYFGGPPLSQKIAPLPCLVPEQDDWLYKEFTWSQYKSSAYKSKL 321
Query: 314 GDSRMELF 321
GD R+ LF
Sbjct: 322 GDYRLGLF 329
>At2g34550 putative gibberellin 2-oxidase
Length = 335
Score = 252 bits (644), Expect = 2e-67
Identities = 135/310 (43%), Positives = 191/310 (61%), Gaps = 19/310 (6%)
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+P++DL S+ IVKA EE+GFFKVINHG+ + + ++E+ FFA K +A
Sbjct: 27 IPVIDLT--DSDAKTQIVKACEEFGFFKVINHGVRPDLLTQLEQEAINFFALHHSLKDKA 84
Query: 79 AP----GYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSI---DPSNFRSTVSAYTE 131
P GYG K IG NGD+G +EY+LLNAN + K+ +I P+ FR V Y +
Sbjct: 85 GPPDPFGYGTKRIGPNGDLGWLEYILLNANL--CLESHKTTAIFRHTPAIFREAVEEYIK 142
Query: 132 AVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYN 191
++ ++ + LE++ E L ++ SRL++ ESDS LR+NHYP E +
Sbjct: 143 EMKRMSSKFLEMVEEELKIEPKEKLSRLVKVKESDSCLRMNHYP--------EKEETPVK 194
Query: 192 NKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRF 251
++GFGEH+DPQ++++LRSND GLQI ++DG W+ V PD +F V VGD L+VMTNGRF
Sbjct: 195 EEIGFGEHTDPQLISLLRSNDTEGLQICVKDGTWVDVTPDHSSFFVLVGDTLQVMTNGRF 254
Query: 252 VSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYKKATYSL 311
SV+HR +TN+ +SR+SM YF PPL I LV + L+ FTW+ YK + Y
Sbjct: 255 KSVKHRVVTNTKRSRISMIYFAGPPLSEKIAPLSCLVPKQDDCLYNEFTWSQYKLSAYKT 314
Query: 312 RLGDSRMELF 321
+LGD R+ LF
Sbjct: 315 KLGDYRLGLF 324
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 147 bits (370), Expect = 1e-35
Identities = 95/298 (31%), Positives = 155/298 (51%), Gaps = 22/298 (7%)
Query: 18 DLPIVDLKAE--KSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQK 75
++P++D+ K E RL+ A EE+GFF+++NHG++H + ++ A FF P+ +K
Sbjct: 47 EIPVLDMNDVWGKPEGLRLVRSACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEK 106
Query: 76 KQAA------PGYGCK-NIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSA 128
++ A GYG + + + + +Y LN SI SK S P R +
Sbjct: 107 RKYANSPDTYEGYGSRLGVVKDAKLDWSDYFFLNYLPSSIRNPSKWPS-QPPKIRELIEK 165
Query: 129 YTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSP 188
Y E VR+L + E ++E LG++ + L + + LR N YP K P
Sbjct: 166 YGEEVRKLCERLTETLSESLGLKPNKLMQALGGGDKVGASLRTNFYP----------KCP 215
Query: 189 SYNNKVGFGEHSDPQILTILRSND-VSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMT 247
+G HSDP +TIL ++ V+GLQ+ DG W+ +K P A VN+GD L++++
Sbjct: 216 QPQLTLGLSSHSDPGGITILLPDEKVAGLQVRRGDG-WVTIKSVPNALIVNIGDQLQILS 274
Query: 248 NGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYK 305
NG + SV H+ + NS R+S+A+F P + LVT RP+L++P + +Y+
Sbjct: 275 NGIYKSVEHQVIVNSGMERVSLAFFYNPRSDIPVGPIEELVTANRPALYKPIRFDEYR 332
>At5g51810 gibberellin 20-oxidase (emb|CAA58294.1)
Length = 378
Score = 140 bits (353), Expect = 9e-34
Identities = 97/306 (31%), Positives = 149/306 (47%), Gaps = 29/306 (9%)
Query: 18 DLPIVDLKAEKS--EVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMP-- 73
++P +DL ++ S E R+I +A ++GFF V+NHG+S IA FF P+
Sbjct: 62 NVPFIDLSSQDSTLEAPRVIAEACTKHGFFLVVNHGVSESLIADAHRLMESFFDMPLAGK 121
Query: 74 QKKQAAPGYGCKNIG-FNGDMGE-------VEYLLLNANTPSIA---QISKSVSIDPSNF 122
QK Q PG C F G + + N N+ S S ++ + F
Sbjct: 122 QKAQRKPGESCGYASSFTGRFSTKLPWKETLSFQFSNDNSGSRTVQDYFSDTLGQEFEQF 181
Query: 123 RSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNN 182
Y EA+ L+ +I+E++ LGV F E+DS++RLNHYPP
Sbjct: 182 GKVYQDYCEAMSSLSLKIMELLGLSLGVNRDY-FRGFFE--ENDSIMRLNHYPPC----- 233
Query: 183 REDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDV 242
++P +G G H DP LTIL + V+GLQ+ + D W ++P+P+AF VN+GD
Sbjct: 234 ---QTPDLT--LGTGPHCDPSSLTILHQDHVNGLQVFV-DNQWQSIRPNPKAFVVNIGDT 287
Query: 243 LEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWA 302
++NG F S HRA+ N +R SMA+F P + P ++ + + FTW+
Sbjct: 288 FMALSNGIFKSCLHRAVVNRESARKSMAFFLCPKKDKVVKPPSDILEKMKTRKYPDFTWS 347
Query: 303 DYKKAT 308
+ + T
Sbjct: 348 MFLEFT 353
>At1g17020 SRG1-like protein
Length = 358
Score = 140 bits (353), Expect = 9e-34
Identities = 98/309 (31%), Positives = 155/309 (49%), Gaps = 26/309 (8%)
Query: 18 DLPIVDLK------AEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKP 71
++PI+D+K SEV +L A +E+GFF+++NHGI + K++ FF P
Sbjct: 52 EIPIIDMKRLCSSTTMDSEVEKLDF-ACKEWGFFQLVNHGIDSSFLDKVKSEIQDFFNLP 110
Query: 72 MPQKKQ------AAPGYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRST 125
M +KK+ G+G + + L + P + P FR T
Sbjct: 111 MEEKKKFWQRPDEIEGFGQAFVVSEDQKLDWADLFFHTVQPVELRKPHLFPKLPLPFRDT 170
Query: 126 VSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNRED 185
+ Y+ V+ +A ++ MA L ++ + +L D++S +R+N+YPP
Sbjct: 171 LEMYSSEVQSVAKILIAKMARALEIKPEEL-EKLFDDVDSVQSMRMNYYPPC-------- 221
Query: 186 KSPSYNNKVGFGEHSDPQILTILRS-NDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLE 244
P + +G HSD LT+L NDV GLQI +DG W+PVKP P AF VN+GDVLE
Sbjct: 222 --PQPDQVIGLTPHSDSVGLTVLMQVNDVEGLQIK-KDGKWVPVKPLPNAFIVNIGDVLE 278
Query: 245 VMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADY 304
++TNG + S+ HR + NS K R+S+A F ++ + LV ++ + F+ T +Y
Sbjct: 279 IITNGTYRSIEHRGVVNSEKERLSIATFHNVGMYKEVGPAKSLVERQKVARFKRLTMKEY 338
Query: 305 KKATYSLRL 313
+S L
Sbjct: 339 NDGLFSRTL 347
>At3g21420 unknown protein
Length = 364
Score = 139 bits (350), Expect = 2e-33
Identities = 102/304 (33%), Positives = 154/304 (50%), Gaps = 34/304 (11%)
Query: 19 LPIVDL----KAEKSEVTRLIVKAS---EEYGFFKVINHGISHETIAKMEEAGFGFFAKP 71
+P++DL K + + I+K S E++GFF+VINHGI E + +EE FF P
Sbjct: 55 IPVIDLSKLSKPDNDDFFFEILKLSQACEDWGFFQVINHGIEVEVVEDIEEVASEFFDMP 114
Query: 72 MPQKKQ------AAPGYGCKNIGFNGD--MGEVEYLLLNANTPSIAQISKSVSIDPSNFR 123
+ +KK+ GYG I F+ D + L + P I K P+ F
Sbjct: 115 LEEKKKYPMEPGTVQGYGQAFI-FSEDQKLDWCNMFALGVHPPQIRN-PKLWPSKPARFS 172
Query: 124 STVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNR 183
++ Y++ +REL +L+ +A LG+++ F + E+ +R+N+YPP
Sbjct: 173 ESLEGYSKEIRELCKRLLKYIAISLGLKEER-FEEMFG--EAVQAVRMNYYPPC------ 223
Query: 184 EDKSPSYNNKVGFGEHSDPQILTILRS--NDVSGLQISLQDGVWIPVKPDPEAFCVNVGD 241
SP +G HSD LT+L+ N GLQI L+D W+PVKP P A +N+GD
Sbjct: 224 --SSPDL--VLGLSPHSDGSALTVLQQSKNSCVGLQI-LKDNTWVPVKPLPNALVINIGD 278
Query: 242 VLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPE-RPSLFRPFT 300
+EV++NG++ SV HRA+TN K R+++ F AP I LV E P +R +
Sbjct: 279 TIEVLSNGKYKSVEHRAVTNREKERLTIVTFYAPNYEVEIEPMSELVDDETNPCKYRSYN 338
Query: 301 WADY 304
DY
Sbjct: 339 HGDY 342
>At1g78550 unknown protein
Length = 356
Score = 139 bits (349), Expect = 3e-33
Identities = 101/331 (30%), Positives = 169/331 (50%), Gaps = 29/331 (8%)
Query: 6 RNPIRSEKIVPGDLPIVDL------KAEKSEVTRLIVKASEEYGFFKVINHGISHETIAK 59
+ I ++ + ++P++D+ A SE+ +L A +++GFF+++NHGI + K
Sbjct: 40 KTEILNDSSLSSEIPVIDMTRLCSVSAMDSELKKLDF-ACQDWGFFQLVNHGIDSSFLEK 98
Query: 60 MEEAGFGFFAKPMPQKKQA------APGYGCKNIGFNGDMGEVEYLLLNANTPSIAQISK 113
+E FF PM +K++ G+G NI + + + P ++ S
Sbjct: 99 LETEVQEFFNLPMKEKQKLWQRSGEFEGFGQVNIVSENQKLDWGDMFILTTEPIRSRKSH 158
Query: 114 SVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNH 173
S P FR T+ Y+ V+ +A + MA L ++ + L D+ +++N+
Sbjct: 159 LFSKLPPPFRETLETYSSEVKSIAKILFAKMASVLEIKHEEM-EDLFDDVWQS--IKINY 215
Query: 174 YPPILNSNNREDKSPSYNNKVGFGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKPDP 232
YPP P + +G +HSD LTIL + N V GLQI +DG W+ VKP
Sbjct: 216 YPPC----------PQPDQVMGLTQHSDAAGLTILLQVNQVEGLQIK-KDGKWVVVKPLR 264
Query: 233 EAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPER 292
+A VNVG++LE++TNGR+ S+ HRA+ NS K R+S+A F +P I LV ++
Sbjct: 265 DALVVNVGEILEIITNGRYRSIEHRAVVNSEKERLSVAMFHSPGKETIIRPAKSLVDRQK 324
Query: 293 PSLFRPFTWADYKKATYSLRL-GDSRMELFR 322
LF+ + +Y A ++ +L G S ++L R
Sbjct: 325 QCLFKSMSTQEYFDAFFTQKLNGKSHLDLMR 355
>At5g63580 flavonol synthase
Length = 310
Score = 138 bits (347), Expect = 5e-33
Identities = 92/294 (31%), Positives = 150/294 (50%), Gaps = 19/294 (6%)
Query: 19 LPIVDLKAEKSE-VTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQ 77
+PI+DL E V +VK SEE+G F V+NHGI + I ++++ G FF P +KK
Sbjct: 19 IPIIDLSNLDEELVAHAVVKGSEEWGIFHVVNHGIPMDLIQRLKDVGTQFFELPETEKKA 78
Query: 78 AAPGYGCKNI-GFNGDMGEVEYLLLNANTPSIAQISKSVSID-----PSNFRSTVSAYTE 131
A G K+ G+ ++ V+ + N ++ D P +R + YT+
Sbjct: 79 VAKQDGSKDFEGYTTNLKYVKGEVWTENLFHRIWPPTCINFDYWPKNPPQYREVIEEYTK 138
Query: 132 AVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYN 191
++L+ IL ++EGLG+ + L + ++ V+R+N+YPP P +
Sbjct: 139 ETKKLSERILGYLSEGLGLPSEALIQGLGGE-STEYVMRINNYPP----------DPKPD 187
Query: 192 NKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRF 251
+G EH+D +TI+ +N+V GLQI +D W+ V P + VN+GD + ++NG++
Sbjct: 188 LTLGVPEHTDIIGITIIITNEVPGLQI-FKDDHWLDVHYIPSSITVNIGDQIMRLSNGKY 246
Query: 252 VSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYK 305
+V HRA+ + RMS F I P L+T + PS F+P T D+K
Sbjct: 247 KNVLHRAIVDKENQRMSWPVFVDANPDVVIGPLPELITGDNPSKFKPITCKDFK 300
>At5g24530 flavanone 3-hydroxylase-like protein
Length = 341
Score = 137 bits (346), Expect = 6e-33
Identities = 113/339 (33%), Positives = 167/339 (48%), Gaps = 40/339 (11%)
Query: 5 SRNPIRSEKIVPGDLPIVDLKA-EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEA 63
S P SE D P++DL + ++S + + I +A +GFF+VINHG++ + I +M
Sbjct: 24 SDRPRLSEVSQLEDFPLIDLSSTDRSFLIQQIHQACARFGFFQVINHGVNKQIIDEMVSV 83
Query: 64 GFGFFAKPMPQKKQAAPGYGCK----NIGFNGDMGEV----EYLLLNANTPSIAQISKSV 115
FF+ M +K + K + FN EV +YL L+ I K V
Sbjct: 84 AREFFSMSMEEKMKLYSDDPTKTTRLSTSFNVKKEEVNNWRDYLRLHCYP-----IHKYV 138
Query: 116 SIDPSN---FRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLN 172
+ PSN F+ VS Y+ VRE+ +I E+++E LG++ + L E + +N
Sbjct: 139 NEWPSNPPSFKEIVSKYSREVREVGFKIEELISESLGLEKDYMKKVLG---EQGQHMAVN 195
Query: 173 HYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKPD 231
+YPP P G H+DP LTIL + V GLQI L DG W V P
Sbjct: 196 YYPPC----------PEPELTYGLPAHTDPNALTILLQDTTVCGLQI-LIDGQWFAVNPH 244
Query: 232 PEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPV----- 286
P+AF +N+GD L+ ++NG + SV HRA+TN+ R+S+A F P C V P
Sbjct: 245 PDAFVINIGDQLQALSNGVYKSVWHRAVTNTENPRLSVASFLCPA--DCAVMSPAKPLWE 302
Query: 287 LVTPERPSLFRPFTWADYKKATYSLRLGDSR-MELFRNN 324
E +++ FT+A+Y K +S L +E F NN
Sbjct: 303 AEDDETKPVYKDFTYAEYYKKFWSRNLDQEHCLENFLNN 341
>At4g25420 gibberellin 20-oxidase - Arabidopsis thaliana
Length = 377
Score = 135 bits (340), Expect = 3e-32
Identities = 94/311 (30%), Positives = 155/311 (49%), Gaps = 35/311 (11%)
Query: 18 DLPIVDLK------AEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKP 71
D+P++DL+ + + +RLI +A +++GFF V+NHGIS E I+ E FF P
Sbjct: 60 DVPLIDLQNLLSDPSSTLDASRLISEACKKHGFFLVVNHGISEELISDAHEYTSRFFDMP 119
Query: 72 MPQKKQAAPGYGCKNIGFNGDM-GEVEYLLLNANTPSIA---QISKSVSIDP-------- 119
+ +K++ G +++G+ G L T S +S+S S+
Sbjct: 120 LSEKQRVLRKSG-ESVGYASSFTGRFSTKLPWKETLSFRFCDDMSRSKSVQDYFCDALGH 178
Query: 120 --SNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPI 177
F Y EA+ L+ +I+E++ LGV+ F E+DS++RLN+YPP
Sbjct: 179 GFQPFGKVYQEYCEAMSSLSLKIMELLGLSLGVKRDY-FREFFE--ENDSIMRLNYYPPC 235
Query: 178 LNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCV 237
+ + +G G H DP LTIL + V+GLQ+ +++ W ++P+P+AF V
Sbjct: 236 IKPDLT----------LGTGPHCDPTSLTILHQDHVNGLQVFVENQ-WRSIRPNPKAFVV 284
Query: 238 NVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFR 297
N+GD ++N R+ S HRA+ NS R S+A+F P + P L+ +
Sbjct: 285 NIGDTFMALSNDRYKSCLHRAVVNSESERKSLAFFLCPKKDRVVTPPRELLDSITSRRYP 344
Query: 298 PFTWADYKKAT 308
FTW+ + + T
Sbjct: 345 DFTWSMFLEFT 355
>At5g08640 flavonol synthase (FLS) (sp|Q96330)
Length = 336
Score = 134 bits (337), Expect = 7e-32
Identities = 95/315 (30%), Positives = 156/315 (49%), Gaps = 32/315 (10%)
Query: 9 IRSEKIVPG---------DLPIVDLK-AEKSEVTRLIVKASEEYGFFKVINHGISHETIA 58
IRSEK P +P+VDL ++ V R +VKASEE+G F+V+NHGI E I
Sbjct: 24 IRSEKEQPAITTFRGPTPAIPVVDLSDPDEESVRRAVVKASEEWGLFQVVNHGIPTELIR 83
Query: 59 KMEEAGFGFFAKPMPQKKQAAP--------GYGCK-NIGFNGDMGEVEYLLLNANTPSIA 109
++++ G FF P +K+ A GYG K G V++L PS
Sbjct: 84 RLQDVGRKFFELPSSEKESVAKPEDSKDIEGYGTKLQKDPEGKKAWVDHLFHRIWPPSCV 143
Query: 110 QISKSVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVL 169
+ +P +R Y V++L+ +L ++++GLG++ + L ++ ++ ++
Sbjct: 144 NY-RFWPKNPPEYREVNEEYAVHVKKLSETLLGILSDGLGLKRDALKEGLGGEM-AEYMM 201
Query: 170 RLNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVK 229
++N+YPP P + +G H+D +T+L N+V GLQ+ +D W +
Sbjct: 202 KINYYPPC----------PRPDLALGVPAHTDLSGITLLVPNEVPGLQV-FKDDHWFDAE 250
Query: 230 PDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVT 289
P A V++GD + ++NGR+ +V HR + K+RMS F PP + P L
Sbjct: 251 YIPSAVIVHIGDQILRLSNGRYKNVLHRTTVDKEKTRMSWPVFLEPPREKIVGPLPELTG 310
Query: 290 PERPSLFRPFTWADY 304
+ P F+PF + DY
Sbjct: 311 DDNPPKFKPFAFKDY 325
>At5g07200 gibberellin 20-oxidase
Length = 380
Score = 134 bits (336), Expect = 9e-32
Identities = 97/321 (30%), Positives = 150/321 (46%), Gaps = 37/321 (11%)
Query: 11 SEKIVPGDLPIVDL-------KAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEA 63
S + P +P++DL SE TRL+ KA+ ++GFF + NHG+ +++
Sbjct: 50 STDVQPLQVPLIDLAGFLSGDSCLASEATRLVSKAATKHGFFLITNHGVDESLLSRAYLH 109
Query: 64 GFGFFAKPMPQKKQA------APGYGCKNIG-FNGDMGEVEYLLLNANTPSIAQ------ 110
FF P +K++A + GY +G F+ + E L +
Sbjct: 110 MDSFFKAPACEKQKAQRKWGESSGYASSFVGRFSSKLPWKETLSFKFSPEEKIHSQTVKD 169
Query: 111 -ISKSVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVL 169
+SK + +F Y EA+ L+ +I+E++ LGV+ F D SDS+
Sbjct: 170 FVSKKMGDGYEDFGKVYQEYAEAMNTLSLKIMELLGMSLGVERRY-FKEFFED--SDSIF 226
Query: 170 RLNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVK 229
RLN+YP + K P +G G H DP LTIL + V GLQ+ + D W +
Sbjct: 227 RLNYYP--------QCKQPEL--ALGTGPHCDPTSLTILHQDQVGGLQVFV-DNKWQSIP 275
Query: 230 PDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVT 289
P+P AF VN+GD +TNGR+ S HRA+ NS + R + A+F P + P LV
Sbjct: 276 PNPHAFVVNIGDTFMALTNGRYKSCLHRAVVNSERERKTFAFFLCPKGEKVVKPPEELVN 335
Query: 290 PERPS--LFRPFTWADYKKAT 308
+ + FTW+ + + T
Sbjct: 336 GVKSGERKYPDFTWSMFLEFT 356
>At1g80340 gibberellin 3 beta-hydroxylase
Length = 347
Score = 132 bits (332), Expect = 3e-31
Identities = 92/303 (30%), Positives = 146/303 (47%), Gaps = 27/303 (8%)
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
LP++DL V L+ A +G F++ NHG+ + +E F P+ +K +A
Sbjct: 50 LPLIDLS--DIHVATLVGHACTTWGAFQITNHGVPSRLLDDIEFLTGSLFRLPVQRKLKA 107
Query: 79 A------PGYGCKNIG--FNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYT 130
A GYG I FN M + ++ + ++ S + + + Y
Sbjct: 108 ARSENGVSGYGVARIASFFNKKMWSEGFTVIGSPLHDFRKLWPSHHL---KYCEIIEEYE 164
Query: 131 EAVRELACEILEVMAEGLGVQDT-LVFSRLIRDIE-SDSVLRLNHYPPILNSNNREDKSP 188
E +++LA +++ LGV++ + ++ D + + +V++LNHYP K P
Sbjct: 165 EHMQKLAAKLMWFALGSLGVEEKDIQWAGPNSDFQGTQAVIQLNHYP----------KCP 214
Query: 189 SYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTN 248
+ +G H+D ++TIL N+ +GLQ+ D W+ P P + VNVGD+L ++TN
Sbjct: 215 EPDRAMGLAAHTDSTLMTILYQNNTAGLQVFRDDVGWVTAPPVPGSLVVNVGDLLHILTN 274
Query: 249 GRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADY--KK 306
G F SV HRA N +SR SMAY PP I P LV P + L+ TW Y K
Sbjct: 275 GIFPSVLHRARVNHVRSRFSMAYLWGPPSDIMISPLPKLVDPLQSPLYPSLTWKQYLATK 334
Query: 307 ATY 309
AT+
Sbjct: 335 ATH 337
>At1g80330 putative gibberellin 3 beta-hydroxylase
Length = 355
Score = 130 bits (327), Expect = 1e-30
Identities = 90/310 (29%), Positives = 146/310 (47%), Gaps = 27/310 (8%)
Query: 15 VPGDLPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQ 74
V +P++DL +VT LI AS+ +G F++ NHGIS + + +E F P +
Sbjct: 45 VEESIPVIDLS--NPDVTTLIGDASKTWGAFQIANHGISQKLLDDIESLSKTLFDMPSER 102
Query: 75 KKQAAP------GYGCKNIG--FNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTV 126
K +AA GYG I F M + + + + + + D + + +
Sbjct: 103 KLEAASSDKGVSGYGEPRISPFFEKKMWSEGFTIADDSYRN--HFNTLWPHDHTKYCGII 160
Query: 127 SAYTEAVRELACEILEVMAEGLGVQ-DTLVFSRLIRDIESD---SVLRLNHYPPILNSNN 182
Y + + +LA +L + LGV + + ++ + S +RLNHYP
Sbjct: 161 QEYVDEMEKLASRLLYCILGSLGVTVEDIEWAHKLEKSGSKVGRGAIRLNHYPVC----- 215
Query: 183 REDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDV 242
P +G H+D ILTIL ++ GLQ+ ++ W+ V+P P VN+GD+
Sbjct: 216 -----PEPERAMGLAAHTDSTILTILHQSNTGGLQVFREESGWVTVEPAPGVLVVNIGDL 270
Query: 243 LEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAP-PVLVTPERPSLFRPFTW 301
+++NG+ SV HRA N +SR+S+AY P +AP L P PSL+R TW
Sbjct: 271 FHILSNGKIPSVVHRAKVNHTRSRISIAYLWGGPAGDVQIAPISKLTGPAEPSLYRSITW 330
Query: 302 ADYKKATYSL 311
+Y + Y +
Sbjct: 331 KEYLQIKYEV 340
>At5g05600 leucoanthocyanidin dioxygenase-like protein
Length = 371
Score = 129 bits (325), Expect = 2e-30
Identities = 92/303 (30%), Positives = 147/303 (48%), Gaps = 28/303 (9%)
Query: 18 DLPIVDLKAEKSE-------VTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAK 70
++PI+DL+ SE + I +A +GFF+V+NHG+ E + E FF
Sbjct: 61 NIPIIDLEGLFSEEGLSDDVIMARISEACRGWGFFQVVNHGVKPELMDAARENWREFFHM 120
Query: 71 PMPQKK------QAAPGYGCK-NIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFR 123
P+ K+ + GYG + + + +Y L+ + +K S P+ R
Sbjct: 121 PVNAKETYSNSPRTYEGYGSRLGVEKGASLDWSDYYFLHLLPHHLKDFNKWPSFPPT-IR 179
Query: 124 STVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNR 183
+ Y E + +L+ I+ V++ LG+++ F + LR+N+YP
Sbjct: 180 EVIDEYGEELVKLSGRIMRVLSTNLGLKEDK-FQEAFGGENIGACLRVNYYP-------- 230
Query: 184 EDKSPSYNNKVGFGEHSDPQILTILRSND-VSGLQISLQDGVWIPVKPDPEAFCVNVGDV 242
K P +G HSDP +TIL +D V GLQ+ +D WI VKP P AF VN+GD
Sbjct: 231 --KCPRPELALGLSPHSDPGGMTILLPDDQVFGLQVR-KDDTWITVKPHPHAFIVNIGDQ 287
Query: 243 LEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWA 302
+++++N + SV HR + NS K R+S+A+F P I LV+ P L+ P T+
Sbjct: 288 IQILSNSTYKSVEHRVIVNSDKERVSLAFFYNPKSDIPIQPLQELVSTHNPPLYPPMTFD 347
Query: 303 DYK 305
Y+
Sbjct: 348 QYR 350
>At3g19000 unknown protein
Length = 352
Score = 129 bits (324), Expect = 2e-30
Identities = 89/315 (28%), Positives = 147/315 (46%), Gaps = 37/315 (11%)
Query: 14 IVPGDLPIVDLKA------EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGF 67
I ++P +DL + +K+ + + I +A + +GFF+VINHG+ ++E+ F
Sbjct: 27 IFSDEIPTIDLSSLEDTHHDKTAIAKEIAEACKRWGFFQVINHGLPSALRHRVEKTAAEF 86
Query: 68 FAKPMPQKKQAAP------GYGCKNIGFN-GDMGEVEYLLLNANT----------PSIAQ 110
F +K++ GY + N D E+ L +T + +
Sbjct: 87 FNLTTEEKRKVKRDEVNPMGYHDEEHTKNVRDWKEIFDFFLQDSTIVPASPEPEDTELRK 146
Query: 111 ISKSVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLR 170
++ +PS+FR Y V +LA +LE+++ LG+ + E S LR
Sbjct: 147 LTNQWPQNPSHFREVCQEYAREVEKLAFRLLELVSISLGLPGDRLTGFFN---EQTSFLR 203
Query: 171 LNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQ-DGVWIPVK 229
NHYPP P+ +G G H D LT+L + V GLQ+S + DG WIPVK
Sbjct: 204 FNHYPPC----------PNPELALGVGRHKDGGALTVLAQDSVGGLQVSRRSDGQWIPVK 253
Query: 230 PDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVT 289
P +A +N+G+ ++V TN + S HR + N+ K R S+ +F P A I L++
Sbjct: 254 PISDALIINMGNCIQVWTNDEYWSAEHRVVVNTSKERFSIPFFFFPSHEANIEPLEELIS 313
Query: 290 PERPSLFRPFTWADY 304
E P ++ + W +
Sbjct: 314 EENPPCYKKYNWGKF 328
>At1g15550 Gibberillin 3 beta-hydroxylase (AtGA3ox1)
Length = 358
Score = 128 bits (321), Expect = 5e-30
Identities = 89/321 (27%), Positives = 156/321 (47%), Gaps = 31/321 (9%)
Query: 18 DLPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQ 77
++P++DL + + T I A +G F++ NHG+ + +E F P+ +K +
Sbjct: 56 NIPLIDL--DHPDATNQIGHACRTWGAFQISNHGVPLGLLQDIEFLTGSLFGLPVQRKLK 113
Query: 78 AA------PGYGCKNIG--FNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAY 129
+A GYG I FN M + + + ++ + N+ V Y
Sbjct: 114 SARSETGVSGYGVARIASFFNKQMWSEGFTITGSPLNDFRKLWPQHHL---NYCDIVEEY 170
Query: 130 TEAVRELACEILEVMAEGLGV-QDTLVFSRLIRDIE-SDSVLRLNHYPPILNSNNREDKS 187
E +++LA +++ + LGV ++ + ++ L D+ + + L+LNHYP
Sbjct: 171 EEHMKKLASKLMWLALNSLGVSEEDIEWASLSSDLNWAQAALQLNHYPVC---------- 220
Query: 188 PSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMT 247
P + +G H+D +LTIL N+ +GLQ+ D W+ V P P + VNVGD+ +++
Sbjct: 221 PEPDRAMGLAAHTDSTLLTILYQNNTAGLQVFRDDLGWVTVPPFPGSLVVNVGDLFHILS 280
Query: 248 NGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADY--K 305
NG F SV HRA N ++R+S+A+ P I P LV+P L++ TW +Y
Sbjct: 281 NGLFKSVLHRARVNQTRARLSVAFLWGPQSDIKISPVPKLVSPVESPLYQSVTWKEYLRT 340
Query: 306 KATYSLRLGDSRMELFRNNND 326
KAT+ + + + RN+ +
Sbjct: 341 KATHF----NKALSMIRNHRE 357
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,556,021
Number of Sequences: 26719
Number of extensions: 321918
Number of successful extensions: 1082
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 772
Number of HSP's gapped (non-prelim): 128
length of query: 327
length of database: 11,318,596
effective HSP length: 100
effective length of query: 227
effective length of database: 8,646,696
effective search space: 1962799992
effective search space used: 1962799992
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0333b.12


