

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0331c.2
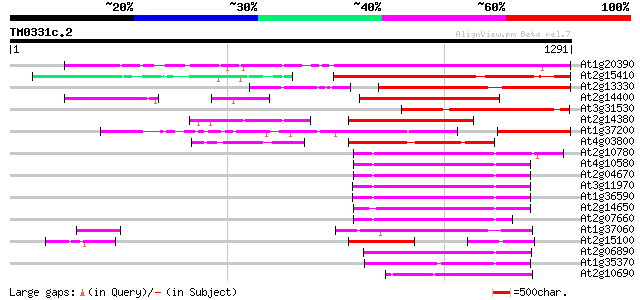
(1291 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 721 0.0
At2g15410 putative retroelement pol polyprotein 487 e-137
At2g13330 F14O4.9 409 e-114
At2g14400 putative retroelement pol polyprotein 384 e-106
At3g31530 hypothetical protein 372 e-103
At2g14380 putative retroelement pol polyprotein 355 7e-98
At1g37200 hypothetical protein 332 1e-90
At4g03800 318 1e-86
At2g10780 pseudogene 223 4e-58
At4g10580 putative reverse-transcriptase -like protein 218 2e-56
At2g04670 putative retroelement pol polyprotein 214 2e-55
At3g11970 hypothetical protein 206 7e-53
At1g36590 hypothetical protein 206 7e-53
At2g14650 putative retroelement pol polyprotein 206 9e-53
At2g07660 putative retroelement pol polyprotein 205 2e-52
At1g37060 Athila retroelment ORF 1, putative 198 2e-50
At2g15100 putative retroelement pol polyprotein 183 5e-46
At2g06890 putative retroelement integrase 182 1e-45
At1g35370 hypothetical protein 174 2e-43
At2g10690 putative retroelement pol polyprotein 171 3e-42
>At1g20390 hypothetical protein
Length = 1791
Score = 721 bits (1861), Expect = 0.0
Identities = 452/1199 (37%), Positives = 652/1199 (53%), Gaps = 116/1199 (9%)
Query: 126 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKM----VIIAASDAVKCRMFPS 181
PF+ + +V+I K + L+SY+G +DPK+ L FN + + I DA +C++F
Sbjct: 156 PFTRRITNVSIRGAQK-IKLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIE 214
Query: 182 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 241
A WF+ L SI +F +S FL ++ + DL++I Q ESL+ ++
Sbjct: 215 HLTGPAHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFV 274
Query: 242 ARYS--AASVKVEDEEPRACALAFKNGL-LPGGLNSKLTRKPARSMEEMRARASTYILDE 298
R+ ++ V DE A +A +N + +T ++E+ RAS +I E
Sbjct: 275 DRFKLVVTNITVPDE---AAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELE 331
Query: 299 EDDAFKRKRAKLEKGDTSPKRAKKDSSGEDKGDGKQQRPGKGKSVFKPTKEQLYPRRDDY 358
E+ K K +++ A KD+ G D
Sbjct: 332 EE-----KLILARKHNSTKTPACKDAVVIKVGPD-----------------------DSN 363
Query: 359 EQRRPWQSKSHRQREETDMVMNTDLSDMLQGARDANLVDEPEA-PKYQPRDANPKKWCEF 417
E R+ R+ T +++T+ D + D P A P Y CE+
Sbjct: 364 EPRQHLDRNPSAGRKPTSFLVSTETPDAKPWNKYIRDADSPAAGPMY----------CEY 413
Query: 418 HRSAGHDTDDCWTLQREIDKLIRAG---YQGNRQGLWRNGGDQNKAHKREEERADTKGKK 474
H+S H T++C LQ + ++G + +R + +N+ R+ TK
Sbjct: 414 HKSRAHSTENCRFLQGLLMAKYKSGGITIECDRPPINNKNQRRNETTARQYLNDQTKPPT 473
Query: 475 KQESAAIATKGADDTFA--QHSGPPVGT-------INTIAGGFGGGGDTHAARKRHVRAV 525
E I + ADD A Q +G + ++ I GG D+ + K++ +
Sbjct: 474 PAEQGIITS--ADDPAAKRQRNGKAIAAEPVVVRQVHVIMGGLQNCSDSVRSIKQYRKKA 531
Query: 526 NSVHEVAFGFV-----HPD----ITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQ 576
V VA+ +P+ I+ + D EG+ +DP+VV+L ++ V RVL+D
Sbjct: 532 EMV--VAWPSSTSTTRNPNQSAPISFTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDT 589
Query: 577 GSSADIIYGDAFDKLGLTDNDLTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKV 636
GSS D+I+ D + +TD + P + L GF G+ VM G I L G + V
Sbjct: 590 GSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTIKLPIFVG----GLIAWV 645
Query: 637 RYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMARECYNNC 696
+++V+ A YNVI+G ++++ A+ ST H VK+P +N
Sbjct: 646 KFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPT------------------HNGI 687
Query: 697 LNLYGKKSALVGHRCYEIEASDENLDPRGEGRVNRPTPIEETKALKFGDRTLKIGTRLTE 756
L K A R YE + L ++ P R + +G ++
Sbjct: 688 FTLRAPKEAKTPSRSYE----ESELCRTEMVNIDESDPT----------RCVGVGAEISP 733
Query: 757 EQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHRLALNPSLKPVS*LRRRLGGDKGKAV 816
L LL N FAWS +DM GIDP H L ++P+ KPV RR+LG ++ +AV
Sbjct: 734 SIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLGPERARAV 793
Query: 817 QQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKWRMCVDYTDLNKACPKDSYPLPSIDS 876
+EV+KLL A I EVKYP WLAN V+VKK NGKWR+CVDYTDLNKACPKDSYPLP ID
Sbjct: 794 NEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPLPHIDR 853
Query: 877 LVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMTARANYCYRTMPFGLKNAGATYQR 936
LV+ SGN LLS MDA+SG +QI MH D++KT+F+T R YCY+ M FGLKNAGATYQR
Sbjct: 854 LVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQR 913
Query: 937 LMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQG 996
++++ A Q+GR +EVY+DDM+VKS++ DH + L + F + + M+LNP KC FGV
Sbjct: 914 FVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVTS 973
Query: 997 GKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFF 1056
G+FLG+++T RGIE NP++ +AI ++ SP N +EVQRLTGRIAAL+RF+ +S D+ PF+
Sbjct: 974 GEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDKCLPFY 1033
Query: 1057 KCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLYFAVSDGALSSVMLQEI 1116
L++ F+W + EEAF +LK+ LS+PPIL KP G L+LY AVSD A+SSV+++E
Sbjct: 1034 NLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHAVSSVLVRED 1093
Query: 1117 DGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARRLRPYFQSFPVKVRTDLPLRQVLQ 1176
GE R +++ S +L AE RY IEKAALAV+ +AR+LRPYFQS + V TD PLR L
Sbjct: 1094 RGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLTDQPLRVALH 1153
Query: 1177 KPDLSGRLVAWSVELSEYSLQYDKRGAVGAQSLADFVVELTPDRFERV-----DTQWTLF 1231
P SGR+ W+VELSEY + + R A+ +Q LADF++EL ER +W+L+
Sbjct: 1154 SPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLADFLIELPLQSAERAVSGNRGEEWSLY 1213
Query: 1232 VDGSSNSSGSGAGVTLEGPGELVLEQSLKFEFKATNNQAEYEALIAGLKLAREVKIRSL 1290
VDGSS++ GSG G+ L P VLEQS + F ATNN AEYE LIAGL+LA ++I ++
Sbjct: 1214 VDGSSSARGSGIGIRLVSPTAEVLEQSFRLRFVATNNVAEYEVLIAGLRLAAGMQITTI 1272
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 487 bits (1253), Expect = e-137
Identities = 253/545 (46%), Positives = 340/545 (61%), Gaps = 55/545 (10%)
Query: 746 RTLKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHRLALNPSLKPVS*LR 805
R + I L E + L L +N FAWS +DMPGID CH L ++P+ KP+ R
Sbjct: 728 RCVGIRIDLPSELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKR 787
Query: 806 RRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKWRMCVDYTDLNKACP 865
R+LG D+ K V +EV KLL A I EV+YP WL N V+VKK NGKWR+C+D+TDLNKACP
Sbjct: 788 RKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACP 847
Query: 866 KDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMTARANYCYRTMPF 925
KDS+PLP ID LV+ +GNELLS MDA+SG +QI MH D +KT F+T + YCY+ MPF
Sbjct: 848 KDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVMPF 907
Query: 926 GLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRL 985
GLKNAGATY RL++++F Q+ +MEVY+DDM+VKS+R +H L + F + ++NM+L
Sbjct: 908 GLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKL 967
Query: 986 NPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGRIAALSRFL 1045
NP KC FGV G+FLG+++T RGIE NP++ AI + SP N +EVQRL GRIAAL+RF+
Sbjct: 968 NPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSPRNTREVQRLIGRIAALNRFI 1027
Query: 1046 PKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLYFAVSD 1105
+S D+ PF++ LR N FEW +CEE G L+LY AVS
Sbjct: 1028 SRSTDKCLPFYQLLRANKRFEWDEKCEE--------------------GETLYLYIAVST 1067
Query: 1106 GALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARRLRPYFQSFPVKV 1165
A+S V+++E GE +++VS TL GAE+RY +EK A AV+++AR+LRPYF+S+ V+V
Sbjct: 1068 SAVSGVLVREDRGEQHPIFYVSKTLDGAELRYPTLEKLAFAVVISARKLRPYFKSYTVEV 1127
Query: 1166 RTDLPLRQVLQKPDLSGRLVAWSVELSEYSLQYDKRGAVGAQSLADFVVELTPDRFERVD 1225
T+ PLR +L P SGRL W+VELSEY + Y R + P R R
Sbjct: 1128 LTNQPLRTILHSPSQSGRLAKWAVELSEYDIAYKNRTCAKSHR--------APTRPYR-- 1177
Query: 1226 TQWTLFVDGSSNSSGSGAGVTLEGPGELVLEQSLKFEFKATNNQAEYEALIAGLKLAREV 1285
+ + F A+NN+AEYEALIAGL+LA +
Sbjct: 1178 -------------------------RSIPCRKVDLARFPASNNEAEYEALIAGLRLAHGI 1212
Query: 1286 KIRSL 1290
+++ +
Sbjct: 1213 EVKKI 1217
Score = 66.6 bits (161), Expect = 9e-11
Identities = 138/634 (21%), Positives = 221/634 (34%), Gaps = 125/634 (19%)
Query: 53 PSPSQVGSL-ERSPENSPAPEQQPAVTQEQWRHLMRSIGNIQQRNEHLQAQL-DFYRREQ 110
P+ VGS+ E + EN+P + +P V R G + + L+ L D +
Sbjct: 158 PASIVVGSIDEHTRENAPNRDNRPNVDTRARRTEDDDTGARDRELQQLRTTLKDTSSKMH 217
Query: 111 RDDGSR-EADSVAEFR---PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMV 166
R S E D V E PF+ + V I ++ + L +Y G DP+ L + +
Sbjct: 218 RAVSSTPEVDKVLEETQRSPFTSCISDVRIR-HISKIKLANYEGLVDPRPFLTSVSIAIG 276
Query: 167 IIAASD----AVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVT 222
SD A C++F A+ WF+ L SI + ++ FL + + +
Sbjct: 277 RAHFSDEDRDAGSCQLFVKHLSGAALTWFSRLEANSIDSVHALTTSFLKNYGVFMEKGAS 336
Query: 223 INDLYNIRQQEGESLKEYMARYSAASVKVEDEEPRACALAFKNGLLPGGLNSKLTR-KPA 281
DL+ + Q ESL+ ++ R+ V + A A A +N L G + T KP
Sbjct: 337 NVDLWTMAQTAKESLRSFIGRFKEIVTSVATPDDAAIA-ALRNALWHGDSSRCTTLCKPF 395
Query: 282 RSMEEMRARASTYILDEEDDAFKRKRAKLEKGDTSPKRAKKDSSGEDKGD-GKQQRPGKG 340
++ AK PK +S D K+++ KG
Sbjct: 396 H---------------------RQGSAKAPIAKEKPKEGHHESRQHYDADYAKEEKLKKG 434
Query: 341 KSVFKPTKEQLYPRRDDYEQRRPWQSKSHRQREETDMVMNTDLSDMLQGARDANLVDEPE 400
S + PR D +PW + + D
Sbjct: 435 TSYYVGDTA---PRSD-----KPWNKWNRSADAKGD------------------------ 462
Query: 401 APKYQPRDANPKKWCEFHRSAGHDTDDCWTLQREIDKLIRAGYQGNRQGLWRNG-GDQNK 459
+K+ EFH+ GH TD+C LQ + + G R G D++
Sbjct: 463 -----------QKYYEFHKIPGHSTDECRQLQTLLLTKFKKGDLDIEPDRRRTGTNDRDN 511
Query: 460 AHKREE--ERADTKGKKKQESA-------------AIATKGADDTFAQHSGPPVG--TIN 502
H+R E +R + K++Q+ A K D Q P N
Sbjct: 512 THRRVENTDRDNQDDKRRQDETDRRAERIHDDDRRAEPHKRNHDAPRQFEDEPASKRRRN 571
Query: 503 TIAGGFGGGGDTHAARKRHVRAVNSVH-----EVAFGFVHPDITISMADFEGIKPHKDDP 557
I GG D+ + K + R + + IT S AD + +DP
Sbjct: 572 MIMGGLTACRDSVRSIKSYRRQGEKQRAWIEKNTSAVECNEPITFSEADI-ALPGSHNDP 630
Query: 558 IVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDNDLTPYAGTLVGFAGEQVMVRG 617
+VV+L++ + ++ +D S D + L GF G+ +M G
Sbjct: 631 LVVELKIGENVLLQLEIDVRSFQDKVQ-------------------PLTGFDGDTIMTVG 671
Query: 618 YIDLDTIFGEDECARVLKVRYLVLQVVASYNVII 651
I L G V + V+ YNVI+
Sbjct: 672 TITLPIYVG----GTVSWFEFAVVDKPIIYNVIL 701
>At2g13330 F14O4.9
Length = 889
Score = 409 bits (1052), Expect = e-114
Identities = 211/445 (47%), Positives = 286/445 (63%), Gaps = 32/445 (7%)
Query: 848 NGKWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADED 907
NGKWR+C+D+ DLNKACPKDS+PLP ID LV+ +E LS MDA+ G +QI M ++
Sbjct: 291 NGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQE 350
Query: 908 KTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDH 967
KTAF+T R Y Y+ MPFGLKNAG TYQRL++R+F Q+G+ +EVY+DDM+VKS DH
Sbjct: 351 KTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDH 410
Query: 968 HQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSN 1027
L E F + K M+LNPEKC F VQ +FL +++T RGIE NP++ A +M SP
Sbjct: 411 VPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSPKM 470
Query: 1028 VKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPI 1087
+EVQRLTGRI AL+ F+ +S ++ PF++ LRK F+W +CE+AF +LK L+ PPI
Sbjct: 471 AREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTEPPI 530
Query: 1088 LSKPIQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAV 1147
L+KP +G PL+LY + R +EK AL V
Sbjct: 531 LAKPEKGEPLYLY------------------------------TNRDKRCPAMEKLALTV 560
Query: 1148 LVTARRLRPYFQSFPVKVRTDLPLRQVLQKPDLSGRLVAWSVELSEYSLQYDKRGAVGAQ 1207
+++ R+LR YFQS P+ V T P+R +L SGRL W++EL EY ++Y R ++ AQ
Sbjct: 561 VMSVRKLRLYFQSHPIVVMTSQPIRTILHSLTQSGRLAKWAIELREYDIEYRTRTSLKAQ 620
Query: 1208 SLADFVVELTPDRFERVDT--QWTLFVDGSSNSSGSGAGVTLEGPGELVLEQSLKFEFKA 1265
LADFV++L + ++ +W L VDGSSN GSG G+ L P V+EQSL+ F A
Sbjct: 621 VLADFVIKLPLADLDGTNSNKKWLLHVDGSSNRQGSGVGIQLTSPTGEVIEQSLQLGFNA 680
Query: 1266 TNNQAEYEALIAGLKLAREVKIRSL 1290
+NN++EYEALIAG+KLA+E IR +
Sbjct: 681 SNNESEYEALIAGIKLAQEKGIREI 705
Score = 87.8 bits (216), Expect = 4e-17
Identities = 75/241 (31%), Positives = 117/241 (48%), Gaps = 21/241 (8%)
Query: 551 KPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDNDLTPYAGTLVGFAG 610
KPH DD +V+++ + ++ + +++D GSS D+++ DAF + G D+ L L GFAG
Sbjct: 59 KPH-DDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTGFAG 117
Query: 611 EQVMVRGYIDLDTIFGEDECARVLK--VRYLVLQVVASYNVIIGRNTLNRLCAVISTAHL 668
+ G I L TI AR ++ +LV+ A +N I+GR L+ + AV ST H
Sbjct: 118 DTTFSIGTIQLPTI------ARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQ 171
Query: 669 AVKYPLSSGKVGKLKVDQKMARECYNNCLNLYGKKSALVGHRCYEIEASD----ENLDPR 724
+K+P G + + Q+ +R+CY K +V E E ++ +LDP
Sbjct: 172 CIKFPSYKG-IAVVYGSQRSSRKCYMGSYEDIKKADPVV--LMIEDELAEMKTVRSLDPS 228
Query: 725 GEGRVNRPTPIEETKALKFGD--RTLKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPG 782
G R + I + + D R + IG L L L EN D FAWS ++ G
Sbjct: 229 QRG--TRKSLITQV-CIDESDPKRCVGIGHDLDLTVREDLITFLKENKDSFAWSSANLQG 285
Query: 783 I 783
I
Sbjct: 286 I 286
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 384 bits (985), Expect = e-106
Identities = 181/322 (56%), Positives = 243/322 (75%)
Query: 805 RRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKWRMCVDYTDLNKAC 864
RR+LG ++ KAV EVDKLL IREV+YP WLAN V+VKK NGK R+C+D+TDLNKAC
Sbjct: 469 RRKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKAC 528
Query: 865 PKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMTARANYCYRTMP 924
PKDS+PLP ID LV+ +GNELLS MDA+SG +QI M+P D++KT F+T R YCY+ MP
Sbjct: 529 PKDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMP 588
Query: 925 FGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMR 984
FGL+NAGATY RL++++F+ VG+ MEVY+DDM++KS++ DH + LEE F + ++ M+
Sbjct: 589 FGLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMK 648
Query: 985 LNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGRIAALSRF 1044
LNP KC FGV G+FLG+++T RGIE NP + A M SP N KEVQRLTGRIAAL+RF
Sbjct: 649 LNPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRF 708
Query: 1045 LPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLYFAVS 1104
+ +S D+S PF++ L+ N F W +CEEAF +LK L+SPP+L KP L+LY +VS
Sbjct: 709 ISRSTDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVS 768
Query: 1105 DGALSSVMLQEIDGEHRIVYFV 1126
+ A+S V+++E GE + +Y++
Sbjct: 769 NHAVSGVLIREDRGEQKPIYYI 790
Score = 81.6 bits (200), Expect = 3e-15
Identities = 63/234 (26%), Positives = 112/234 (46%), Gaps = 20/234 (8%)
Query: 126 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVII----AASDAVKCRMFPS 181
PF+ + S+ I D+ K L L+SY+G DPK +L F + A D C++F
Sbjct: 144 PFTPQITSLRIRDSRK-LNLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSE 202
Query: 182 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 241
A+ WFT L GSISNF + S FL Q+S ++ ++ DL+N+ Q E+L+ ++
Sbjct: 203 NLCGQALMWFTQLEPGSISNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFI 262
Query: 242 ARYSAASVKVEDEEPRACALAFKNGL-LPGGLNSKLTRKPARSMEEMRARASTYI-LDEE 299
++ K+ ++ A + GL L ++++ RA+ ++ +++E
Sbjct: 263 TKFKYVLSKLSRISQQSALSALRKGLWYDSRFKEDLILHKLDTIQDALFRANNWMEVEDE 322
Query: 300 DDAFKRKRAKLEKGDTSPKRAKKDSSGEDKGDGK-----------QQRPGKGKS 342
++F ++ + + T P KK E++G K +Q GKG+S
Sbjct: 323 KESFAKRDKQAKPAVTFP--PKKFEPRENQGPRKFGLLPFNNNVGKQFQGKGRS 374
Score = 43.5 bits (101), Expect = 8e-04
Identities = 36/148 (24%), Positives = 66/148 (44%), Gaps = 13/148 (8%)
Query: 464 EEERADTKGKKKQESAAIATKGADDTFAQHSGP-PVGTI---NTIAGGFGGGG------- 512
E+E+ + KQ A+ ++ GP G + N + F G G
Sbjct: 320 EDEKESFAKRDKQAKPAVTFPPKKFEPRENQGPRKFGLLPFNNNVGKQFQGKGRSNTWIR 379
Query: 513 DTHAARKRHVRAVNSVHEVA--FGFVHPDITISMADFEGIKPHKDDPIVVQLRMNSFNVR 570
D + K+H R V+ ++ F I + + ++ DD +VV L + +F V
Sbjct: 380 DESSVIKKHSRNVHLKLSLSEEVDFQSTSILFDEKETQHLERSHDDALVVTLDVANFEVS 439
Query: 571 RVLLDQGSSADIIYGDAFDKLGLTDNDL 598
R+L+D GSS D+I+ +++G++ D+
Sbjct: 440 RILIDTGSSVDLIFLSTLERMGISRADV 467
>At3g31530 hypothetical protein
Length = 831
Score = 372 bits (955), Expect = e-103
Identities = 191/390 (48%), Positives = 262/390 (66%), Gaps = 26/390 (6%)
Query: 901 MHPADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVK 960
M+P D++KTAF T + +CYR MPFGLKNAGATY+R ++++FA Q+G+ MEVY+DDM+VK
Sbjct: 96 MNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKIFALQIGKTMEVYIDDMLVK 155
Query: 961 SVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQ 1020
S+ DH L E F ++ +N++LNP KC FGV+ G IE NP++ +A+
Sbjct: 156 SMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRSG-----------IEANPKQIEALL 204
Query: 1021 QMKSPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKE 1080
M SP N +EVQ LTGR+AAL+RF+ +S ++ F+ LR+N FEWT CEEAF LK+
Sbjct: 205 GMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRRNKKFEWTTRCEEAFQELKK 264
Query: 1081 LLSSPPILSKPIQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKI 1140
L++PPIL+KP+ G P +LY AVSD A+S +++E GE +++++VS T AE RY ++
Sbjct: 265 YLATPPILAKPVIGEPQYLYVAVSDTAVSGELVREDRGEQKLIFYVSQTFTSAESRYPQM 324
Query: 1141 EKAALAVLVTARRLRPYFQSFPVKVRTDLPLRQVLQKPDLSGRLVAWSVELSEYSLQYDK 1200
EK ALAV+++A++LRPYFQS + V +PLR +L P SGRL W++ELSEY ++Y
Sbjct: 325 EKLALAVVMSAQKLRPYFQSHSIIVMGSMPLRVILHSPSQSGRLAKWTIELSEYDIEYQN 384
Query: 1201 RGAVGAQSLADFVVELTPD--RFERVDTQWTLFVDGSSNSSGSGAGVTLEGP-GELVLEQ 1257
+ +Q LADF+VEL R +DT W L VDGSS+ GSG G+ L P GE
Sbjct: 385 KTCAKSQVLADFIVELPTKEARENPLDTTWLLHVDGSSSKQGSGVGIRLTSPTGE----- 439
Query: 1258 SLKFEFKATNNQAEYEALIAGLKLAREVKI 1287
ATNN AEYEAL+AGL LA +KI
Sbjct: 440 -------ATNNVAEYEALVAGLNLAWGLKI 462
Score = 35.8 bits (81), Expect = 0.16
Identities = 21/51 (41%), Positives = 27/51 (52%)
Query: 647 YNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMARECYNNCL 697
YN IIG LN+ AV ST HL +K+P S KL ++ A + N L
Sbjct: 30 YNAIIGTPWLNQFRAVASTYHLCLKFPTSDSVWEKLGHERAEAVKAENQLL 80
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 355 bits (912), Expect = 7e-98
Identities = 169/289 (58%), Positives = 217/289 (74%), Gaps = 1/289 (0%)
Query: 779 DMPGIDPNFICHRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWL 838
DM GIDP CH L ++P+ K V RR+LG ++ KAV EVDKLL A I EVKYP WL
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 839 ANVVIVKKANGKWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQ 898
AN V+VKK N KWR+C+D+TDLNKACPKDS+PLP ID +V+ +GNELLS MDA+SG +Q
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 899 IRMHPADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMI 958
I MH D++KT+F+ R YCY+ MPFGLKN GA YQRL++++FA Q+G+ MEVY+DDM+
Sbjct: 596 IPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDML 655
Query: 959 VKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKA 1018
VKS R DH L+ F + K+NM+LNP KC FGV G+FLG+++T RGIE NP++ +A
Sbjct: 656 VKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRA 715
Query: 1019 IQQMKSPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRK-NVAFE 1066
I ++SP N KEVQRLTGRIA L+RF+ +S D+S PF++ LR N FE
Sbjct: 716 ILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLRSANKTFE 764
Score = 93.6 bits (231), Expect = 7e-19
Identities = 73/298 (24%), Positives = 131/298 (43%), Gaps = 25/298 (8%)
Query: 413 KWCEFHRSAGHDTDDCWTLQR---------EIDKLIRAGYQGNRQGLWRNGGDQNKA--- 460
++C+FH+ +GH T C LQ +I+ R N R G D N
Sbjct: 185 QYCDFHKRSGHSTAACRHLQSILLNKYKKGDIEVQHRQYKSHNNTYAARGGRDGNNVFHR 244
Query: 461 ---HKREEERADTKGKKKQESAAIATKGADDTFAQHSGPPVGT--INTIAGGFGGGGDTH 515
H ++ A ++++ K + QH+ PV +N I GG D+
Sbjct: 245 LGPHTGRQQEAPPANEEERHPDMEPPKKNRENDQQHNDAPVPRRRVNMIMGGLTACRDSF 304
Query: 516 AARKRHVR--AVNSVHEVAFGFVHPDITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVL 573
+ K +++ A A + P +T + D G+ +DP+V++L + V R+L
Sbjct: 305 RSIKEYIKSGAATLWSSPATKEMTP-LTFTSEDLFGVDLPHNDPLVIELHIGESEVTRIL 363
Query: 574 LDQGSSADIIYGDAFDKLGLTDNDLTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECARV 633
+D GSS ++++ D K+ + D + P L GF G +M G I L G
Sbjct: 364 IDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTNGTIKLPIYLG----GAA 419
Query: 634 LKVRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMARE 691
+++V+ YN+I+G ++ + A+ S+ H +K P S G + ++ +Q +A +
Sbjct: 420 TWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG-IETIRGNQNLAHD 476
Score = 32.7 bits (73), Expect = 1.4
Identities = 34/137 (24%), Positives = 57/137 (40%), Gaps = 15/137 (10%)
Query: 30 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVTQEQWRHLMRSI 89
+P+ V++ P + P P P G R PA P LMR+I
Sbjct: 39 NPIEVNPVEN-EPTLNTPTPTVGLEPGVQGMEGRPALTPPARPDDPGA-------LMRNI 90
Query: 90 -GNIQQRNEHLQAQLDFYRREQRDDGSREADSVAEFR---PFSEDVESVAIPDNMKTLVL 145
IQ+ +QA + Q + E + + E PF+ + +V++ + K + L
Sbjct: 91 LAQIQELRSTIQAMSQ--KVHQATSSTPELEQILEKNQKTPFAVHISNVSVRHDNK-IKL 147
Query: 146 DSYSGDSDPKDHLLYFN 162
SY G++DP+ L F+
Sbjct: 148 KSYEGNTDPQQFLSSFS 164
>At1g37200 hypothetical protein
Length = 1564
Score = 332 bits (850), Expect = 1e-90
Identities = 254/848 (29%), Positives = 389/848 (44%), Gaps = 138/848 (16%)
Query: 209 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASVKVEDEEPRACALAFKNGL- 267
FL Q S Q + DL+++ + ES + Y+ ++ A K+ + A +NGL
Sbjct: 176 FLKQDSVLIEQRTSEADLWSLTLLQNESHRSYVEKFKAIKSKIANLNEEVAIAALRNGLW 235
Query: 268 LPGGLNSKLTRKPARSMEEMRARASTYILDEEDDAFKRKRAKLEKGDTSPKRAKKDSSGE 327
+LT + S+++ +A + EE+ A +
Sbjct: 236 FSSRFREELTVRQPISLDDALHKALHFAKGEEELAVLALK-------------------- 275
Query: 328 DKGDGKQQRPGKGKSVFKPTKEQLYPRRDDYEQRRPWQSKSHRQREETDMVMNTDLSDML 387
FK +K Q P + P++ ++ Q + T +
Sbjct: 276 ----------------FKESKTQNAP----LAIKTPFKKENQTQGQHTLFAIE------- 308
Query: 388 QGARDANLVDEPEAPKYQPRDANPKKWCEFHRSAGHDTDDCWTLQREIDKLIRAGYQGNR 447
EA + + + + K+C++H GH T++C R + KLI AG + +
Sbjct: 309 ------------EAAEDESPELDLGKYCKYHNKRGHSTEEC----RAVKKLIAAGGKTKK 352
Query: 448 QGLWRNGGDQNKAHKREEERADTKGKKKQESAAIATKGADDTFAQHSGPPVGT--INTIA 505
G K + + + KQ+ +G D S PP I+ +
Sbjct: 353 -------GSNPKVETPPPDEQEEEQTPKQKKRERTLEGGD------SPPPARRERIDLVF 399
Query: 506 GGFGGGGDTHAARKRHVRAVNSV-HEVAFGFVHPDITISMADFEGIKPHKDDPIVVQLRM 564
GG T R V S H+ F + E + P + D I+ ++
Sbjct: 400 TELDLGGKTG----RSVTTPPSAPHKKNMRFPLRLSKFCRSATEHLSPKRIDFIMGGSQL 455
Query: 565 NSFNVRRVLLDQGSSADIIYGDAF-----DKLGLTDNDLTPYAGTLVGFAGEQVMVRGYI 619
+ ++ + Q + G + K+ L +++ T
Sbjct: 456 CNDSINSIKTHQRKADSYTKGKSLMMGPDHKITLWESETT-------------------- 495
Query: 620 DLDTIFGEDECARVLKVRYLVLQVV---------ASYNVIIGRNTLNRLCAVISTAHLAV 670
DLD + R+ Y + +++ A +N I+GR L+ + AV ST H +
Sbjct: 496 DLDKPHDDAIVIRIDVGNYKLSRIMIDTGSSVDKAPFNAILGRPWLHAMKAVPSTYHQCI 555
Query: 671 KYPLSSGKVGKLKVDQKMARECYNNCLNLYGKKSALVGHRCYEIEASDENLDPRGEGRVN 730
K+P G + + Q+ +R CY L KK+ LV + ++ L R +
Sbjct: 556 KFPSEKG-IAVIYGSQRSSRRCYMGSYELI-KKAELV------VLMIEDKLAEMKTVRSS 607
Query: 731 RPT---PIEETKALKFGDRT-----LKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPG 782
P+ P + + + D + + +G L L EN D FAWS ++ G
Sbjct: 608 DPSQCGPQKSSITQVYIDESDPKQCVGVGQDLDPAIREDFITFLKENKDSFAWSSANLQG 667
Query: 783 IDPNFICHRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVV 842
I H L ++P+ +P+ RR+LG ++ KAVQ EVD+LL IREVKYP WLAN V
Sbjct: 668 ISLEVTSHELNVDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPDWLANPV 727
Query: 843 IVKKANGKWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMH 902
+VK NGKWR+C+D+ DLNKACPKDS+PLP ID LV +G+ELLS MDA+SG +QI M
Sbjct: 728 VVKNKNGKWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQILMR 787
Query: 903 PADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSV 962
P D++KTAF+T CY+ MPFGLKN GATYQRL++R+FA Q+G+ ME+Y+DDM+VKS
Sbjct: 788 PDDQEKTAFITD----CYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVKSA 843
Query: 963 RGLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQM 1022
DH L E F + K M+LNPEKC FGV G+FLG+++T RGIE NP++ A M
Sbjct: 844 HEKDHLPQLRECFKILNKFEMKLNPEKCSFGVPSGEFLGYLVTQRGIEANPKQIAAFIDM 903
Query: 1023 KSPSNVKE 1030
SP ++
Sbjct: 904 PSPKMARD 911
Score = 156 bits (395), Expect = 6e-38
Identities = 81/170 (47%), Positives = 113/170 (65%), Gaps = 2/170 (1%)
Query: 1123 VYFVSHTLQGAEVRYQKIEKAALAVLVTARRLRPYFQSFPVKVRTDLPLRQVLQKPDLSG 1182
+Y+VS TL AE Y+ +EK ALAV+++AR+LR Y QS P+ V T P+R +L S
Sbjct: 934 IYYVSKTLIDAETHYRAMEKLALAVVMSARKLRQYLQSHPIVVMTSQPIRTILHSTTQSS 993
Query: 1183 RLVAWSVELSEYSLQYDKRGAVGAQSLADFVVELTPDRFERVDT--QWTLFVDGSSNSSG 1240
RL W++ELSEY ++Y R + AQ LADFV+EL + ++ +W L VDGSSN G
Sbjct: 994 RLAKWAIELSEYDIEYRTRTSFKAQVLADFVIELPLADLDGTNSNKKWLLHVDGSSNRQG 1053
Query: 1241 SGAGVTLEGPGELVLEQSLKFEFKATNNQAEYEALIAGLKLAREVKIRSL 1290
SG G+ L P V+EQS + F A+NN++EYEALI G+KLA+ ++IR +
Sbjct: 1054 SGEGIQLTSPTGEVIEQSFRLGFNASNNESEYEALIDGIKLAQGMRIRDI 1103
>At4g03800
Length = 637
Score = 318 bits (815), Expect = 1e-86
Identities = 163/336 (48%), Positives = 227/336 (67%), Gaps = 21/336 (6%)
Query: 780 MPGIDPNFICHRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLA 839
MP IDP+ ICH L ++P KP+ RR+LG ++ KAV ++DKLL IREV+YP W+A
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 840 NVVIVKKANGKWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQI 899
V+VKK NGK R+C+D+TDLNKACPKDS+PLP ID LV+ +GNELL+ MDA+ G +QI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 900 RMHPADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIV 959
M+P D++KT+F+T R GATYQ L++++F + + MEV +DD +V
Sbjct: 441 MMNPEDQEKTSFITDR---------------GATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 960 KSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAI 1019
KS++ DH + L E F + ++ M+LN KC FGV G+FLG+++T RGIE NP + A
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINAF 545
Query: 1020 QQMKSPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLK 1079
+ S N KEVQRLTGRIA+ +S D+S PF++ L+ N F W +CEEAF +LK
Sbjct: 546 LKTPSLRNFKEVQRLTGRIAS------RSTDKSLPFYQILKGNNGFLWDEKCEEAFRQLK 599
Query: 1080 ELLSSPPILSKPIQGHPLHLYFAVSDGALSSVMLQE 1115
L++PP+LSKP L+LY VS+ A+S V+++E
Sbjct: 600 AYLTTPPVLSKPEADEKLYLYVFVSNHAVSGVLVRE 635
Score = 63.9 bits (154), Expect = 6e-10
Identities = 67/262 (25%), Positives = 115/262 (43%), Gaps = 37/262 (14%)
Query: 419 RSAGHDTDDCWTLQREIDKLIRAGYQGNRQGLWRNGGDQNKAHKREEERADTKGKKKQES 478
R H T DC L+R + +L +G N D K + + ++T+ K+
Sbjct: 77 RVTEHLTKDCIVLKRYLAELWASGDLSKF-----NIDDFVKQYHEARDNSETQNSKRPRL 131
Query: 479 AAIATKGADDTFAQHSGPPVGTINTIAGGFGGGGDTHAARKRH---VRAVNSVHEVAFGF 535
T + G IN I GG ++ +A K+H V +S+ E +
Sbjct: 132 ----------TNEEEPRSSKGKINVILGGSKLCHNSVSAIKKHRPNVLLKSSLSEEV-NY 180
Query: 536 VHPDITISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTD 595
++ + I+ DD +++ L + +F + R+L+D GSS D+I+
Sbjct: 181 QCSSVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLIF----------- 229
Query: 596 NDLTPYAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKVRYLVLQVVASYNVIIGRNT 655
L P LV F E M G I L + G + +++ V ++V A+YN+I+G
Sbjct: 230 --LGP-PSPLVAFTSESAMSLGTIKLPVLAG--KMSKI--VDFVVFDKPATYNIILGTPW 282
Query: 656 LNRLCAVISTAHLAVKYPLSSG 677
+ ++ AV ST H +K+P S+G
Sbjct: 283 IFQMKAVPSTYHQWLKFPTSNG 304
>At2g10780 pseudogene
Length = 1611
Score = 223 bits (569), Expect = 4e-58
Identities = 147/494 (29%), Positives = 245/494 (48%), Gaps = 18/494 (3%)
Query: 792 LALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKW 851
+ L P P+S R+ + +++++++LL FIR P W A V+ VKK +G +
Sbjct: 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSF 709
Query: 852 RMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAF 911
R+C+DY LNK K+ YPLP ID L+D G + S +D SG HQI + P D KTAF
Sbjct: 710 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 769
Query: 912 MTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 971
T ++ + MPFGL NA A + ++M+ VF + + ++++D++V S H + L
Sbjct: 770 RTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHL 829
Query: 972 EEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEV 1031
+R+H + KC F + FLG +I+ +G+ ++PEK ++I++ P N E+
Sbjct: 830 RAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 889
Query: 1032 QRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKP 1091
+ G RF+ + P + K+ AF W+ ECE++F+ LK +L++ P+L P
Sbjct: 890 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLP 949
Query: 1092 IQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTA 1151
+G P +Y S L V++Q + ++ + S L+ E Y + AV+
Sbjct: 950 EEGEPYTVYTDASIVGLGCVLMQ----KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFL 1005
Query: 1152 RRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSEYSLQYDKRGAVGAQSLA 1210
+ R Y V++ TD L+ + +P+L+ R W +++Y+L A +A
Sbjct: 1006 KIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVADYNLDIAYHPG-KANQVA 1064
Query: 1211 DFV------VELTPDRFERVDTQWTLFVDGSSNS---SGSGAGVTLEGPGELVLEQSLKF 1261
D + VE + + V+ TL V+ S G GA + + L Q
Sbjct: 1065 DALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLGLGAADQADLLSRIRLAQERDE 1124
Query: 1262 EFK--ATNNQAEYE 1273
E K A NN+ EY+
Sbjct: 1125 EIKGWAQNNKTEYQ 1138
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 218 bits (554), Expect = 2e-56
Identities = 131/407 (32%), Positives = 209/407 (51%), Gaps = 6/407 (1%)
Query: 792 LALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKW 851
+ L P P+S R+ + +++++ LL FIR P W A V+ VKK +G +
Sbjct: 483 IELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 541
Query: 852 RMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAF 911
R+C+DY +LN+ K+ YPLP ID L+D G S +D SG HQI + AD KTAF
Sbjct: 542 RLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 601
Query: 912 MTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 971
T ++ + MPFGL NA A + RLM+ VF + + +++DD++V S + L
Sbjct: 602 RTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHL 661
Query: 972 EEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEV 1031
++R+ + KC F + FLG ++++ G+ ++PEK +AI+ P+N E+
Sbjct: 662 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 721
Query: 1032 QRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKP 1091
+ G RF+ + P K K+V F W+ ECEE FV LKE+L+S P+L+ P
Sbjct: 722 RSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALP 781
Query: 1092 IQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTA 1151
G P +Y S L V++Q +++ + S L E Y + AV+
Sbjct: 782 EHGQPYMVYTDASRVGLGCVLMQ----HGKVIAYASRQLMKHEGNYPTHDLEMAAVIFAL 837
Query: 1152 RRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSEYSLQ 1197
+ R Y V+V TD L+ + +P+L+ R W +++Y L+
Sbjct: 838 KIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLE 884
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 214 bits (546), Expect = 2e-55
Identities = 129/407 (31%), Positives = 208/407 (50%), Gaps = 6/407 (1%)
Query: 792 LALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKW 851
+ L P P+S R+ + ++++++ LL FIR P W A V+ VKK +G +
Sbjct: 509 IELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 567
Query: 852 RMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAF 911
R+C+DY LN K+ YPLP ID L+D G S +D SG HQI + AD KTAF
Sbjct: 568 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 627
Query: 912 MTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 971
T ++ + MPF L NA A + RLM+ VF + + +++DD++V S +H L
Sbjct: 628 RTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHL 687
Query: 972 EEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEV 1031
++R+ + KC F + FLG ++++ G+ ++PEK +AI+ P+N E+
Sbjct: 688 RRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 747
Query: 1032 QRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKP 1091
+ RF+ + P K K+V F W+ ECEE FV LKE+L+S P+L+ P
Sbjct: 748 RSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 807
Query: 1092 IQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTA 1151
G P +Y S L V++Q +++ + S L+ E Y + V+
Sbjct: 808 EHGQPYMVYTDASRVGLGCVLMQ----RGKVIAYASRQLRKHEGNYPTHDLEMAVVIFAL 863
Query: 1152 RRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSEYSLQ 1197
+ R Y V+V TD L+ + +P+L+ R + W +++Y L+
Sbjct: 864 KIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADYDLE 910
>At3g11970 hypothetical protein
Length = 1499
Score = 206 bits (524), Expect = 7e-53
Identities = 134/412 (32%), Positives = 215/412 (51%), Gaps = 9/412 (2%)
Query: 790 HRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANG 849
H++ L PV+ R + + + V+ LL ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 850 KWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKT 909
WR+CVDY +LN KDS+P+P I+ L+D G + S +D +G HQ+RM P D KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 910 AFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQ 969
AF T ++ Y MPFGL NA AT+Q LM+ +F + + + V+ DD++V S +H Q
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 970 DLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVK 1029
L++ F +R + + KC F V ++LG I+++GIE +P K KA+++ P+ +K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 1030 EVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILS 1089
+++ G RF+ G + P L K AFEWTA ++AF LK L P+LS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 1090 KPIQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLV 1149
P+ + + +V++QE H + Y +S L+G ++ EK LAV+
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQE---GHPLAY-ISRQLKGKQLHLSIYEKELLAVIF 944
Query: 1150 TARRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSE--YSLQY 1198
R+ R Y ++TD L+ +L++ + W +L E Y +QY
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY 996
>At1g36590 hypothetical protein
Length = 1499
Score = 206 bits (524), Expect = 7e-53
Identities = 134/412 (32%), Positives = 215/412 (51%), Gaps = 9/412 (2%)
Query: 790 HRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANG 849
H++ L PV+ R + + + V+ LL ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 850 KWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKT 909
WR+CVDY +LN KDS+P+P I+ L+D G + S +D +G HQ+RM P D KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 910 AFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQ 969
AF T ++ Y MPFGL NA AT+Q LM+ +F + + + V+ DD++V S +H Q
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 970 DLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVK 1029
L++ F +R + + KC F V ++LG I+++GIE +P K KA+++ P+ +K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 1030 EVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILS 1089
+++ G RF+ G + P L K AFEWTA ++AF LK L P+LS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 1090 KPIQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLV 1149
P+ + + +V++QE H + Y +S L+G ++ EK LAV+
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQE---GHPLAY-ISRQLKGKQLHLSIYEKELLAVIF 944
Query: 1150 TARRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSE--YSLQY 1198
R+ R Y ++TD L+ +L++ + W +L E Y +QY
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY 996
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 206 bits (523), Expect = 9e-53
Identities = 126/407 (30%), Positives = 205/407 (49%), Gaps = 18/407 (4%)
Query: 792 LALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKW 851
+ L P P+S R+ + ++++++ LL A V+ VKK +G +
Sbjct: 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAP-------------VLFVKKKDGSF 516
Query: 852 RMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAF 911
R+C+DY LN K+ YPLP ID L+D G S +D SG H I + AD KTAF
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 912 MTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 971
T ++ + MPFGL NA A + RLM+ VF + + +++DD++V S +H L
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 972 EEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEV 1031
++R+ + KC F + FLG ++++ G+ ++PEK +AI+ +P+N E+
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEI 696
Query: 1032 QRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKP 1091
+ G RF+ + P K K+V F W+ ECEE FV LKE+L+S P+L+ P
Sbjct: 697 RSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 756
Query: 1092 IQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTA 1151
G P +Y S L V++Q +++ + S L+ E Y + AV+
Sbjct: 757 EHGEPYMVYTDASGVGLGCVLMQ----RGKVIAYASRQLRKHEGNYPTHDLEMAAVIFAL 812
Query: 1152 RRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSEYSLQ 1197
+ R Y V+V TD L+ + +P+L+ R W +++Y L+
Sbjct: 813 KIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLE 859
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 205 bits (521), Expect = 2e-52
Identities = 116/366 (31%), Positives = 191/366 (51%), Gaps = 5/366 (1%)
Query: 792 LALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKW 851
+ L P P+S R+ + +++++++LLA FIR P W A V+ VKK +G +
Sbjct: 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSF 204
Query: 852 RMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAF 911
R+C+DY LNK K+ YPLP ID L+D G + S +D SG HQI + P D KTAF
Sbjct: 205 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 264
Query: 912 MTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 971
T ++ + MPFGL NA A + ++M+ VF + + +++DD++V S H + L
Sbjct: 265 RTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHL 324
Query: 972 EEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEV 1031
+R+H + K F + FLG +I+ +G+ ++PEK ++I++ P N E+
Sbjct: 325 RAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 384
Query: 1032 QRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKP 1091
+ G RF+ + P + K+ AF W+ ECE++F+ LK +L + P+L P
Sbjct: 385 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLP 444
Query: 1092 IQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTA 1151
+G P +Y S L V++Q + ++ + S L+ E Y + AV+
Sbjct: 445 EEGEPYTVYTDASIVGLGCVLMQ----KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFAL 500
Query: 1152 RRLRPY 1157
+ R Y
Sbjct: 501 KIWRSY 506
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 198 bits (503), Expect = 2e-50
Identities = 140/477 (29%), Positives = 213/477 (44%), Gaps = 56/477 (11%)
Query: 750 IGTRLTEEQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHRLALN----PSLKPVS*LR 805
+ LT +Q L L + +S D+ GI P HR+ L S++P +
Sbjct: 849 VNDELTADQVNLLITELMKYRKAIGYSLDDIKGISPTLCTHRIHLENESYSSIEP----Q 904
Query: 806 RRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANGKW-------------- 851
RRL + + V++E+ KLL A I + TW++ V V K G
Sbjct: 905 RRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRT 964
Query: 852 ----RMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADED 907
RMC++Y LN A K+ +PLP ID +++ + + +D+YSG QI +HP D+
Sbjct: 965 ITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQG 1024
Query: 908 KTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDH 967
KT F + Y+ MPFGL NA AT+QR M +F+ + +EV++DD V
Sbjct: 1025 KTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSC 1084
Query: 968 HQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSN 1027
+L + N+ LN EKC+F V+ G LG I+ GIE++ K + Q++ P
Sbjct: 1085 LLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKT 1144
Query: 1028 VKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPI 1087
VK+++ G F+ + P + L K F + EC AF +KE L + PI
Sbjct: 1145 VKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEALITAPI 1204
Query: 1088 LSKPIQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAV 1147
+ P P + T+ A+VRY EK LAV
Sbjct: 1205 VQAPNWDFPFEII----------------------------TMDDAQVRYATTEKELLAV 1236
Query: 1148 LVTARRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSEYSLQ-YDKRG 1202
+ + R Y V + TD LR + K D RL+ W + L E+ ++ DK+G
Sbjct: 1237 VFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFDMEIVDKKG 1293
Score = 50.4 bits (119), Expect = 6e-06
Identities = 30/107 (28%), Positives = 46/107 (42%), Gaps = 4/107 (3%)
Query: 153 DPKDHLLYFNTKMVII----AASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 208
DP DHL F+ + + D K R+FP + A W +LP+GSI+++ D
Sbjct: 61 DPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKA 120
Query: 209 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASVKVEDEE 255
FL +F +N ND+ Q E+ E R+ + E
Sbjct: 121 FLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERFKGYQTQCPHHE 167
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 183 bits (465), Expect = 5e-46
Identities = 88/151 (58%), Positives = 111/151 (73%)
Query: 780 MPGIDPNFICHRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLA 839
M GI I H L ++P+ KPV RR+LG D+ +AV EV +LL IREVKYP WLA
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 840 NVVIVKKANGKWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQI 899
N V+VKK NGKWR+CVD+TDLNKAC KD +PLP ID LV+ +G+E+LS MDA+SG +QI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 900 RMHPADEDKTAFMTARANYCYRTMPFGLKNA 930
M+P D++KT+F+T Y Y+ MPFGLKNA
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
Score = 121 bits (304), Expect = 2e-27
Identities = 66/155 (42%), Positives = 93/155 (59%), Gaps = 8/155 (5%)
Query: 1053 FPFFKCLRK-NVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLYFAVSDGALSSV 1111
FPF K + F W CEEAF +LK LS P +L+K G L LY AVS+ A++ V
Sbjct: 616 FPFLHFTAKISKGFIWNESCEEAFKQLKRYLSEPSVLAKREFGEQLFLYIAVSESAVTGV 675
Query: 1112 MLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARRLRPYFQSFPVKVRTDLPL 1171
++ + R +++V + RY +EK ALAV+ AR+LRPYFQS P+ V T LPL
Sbjct: 676 QVRVERSDKRPIFYV-------KTRYPMMEKLALAVVTAARKLRPYFQSHPIVVLTSLPL 728
Query: 1172 RQVLQKPDLSGRLVAWSVELSEYSLQYDKRGAVGA 1206
R +L P SGRL W++ELSE+ L++ R ++ +
Sbjct: 729 RTILHSPTQSGRLGKWAIELSEFDLEFRARTSLNS 763
Score = 66.2 bits (160), Expect = 1e-10
Identities = 49/174 (28%), Positives = 77/174 (44%), Gaps = 18/174 (10%)
Query: 83 RHLMRSIGNIQQRNEHLQAQLDFYRRE--QRDDGSREADSVAEFR---PFSEDVESVAIP 137
RH +R + + ++LQ Q+ + Q + E + V E PF+ + + I
Sbjct: 109 RHRLRILSPLNSELQNLQDQIWAMNAKVHQATTSAPEVEKVIEATRRTPFTPRISKLRIR 168
Query: 138 DNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAA--------SDAVKCRMFPSTFKSTAMA 189
+ + L Y+G D K+HL F +IA DA C++F A+
Sbjct: 169 E-FRDFKLPVYNGKGDLKEHLTSFQ----VIAGRVPLEPHEEDAGLCKLFSENLFGLALT 223
Query: 190 WFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMAR 243
WFT L GSI NF+ S+ F+ Q+ N +T L+N Q E L+ Y+ R
Sbjct: 224 WFTQLEEGSIDNFKQLSTAFIKQYEYFINSDITEAHLWNFSQSADEPLRTYIYR 277
Score = 34.3 bits (77), Expect = 0.48
Identities = 19/47 (40%), Positives = 25/47 (52%)
Query: 633 VLKVRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKV 679
V V + V A+YNVI+G L + V ST H VK+P GK+
Sbjct: 366 VKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKM 412
>At2g06890 putative retroelement integrase
Length = 1215
Score = 182 bits (461), Expect = 1e-45
Identities = 115/391 (29%), Positives = 193/391 (48%), Gaps = 8/391 (2%)
Query: 814 KAVQQEVDKLLAAEFIREVKYPTW-LANVVIVKKANGKWRMCVDYTDLNKACPKDSYPLP 872
+ ++ ++D + A Y T + + ++ +G WRMC D +N K +P+P
Sbjct: 431 RGIEHQIDFVPGASLPNRPAYRTNPVETKELQRQKDGSWRMCFDCRAINNVTVKYCHPIP 490
Query: 873 SIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMTARANYCYRTMPFGLKNAGA 932
+D ++D G+ + S +D SG HQIRM+ DE KTAF T Y + MPFGL +A +
Sbjct: 491 RLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPS 550
Query: 933 TYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYF 992
T+ RLM+ V +G + VY DD++V S +H + L+ +RK + N +KC F
Sbjct: 551 TFMRLMNHVLRAFIGIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTF 610
Query: 993 GVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGRIAALSRFLPKSGDRS 1052
FLGF++++ G++++ EK KAI+ SP V EV+ G RF
Sbjct: 611 CTDNLVFLGFVVSADGVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIV 670
Query: 1053 FPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLYFAVSDGALSSVM 1112
P + ++K+V F+W EEAF LK+ L++ P+L + S + +V+
Sbjct: 671 APLTEVMKKDVGFKWEKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVL 730
Query: 1113 LQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARRLRPYFQSFPVKVRTD-LPL 1171
+Q + +++ F S L GA + Y +K A++ +R + Y + TD L
Sbjct: 731 MQ----DQKLIAFFSEKLGGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESL 786
Query: 1172 RQVLQKPDLSGRLVAW--SVELSEYSLQYDK 1200
+ + + L+ R W +E Y ++Y K
Sbjct: 787 KHLKGQQKLNKRHARWVEFIETFAYVIKYKK 817
>At1g35370 hypothetical protein
Length = 1447
Score = 174 bits (442), Expect = 2e-43
Identities = 120/389 (30%), Positives = 200/389 (50%), Gaps = 21/389 (5%)
Query: 817 QQEVDKLLAAEFIR----EVKYPTWLANVVIVKKANGKWRMCVDYTDLNKACPKDSYPLP 872
+ E+DK++ + I+ +V + + VV+VKK +G WR+CVDYT+LN KD + +P
Sbjct: 593 KDEIDKIVQ-DMIKSGTIQVSSSPFASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIP 651
Query: 873 SIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMTARANYCYRTMPFGLKNAGA 932
I+ L+D G+ + S +D +G HQ+RM P D KTAF T ++ Y M FGL NA A
Sbjct: 652 LIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPA 711
Query: 933 TYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYF 992
T+Q LM+ VF + + + V+ DD+++ S +H + L F +R H + F
Sbjct: 712 TFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHKL--------F 763
Query: 993 GVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGRIAALSRFLPKSGDRS 1052
+ LG I++R IE +P K +A+++ +P+ VK+V+ G RF+ G +
Sbjct: 764 AKGSKEHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIA 823
Query: 1053 FPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLYFAVSDGALSSVM 1112
P L K F W+ E + AF LK +L + P+L+ P+ + + +V+
Sbjct: 824 GP-LHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVL 882
Query: 1113 LQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARRLRPYFQSFPVKVRTD-LPL 1171
+Q+ H + Y +S L+G ++ EK LA + R+ R Y ++TD L
Sbjct: 883 MQK---GHPLAY-ISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSL 938
Query: 1172 RQVLQKPDLSGRLVAWSVELSE--YSLQY 1198
+ +L++ + W +L E Y +QY
Sbjct: 939 KYLLEQRLNTPVQQQWLPKLLEFDYEIQY 967
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 171 bits (432), Expect = 3e-42
Identities = 111/344 (32%), Positives = 173/344 (50%), Gaps = 11/344 (3%)
Query: 865 PKDSYPLPSIDSLVDGASGNELLSLMDAYSG----NHQIRMHPADEDKTAFMTARANYCY 920
P D+YP+ D L D N+LL+ + Y G + +++ + KT F + Y
Sbjct: 119 PNDTYPVIINDGLSD-EQVNQLLNELRKYRGQLVVSFKLQFTLMIKKKTTFTCPNGTFAY 177
Query: 921 RTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRK 980
+ M FGL NA T+QR M +F+ + MEV++DD S L+ + LE +
Sbjct: 178 KRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDDFSGFSSCLLNLCRVLERC----EE 233
Query: 981 HNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGRIAA 1040
N+ LN EKC+F V G LG I+ +GIE++ K + Q++ P VK+++ G
Sbjct: 234 TNLVLNWEKCHFMVHEGIVLGHKISGKGIEVDKAKIDVMIQLQPPKTVKDIRSFLGHAEF 293
Query: 1041 LSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLY 1100
RF+ + P + L K F + +C +AF +KE L S PI P HP +
Sbjct: 294 YRRFIKDFSKIARPLTRLLCKETEFNFDEDCLKAFHLIKETLVSAPIFQAPNWDHPFEIM 353
Query: 1101 FAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARRLRPYFQS 1160
D A+ +V+ Q+ID + ++Y+ S T+ A+ RY IEK LAV+ ++R Y
Sbjct: 354 CDAFDYAVGAVLDQKIDDKLHVIYYASRTMDEAQTRYATIEKELLAVVFAFEKIRSYLVG 413
Query: 1161 FPVKVRTD-LPLRQVLQKPDLSGRLVAWSVELSEYSLQ-YDKRG 1202
F VKV D LR + K + RL+ W + L E+ ++ DK+G
Sbjct: 414 FKVKVYIDHAALRHIYAKKETKSRLLRWILLLQEFDMEIIDKKG 457
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,145,221
Number of Sequences: 26719
Number of extensions: 1423034
Number of successful extensions: 15412
Number of sequences better than 10.0: 406
Number of HSP's better than 10.0 without gapping: 235
Number of HSP's successfully gapped in prelim test: 188
Number of HSP's that attempted gapping in prelim test: 7534
Number of HSP's gapped (non-prelim): 2714
length of query: 1291
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1180
effective length of database: 8,352,787
effective search space: 9856288660
effective search space used: 9856288660
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0331c.2


