

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0330.6
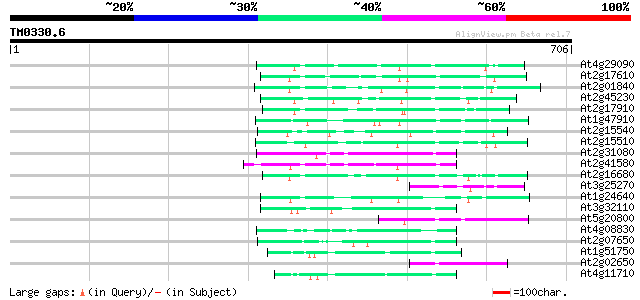
(706 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g29090 putative protein 83 5e-16
At2g17610 putative non-LTR retroelement reverse transcriptase 83 5e-16
At2g01840 putative non-LTR retroelement reverse transcriptase 81 2e-15
At2g45230 putative non-LTR retroelement reverse transcriptase 80 3e-15
At2g17910 putative non-LTR retroelement reverse transcriptase 70 3e-12
At1g47910 reverse transcriptase, putative 70 5e-12
At2g15540 putative non-LTR retroelement reverse transcriptase 69 9e-12
At2g15510 putative non-LTR retroelement reverse transcriptase 69 1e-11
At2g31080 putative non-LTR retroelement reverse transcriptase 67 3e-11
At2g41580 putative non-LTR retroelement reverse transcriptase 64 2e-10
At2g16680 putative non-LTR retroelement reverse transcriptase 61 2e-09
At3g25270 hypothetical protein 59 1e-08
At1g24640 hypothetical protein 56 6e-08
At3g32110 non-LTR reverse transcriptase, putative 56 8e-08
At5g20800 putative protein 54 3e-07
At4g08830 putative protein 54 3e-07
At2g07650 putative non-LTR retrolelement reverse transcriptase 54 4e-07
At1g51750 hypothetical protein 53 7e-07
At2g02650 putative reverse transcriptase 51 2e-06
At4g11710 putative protein 50 3e-06
>At4g29090 putative protein
Length = 575
Score = 83.2 bits (204), Expect = 5e-16
Identities = 87/366 (23%), Positives = 149/366 (39%), Gaps = 48/366 (13%)
Query: 311 EADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGE----------SSW 360
+A+ I KD FN+ALLGK WR L+ P+SL +V K+ + S
Sbjct: 51 KAEGGIGFKDIEAFNLALLGKQMWRMLSRPESLMAKVFKSRYFHKSDPLNAPLGSRPSFV 110
Query: 361 WKEIQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGS--GSLELRFRRLYNLSCQKR 418
WK I + ++ + V G W W S S LR +R + Q+
Sbjct: 111 WKSIHA-----SQEILRQGARAVVGNGEDIIIWRHKWLDSKPASAALRMQR---VPPQEY 162
Query: 419 EQVSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELMV--ILQGIRLVENKKDTWRWKLEG 476
VS++ K + + + +WR+++ E ++ + G R + D++ W
Sbjct: 163 ASVSSILKVSDLIDESGREWRKDVIEMLFPEVERKLIGELRPGGRRI---LDSYTWDYTS 219
Query: 477 EGGFSFNSSFVFL--------QDQYLEEP--DPVFNWIWMVPVPSNIKAFVWRVLLGRIQ 526
G ++ S + L Q + EP +P++ IW I+ F+W+ L +
Sbjct: 220 SGDYTVKSGYWVLTQIINKRSSPQEVSEPSLNPIYQKIWKSQTSPKIQHFLWKCLSNSLP 279
Query: 527 TRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDP 586
L H + E+ C C +E+ +HLLF C+ + L + +
Sbjct: 280 VAGAL---AYRHLSKESACIRCPSCKETVNHLLFKCTFARLTWAISSIPIPLGGEWADSI 336
Query: 587 KVHLLQFSSIG-----WSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQ 641
V+L ++G W KA +L W+ LW +W RN +VF G E + ++L + +
Sbjct: 337 YVNLYWVFNLGNGNPQWEKASQL---VPWL--LWRLWKNRNELVFRGREFNAQEVLRRAE 391
Query: 642 VTAWRW 647
W
Sbjct: 392 DDLEEW 397
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 83.2 bits (204), Expect = 5e-16
Identities = 90/362 (24%), Positives = 141/362 (38%), Gaps = 54/362 (14%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKA---------NCNSCGESSW-WKEIQ 365
++I+D + FN+ALL K WR L +P SL RV KA + + +SS+ WK I
Sbjct: 308 LAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKATSQSSYAWKSIL 367
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQK-REQVSNM 424
+G ++ GN Q W +NW L L R +C Q+
Sbjct: 368 -----HGTKLISRGLKYIAGNGNNIQLWKDNW-----LPLNPPRPPVGTCDSIYSQLKVS 417
Query: 425 GKWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNS 484
+ G W + + + ++ + + G D W +G +S S
Sbjct: 418 DLLIEGRWNEDLLCKL-IHQNDIPHIRAIRPSITGAN------DAITWIYTHDGNYSVKS 470
Query: 485 SFVFL----QDQYLEEPDP-------VFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN 533
+ L Q Q+ P P VF IW P IK F WR + T NL
Sbjct: 471 GYHLLRKLSQQQHASLPSPNEVSAQTVFTNIWKQNAPPKIKHFWWRSAHNALPTAGNLKR 530
Query: 534 *QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQF 593
+++ + C C A E +HLLF C S I + H L + +S+ L+
Sbjct: 531 RRLI---TDDTCQRCGEASEDVNHLLFQCRVSKEIWEQAHIKLCPGDSLMSNSFNQNLE- 586
Query: 594 SSIGWSKAQKLGESA-----IWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRWL 648
QKL +SA ++ + W IW +RN ++FN +++ + +W
Sbjct: 587 ------SIQKLNQSARKDVSLFPFIGWRIWKMRNDLIFNNKRWSIPDSIQKALIDQQQWK 640
Query: 649 RS 650
S
Sbjct: 641 ES 642
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 81.3 bits (199), Expect = 2e-15
Identities = 89/395 (22%), Positives = 156/395 (38%), Gaps = 72/395 (18%)
Query: 309 GAEADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKA---------NCNSCGESS 359
G E + + KD FN ALL K WR LT P SL R+ K N G +S
Sbjct: 1190 GKENEGDLGFKDLHQFNRALLAKQAWRILTNPQSLLARLYKGLYYPNTTYLRANKGGHAS 1249
Query: 360 W-WKEIQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKR 418
+ W IQ G Q+ ++ + G T+ W + W + L R +
Sbjct: 1250 YGWNSIQE-----GKLLLQQGLRVRLGDGQTTKIWEDPWLPT----LPPRPARGPILDED 1300
Query: 419 EQVSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENK------------ 466
+V+++ WR N + E VI +G+ E++
Sbjct: 1301 MKVADL-------------WRENKR-------EWDPVIFEGVLNPEDQQLAKSLYLSNYA 1340
Query: 467 -KDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPD---------PVFNWIWMVPVPSNIKAF 516
+D+++W ++ S + L E + P+ IW + + IK F
Sbjct: 1341 ARDSYKWAYTRNTQYTVRSGYWVATHVNLTEEEIINPLEGDVPLKQEIWRLKITPKIKHF 1400
Query: 517 VWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWL 576
+WR L G + T L N + + C CC A+E+ +H++F+CS + ++ + +
Sbjct: 1401 IWRCLSGALSTTTQLRNRNI---PADPTCQRCCNADETINHIIFTCSYAQVV-WRSANFS 1456
Query: 577 GVSTAQVSDPKVHLLQFSSIGWSKAQKL----GESAIWMSVLWSIWCLRNMVVFNGGELD 632
G + +D ++ G K Q L G W ++W +W RN +F +
Sbjct: 1457 GSNRLCFTDNLEENIRLILQG-KKNQNLPILNGLMPFW--IMWRLWKSRNEYLFQQLDRF 1513
Query: 633 KDQLLEQIQVTAWRWLRSRLDDF*YSWYEWKSNPR 667
++ ++ + A W+ + ++D S +SN R
Sbjct: 1514 PWKVAQKAEQEATEWVETMVNDTAISHNTAQSNDR 1548
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 80.5 bits (197), Expect = 3e-15
Identities = 84/358 (23%), Positives = 137/358 (37%), Gaps = 62/358 (17%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGE----------SSWWKEIQ 365
+ K+ FN+ALLGK WR +TE DSL +V K+ S + S WK I
Sbjct: 853 LGFKEIEAFNIALLGKQLWRMITEKDSLMAKVFKSRYFSKSDPLNAPLGSRPSFAWKSIY 912
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLEL-----RFRRLYNLSCQKREQ 420
V ++ ++ + G W + W G+ + R + +
Sbjct: 913 EAQV-----LIKQGIRAVIGNGETINVWTDPWIGAKPAKAAQAVKRSHLVSQYAANSIHV 967
Query: 421 VSNMGKWVNGVWQWEFK---WRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGE 477
V ++ W W + N Q E ++ + G + +D + W+
Sbjct: 968 VKDLLLPDGRDWNWNLVSLLFPDNTQ-------ENILALRPGGK---ETRDRFTWEYSRS 1017
Query: 478 GGFSFNSSFVFLQD--------QYLEEP--DPVFNWIWMVPVPSNIKAFVWRVLLGRIQT 527
G +S S + + + Q + +P DP+F IW + VP I F+WR + +
Sbjct: 1018 GHYSVKSGYWVMTEIINQRNNPQEVLQPSLDPIFQQIWKLDVPPKIHHFLWRCVNNCLSV 1077
Query: 528 RDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWL--------GVS 579
NL H A E C C E+ +HLLF C + L W G
Sbjct: 1078 ASNL---AYRHLAREKSCVRCPSHGETVNHLLFKCPFARL------TWAISPLPAPPGGE 1128
Query: 580 TAQVSDPKVHLLQFSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLL 637
A+ +H S+ S+ ++ A+ +LW +W RN +VF G E Q++
Sbjct: 1129 WAESLFRNMH--HVLSVHKSQPEESDHHALIPWILWRLWKNRNDLVFKGREFTAPQVI 1184
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 70.5 bits (171), Expect = 3e-12
Identities = 78/334 (23%), Positives = 134/334 (39%), Gaps = 47/334 (14%)
Query: 319 KD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGE----------SSWWKEIQSIT 368
KD + FN ALL K WR L E SL+ RV ++ S + S W+ I
Sbjct: 834 KDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILF-- 891
Query: 369 VDNGWPWFQESVQKTVFAGNATQFWAENW--TGSGSLELRFRRLYNLSCQKREQVSNMGK 426
G + ++ + G T W + W GS L RR N+ + + + +
Sbjct: 892 ---GRELLMQGLRTVIGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSR 948
Query: 427 WVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSF 486
W N+ W + +++ Q R + K+D++ W G +S + +
Sbjct: 949 ----------NWNLNMLRDLFPWKDVEIILKQ--RPLFFKEDSFCWLHSHNGLYSVKTGY 996
Query: 487 VFLQDQ-----YLE-----EPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QV 536
FL Q Y E + +F+ IW + I+ F+W+ L G I D L +
Sbjct: 997 EFLSKQVHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALHGAIPVEDRLRTRGI 1056
Query: 537 LHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSI 596
+ C +C E+ +H+LF C + + H L + ++ S+ V+ I
Sbjct: 1057 ---RSDDGCLMCDTENETINHILFECPLARQVWAITH--LSSAGSEFSN-SVYTNMSRLI 1110
Query: 597 GWSKAQKLGESAIWMS--VLWSIWCLRNMVVFNG 628
++ L ++S +LW +W RN ++F G
Sbjct: 1111 DLTQQNDLPHHLRFVSPWILWFLWKNRNALLFEG 1144
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 69.7 bits (169), Expect = 5e-12
Identities = 82/369 (22%), Positives = 144/369 (38%), Gaps = 54/369 (14%)
Query: 310 AEADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITV 369
+++D + ++ FN ALL K WR +T PDSL+ +V K +S+ I+S +
Sbjct: 631 SKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRYFR--KSNPLDSIKSYSP 688
Query: 370 DNGW-------PWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVS 422
GW + + K V +G + W + W + Q
Sbjct: 689 SYGWRSMISARSLVYKGLIKRVGSGASISVWNDPWIPA-------------------QFP 729
Query: 423 NMGKWVNGVWQWEFKWRRNLQGREVGWFEELMVIL---QGIRLVE-------NKKDTWRW 472
K+ + K + + R W +L+ L + + L+ N +DT W
Sbjct: 730 RPAKYGGSIVDPSLKVKSLIDSRSNFWNIDLLKELFDPEDVPLISALPIGNPNMEDTLGW 789
Query: 473 KLEGEGGFSFNSSFVFL-----QDQYLEEPD--PVFNWIWMVPVPSNIKAFVWRVLLGRI 525
G ++ S + + L PD + +IW V P ++ F+W++L G +
Sbjct: 790 HFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWKVQCPPKLRHFLWQILSGCV 849
Query: 526 QTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSD 585
+NL +L + C C +EES +H LF C + I A + TA
Sbjct: 850 PVSENLRKRGIL---CDKGCVSCGASEESINHTLFQCHPARQI----WALSQIPTAPGIF 902
Query: 586 PKVHLL-QFSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTA 644
P + + W + +SA + ++W IW RN VF + D ++L A
Sbjct: 903 PSNSIFTNLDHLFWRIPSGV-DSAPYPWIIWYIWKARNEKVFENVDKDPMEILLLAVKEA 961
Query: 645 WRWLRSRLD 653
W ++++
Sbjct: 962 QSWQEAQVE 970
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 68.9 bits (167), Expect = 9e-12
Identities = 83/355 (23%), Positives = 133/355 (37%), Gaps = 78/355 (21%)
Query: 312 ADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVV--------------KANCNSCGE 357
+D + + FN+ LL K WR + P+SL RV+ KAN S G
Sbjct: 729 SDGGLGFRTLEEFNLVLLAKQLWRLIRFPNSLLSRVLRGRYFRYSDPIQIGKANRPSFG- 787
Query: 358 SSWWKEIQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGS-------GSLELRFRRL 410
W+ I + P +++T+ +G T+ W + W S L +R L
Sbjct: 788 ---WRSIMAAK-----PLLLSGLRRTIGSGMLTRVWEDPWIPSFPPRPAKSILNIRDTHL 839
Query: 411 YNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELM-----VILQGIRLVEN 465
Y V+++ V W+ +G +EL+ ++ GIR
Sbjct: 840 Y---------VNDLIDPVTKQWK-------------LGRLQELVDPSDIPLILGIRPSRT 877
Query: 466 -KKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDPVFNW---------IWMVPVPSNIKA 515
K D + W G ++ S + +D D F +W + K
Sbjct: 878 YKSDDFSWSFTKSGNYTVKSGYWAARDLSRPTCDLPFQGPSVSALQAQVWKIKTTRKFKH 937
Query: 516 FVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAW 575
F W+ L G + T L + H E +CP C EES +HLLF C S I A
Sbjct: 938 FEWQCLSGCLATNQRLFS---RHIGTEKVCPRCGAEEESINHLLFLCPPSRQI----WAL 990
Query: 576 LGVSTAQVSDPKVHLLQFSSIGWSKAQKLGES----AIWMSVLWSIWCLRNMVVF 626
+ +++ P+ L S+ ++ + I+ +LW IW RN +F
Sbjct: 991 SPIPSSEYIFPRNSLFYNFDFLLSRGKEFDIAEDIMEIFPWILWYIWKSRNRFIF 1045
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 68.6 bits (166), Expect = 1e-11
Identities = 80/372 (21%), Positives = 145/372 (38%), Gaps = 58/372 (15%)
Query: 310 AEADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITV 369
A+ + +D FN ALL K WR L P L+ R K S ++ + + +
Sbjct: 623 AKESGGLGFRDIESFNQALLAKQSWRILQYPSFLFARFFK--------SRYFDDEEFLEA 674
Query: 370 D-------------NGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQ 416
D G + ++K V G + W + W + L +R +++
Sbjct: 675 DLGVRPSYACCSILFGRELLAKGLRKEVGNGKSLNVWMDPWIFDIAPRLPLQRHFSV--- 731
Query: 417 KREQVSNMGKWVNGVWQWEFK-WRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLE 475
N+ VN + +E + W+R+L E ++ + ++ V N D W W
Sbjct: 732 ------NLDLKVNDLINFEDRCWKRDL--LEELFYPTDVELITKRNPVVNMDDFWVWLHA 783
Query: 476 GEGGFSFNSSF----------VFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRI 525
G +S S + + Q L + + +W IK F+WR+L +
Sbjct: 784 KTGEYSVKSGYWLAFQSNKLDLIQQANLLPSTNGLKEQVWSTKSSPKIKMFLWRILSAAL 843
Query: 526 QTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWLGVSTAQVSD 585
D ++ + +++ C +C ES +H+LF+CS + + A GV T +
Sbjct: 844 PVADQIIRRGM---SIDPRCQICGDEGESTNHVLFTCSMAR----QVWALSGVPTPEFGF 896
Query: 586 PKVHLLQFSSIGW---SKAQKLGESAI---WMSVLWSIWCLRNMVVFNGGELDKDQLLEQ 639
+ F++I + K L + W VLW +W RN + F+G + +
Sbjct: 897 QNASI--FANIQFLFELKKMILVPDLVKRSWPWVLWRLWKNRNKLFFDGITFCPLNSIVK 954
Query: 640 IQVTAWRWLRSR 651
IQ W +++
Sbjct: 955 IQEDTLEWFQAQ 966
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 67.4 bits (163), Expect = 3e-11
Identities = 67/262 (25%), Positives = 108/262 (40%), Gaps = 21/262 (8%)
Query: 311 EADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCG--ESSWWKEIQ--S 366
+A+ I ++ R N AL+ K WR L + +SLW RVV+ G ++SW K S
Sbjct: 711 KAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARVVRKKYKVGGVQDTSWLKPQPRWS 770
Query: 367 ITVDNGWPWFQESVQKTV----FAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVS 422
T + +E V K V G +FW + W L+ L + E++
Sbjct: 771 STWRSVAVGLREVVVKGVGWVPGDGCTIRFWLDRWL----LQEPLVELGTDMIPEGERI- 825
Query: 423 NMGKWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSF 482
K W W + G + + ++ +++ D WK +G F+
Sbjct: 826 ---KVAADYWLPGSGWNLEILGLYLPETVKRRLLSVVVQVFLGNGDEISWKGTQDGAFTV 882
Query: 483 NSSFVFLQDQYLEEPD--PVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNA 540
S++ LQ + P+ FN IW + P ++ F+W V I T + H +
Sbjct: 883 RSAYSLLQGDVGDRPNMGSFFNRIWKLITPERVRVFIWLVSQNVIMTNVERVR---RHLS 939
Query: 541 LEAICPLCCLAEESGSHLLFSC 562
AIC +C AEE+ H+L C
Sbjct: 940 ENAICSVCNGAEETILHVLRDC 961
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 64.3 bits (155), Expect = 2e-10
Identities = 69/288 (23%), Positives = 120/288 (40%), Gaps = 45/288 (15%)
Query: 295 LGCCQFVLERFL*GGAEADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKAN--- 351
+G + +L +FL G KD + FN ALL K R T+ DSL +++K+
Sbjct: 568 IGAQKLMLPKFLGG-------FGFKDLQCFNQALLAKQASRLHTDSDSLLSQILKSRYYM 620
Query: 352 -------CNSCGESSWWKEIQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLE 404
S W+ I G ++K + G T W +NW +
Sbjct: 621 NSDFLSATKGTRPSYAWQSILY-----GRELLVSGLKKIIGNGENTYVWMDNWI----FD 671
Query: 405 LRFRRLYNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVE 464
+ RR +L Q+ K + + W N+ R++ ++E+ +I Q R +
Sbjct: 672 DKPRRPESLQIMVDIQL----KVSQLIDPFSRNWNLNML-RDLFPWKEIQIICQQ-RPMA 725
Query: 465 NKKDTWRWKLEGEGGFSFNSSF----------VFLQDQYLEEPDPVFNWIWMVPVPSNIK 514
+++D++ W G ++ S + +F + + +P+F IW + IK
Sbjct: 726 SRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKIWNLNSAPKIK 785
Query: 515 AFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSC 562
F+W+VL G + D L VL +E C +C E+ +H+LF C
Sbjct: 786 VFLWKVLKGAVAVEDRLRTRGVL---IEDGCSMCPEKNETLNHILFQC 830
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 60.8 bits (146), Expect = 2e-09
Identities = 71/364 (19%), Positives = 137/364 (37%), Gaps = 61/364 (16%)
Query: 319 KD*RLFNMALLGKWKWRFLTEPDSLWCRVVKA---------NCNSCGESSW-WKEIQSIT 368
KD + FN ALL K WR ++ S+ ++ K+ N S+ W+ I
Sbjct: 808 KDLQCFNQALLAKQAWRLFSDSKSIVSQIFKSRYFMNTDFLNARQGTRPSYTWRSILY-- 865
Query: 369 VDNGWPWFQESVQKTVFAGNATQFWAENWT--GSGSLELRFRRLYNLSCQKREQVSNMGK 426
G +++ + G T W + W G + L N+ + + + +
Sbjct: 866 ---GRELLNGGLKRLIGNGEQTNVWIDKWLFDGHSRRPMNLHSLMNIHMKVSHLIDPLTR 922
Query: 427 WVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSF 486
W K L + E+ + ++ R + + +D++ W G ++ S +
Sbjct: 923 ------NWNLKKLTEL------FHEKDVQLIMHQRPLISSEDSYCWAGTNNGLYTVKSGY 970
Query: 487 ----------VFLQDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QV 536
+F + +P+F+ +W + IK F+W+ L G + D L + +
Sbjct: 971 ERSSRETFKNLFKEADVYPSVNPLFDKVWSLETVPKIKVFMWKALKGALAVEDRLRSRGI 1030
Query: 537 LHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAW---------LGVSTAQVSDPK 587
C C E+ +HLLF C + W G T+ S+
Sbjct: 1031 ---RTADGCLFCKEEIETINHLLFQCP------FARQVWALSLIQAPATGFGTSIFSNIN 1081
Query: 588 VHLLQFSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLLEQIQVTAWRW 647
H++Q +S + + + + W+ LW IW RN +F G L +++ + W
Sbjct: 1082 -HVIQ-NSQNFGIPRHMRTVSPWL--LWEIWKNRNKTLFQGTGLTSSEIVAKAYEECNLW 1137
Query: 648 LRSR 651
+ ++
Sbjct: 1138 INAQ 1141
>At3g25270 hypothetical protein
Length = 343
Score = 58.5 bits (140), Expect = 1e-08
Identities = 40/152 (26%), Positives = 64/152 (41%), Gaps = 20/152 (13%)
Query: 504 IWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCS 563
IW + IK F+W++L G + T DNL + ++ C CC +E+ HL F C
Sbjct: 18 IWKLKTAPKIKHFLWKLLSGALATGDNLKRRHIRNHPQ---CHRCCQEDETSQHLFFDCF 74
Query: 564 NSMLI*YECHAWLGV--------STAQVSDPKVHLLQFSSIGWSKAQKLGESAIWMSVLW 615
Y W +T + K+ LL S + ++ +L AIW +LW
Sbjct: 75 ------YAQQVWRASGIPHQELRTTGITMETKMELLLSSCLA-NRQPQLFNLAIW--ILW 125
Query: 616 SIWCLRNMVVFNGGELDKDQLLEQIQVTAWRW 647
+W RN +VF + L++ + W
Sbjct: 126 RLWKSRNQLVFQQKSISWQNTLQRARNDVQEW 157
>At1g24640 hypothetical protein
Length = 1270
Score = 56.2 bits (134), Expect = 6e-08
Identities = 80/377 (21%), Positives = 128/377 (33%), Gaps = 98/377 (25%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKAN----------CNSCGESSWWKEIQ 365
+ +D + FN ALL K WR L PD L R++K+ S S W+ I
Sbjct: 816 LGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKSRYFDATDFLDAALSQRPSFGWRSIL 875
Query: 366 SITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMG 425
G + +QK V G + W + W +R+
Sbjct: 876 F-----GRELLSKGLQKRVGDGASLFVWIDPWIDDNGFRAPWRK---------------- 914
Query: 426 KWVNGVWQWEFKWRRNLQGREVGWFEELM---------VILQGIRLVENKKDTWRWKLEG 476
N ++ K + L R W EE++ + ++ I+ V ++ D + WKL
Sbjct: 915 ---NLIYDVTLKVKALLNPRTGFWDEEVLHDLFLPEDILRIKAIKPVISQADFFVWKLNK 971
Query: 477 EGGFSFNSSFVF--------LQDQYLEEPDP--VFNWIWMVPVPSNIKAFVWRVLLGRIQ 526
G FS S++ L+ + +P + +W + IK F+W+V
Sbjct: 972 SGDFSVKSAYWLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTDPKIKIFLWKV------ 1025
Query: 527 TRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWL---------G 577
C ES +H LF C S I W G
Sbjct: 1026 ---------------------CGELGESTNHTLFLCPLSRQI------WALSDYPFPPDG 1058
Query: 578 VSTAQVSDPKVHLLQFSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQLL 637
S + HLL+ +K + I+ +LW IW RN +F G +
Sbjct: 1059 FSNGSIYSNINHLLENKD---NKEWPINLRKIFPWILWRIWKNRNSFIFEGISYPATDTV 1115
Query: 638 EQIQVTAWRWLRSRLDD 654
+I+ W ++ D
Sbjct: 1116 IKIRDDVVEWFEAQCLD 1132
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 55.8 bits (133), Expect = 8e-08
Identities = 54/264 (20%), Positives = 97/264 (36%), Gaps = 35/264 (13%)
Query: 316 ISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCN---------SCGESSW---WKE 363
+ I+ N AL+ K WR L + SLW +VV++ + +S+W W+
Sbjct: 1392 LGIRSATAMNKALIAKVGWRVLNDGSSLWAQVVRSKYKVGDVHDRNWTVAKSNWSSTWRS 1451
Query: 364 IQSITVDNGWPWFQESVQKTVFAGNATQFWAENWTGSGSL---ELRFRRLYNLSCQKREQ 420
+ D W + G +FW + W + + L + C R+
Sbjct: 1452 VGVGLRDVIW----REQHWVIGDGRQIRFWTDRWLSETPIADDSIVPLSLAQMLCTARD- 1506
Query: 421 VSNMGKWVNGVWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGF 480
+W+ W + V + L ++ + V D W + +G F
Sbjct: 1507 ----------LWRDGTGWDMSQIAPFVTDNKRLDLLAVIVDSVTGAHDRLAWGMTSDGRF 1556
Query: 481 SFNSSFVFL--QDQYLEEPDPVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLH 538
+ S+F L D ++ ++ +W V P ++ F+W V+ I T + H
Sbjct: 1557 TVKSAFAMLTNDDSPRQDMSSLYGRVWKVQAPERVRVFLWLVVNQAIMTNSER---KRRH 1613
Query: 539 NALEAICPLCCLAEESGSHLLFSC 562
+C +C ES H+L C
Sbjct: 1614 LCDSDVCQVCRGGIESILHVLRDC 1637
>At5g20800 putative protein
Length = 359
Score = 53.9 bits (128), Expect = 3e-07
Identities = 42/197 (21%), Positives = 83/197 (41%), Gaps = 15/197 (7%)
Query: 465 NKKDTWRWKLEGEGGFSFNSSFVFLQDQYLE--------EPDPVFNWIWMVPVPSNIKAF 516
NK+DT W G ++F S + + +Y++ E + + W V P ++ F
Sbjct: 80 NKEDTLGWHFTKSGKYTFKSGYHTSRIEYIDTNLSFIGPEITLLKAYSWKVQCPPKLRHF 139
Query: 517 VWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCSNSMLI*YECHAWL 576
+W++L + R+NL + + C C E++ +H LF C + I A
Sbjct: 140 LWQILASCVPVRENLRKRGI---NCDIGCARCGATEKTINHTLFQCHPARQI----WALS 192
Query: 577 GVSTAQVSDPKVHLLQFSSIGWSKAQKLGESAIWMSVLWSIWCLRNMVVFNGGELDKDQL 636
+ T P + + + + Q +S + ++W IW RN +F+ + D +
Sbjct: 193 QIPTVIGIFPSDSIYENLDHLFWRVQSAVDSFSYPWIIWYIWKARNAKLFDNVDKDPLDI 252
Query: 637 LEQIQVTAWRWLRSRLD 653
L + A W ++++
Sbjct: 253 LSLAEKEAQSWKSAQVE 269
>At4g08830 putative protein
Length = 947
Score = 53.9 bits (128), Expect = 3e-07
Identities = 50/254 (19%), Positives = 101/254 (39%), Gaps = 45/254 (17%)
Query: 311 EADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITVD 370
+++ + I+ + N AL+ K WR + + SLW R+++ S + ++ + V
Sbjct: 525 KSEGGLGIRTSKCMNKALVSKVGWRLINDRYSLWARILR--------SKYRVGLREV-VS 575
Query: 371 NGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNG 430
G W V G FW++NW +L + + + +K +V ++ W NG
Sbjct: 576 RGSRW-------VVGNGRDILFWSDNWLSHEAL-INRAVIEIPNSEKELRVKDL--WANG 625
Query: 431 VWQWEFKWRRNLQGREVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQ 490
+ W+ + + + L + + V +D W +G F+ S++ L
Sbjct: 626 L-----GWKLDKIEPYISYHTRLELAAVVVDSVTGARDRLSWGYSADGVFTVKSAYRLLT 680
Query: 491 DQYLEEPD--PVFNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLC 548
+ + P+ F+ +W V +K F+W H ++C +C
Sbjct: 681 EDHDPRPNMAAFFDRLWRVVALERVKTFLW-------------------HIGDTSVCQVC 721
Query: 549 CLAEESGSHLLFSC 562
+E+ H+L C
Sbjct: 722 KGGDETILHVLKDC 735
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 53.5 bits (127), Expect = 4e-07
Identities = 58/276 (21%), Positives = 103/276 (37%), Gaps = 51/276 (18%)
Query: 312 ADNKISIKD*RLFNMALLGKWKWRFLTEPDSLWCRVVKANCNSCG-ESSWWKEIQSITVD 370
++ + I+ + N ALL K WR + + SLW R+++ N W +++S+
Sbjct: 374 SEGGLGIRKAQDMNKALLSKVGWRLIQDYHSLWARIMRCNYRVQDVRDGAWTKVRSVC-S 432
Query: 371 NGWPWFQESVQKTVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNG 430
+ W +++ V G + W G G RE + M KW+
Sbjct: 433 STWRSVALGMREVVIPGLS---WV---IGDG----------------REILFWMDKWLTN 470
Query: 431 V-----------WQWEFKWRRNLQGREVGW-----------FEELMVILQGIRLVENKKD 468
+ W+ R+L+ +GW + L + + + +D
Sbjct: 471 IPLAELLVQEPPAGWKGMRARDLRRNGIGWDMATIAPYISSYNRLQLQSFVLDDITGARD 530
Query: 469 TWRWKLEGEGGFSFNSSFVFLQDQYLEEPDP--VFNWIWMVPVPSNIKAFVWRVLLGRIQ 526
W +G F ++++ FL D F+ +W V P ++ F+W V I
Sbjct: 531 RISWGESQDGRFKVSTTYSFLTRDEAPRQDMKRFFDRVWSVTAPERVRLFLWLVAQQAIM 590
Query: 527 TRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSC 562
T H + IC +C A+ES H+L C
Sbjct: 591 TNQER---HRRHLSTTTICQVCKGAKESIIHVLRDC 623
>At1g51750 hypothetical protein
Length = 629
Score = 52.8 bits (125), Expect = 7e-07
Identities = 59/258 (22%), Positives = 103/258 (39%), Gaps = 32/258 (12%)
Query: 325 NMALLGKWKWRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITVDNGWPW-----FQES 379
N+ + K WR + DSLW + K N + S+W + ++ + W W ++E+
Sbjct: 288 NVVSVLKLIWRVTSNDDSLWVKWSKMNLLK--QESFWSLTPNSSLGS-WMWKKMLKYRET 344
Query: 380 VQK----TVFAGNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWE 435
+ V G T FW +NW+G G L +++ Q+ + + + W
Sbjct: 345 AKPFSRVEVNNGARTSFWFDNWSGMG-------HLMDVTGQRGQIDLGISRNKTVAEAWS 397
Query: 436 FKWRRNLQGREVGWFEE-LMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQ--DQ 492
+ RR + ++ E L Q L+ WR K G F +SF +Q
Sbjct: 398 NRRRRKHRTEQLNDIEAALNQKYQTRNLLREDATLWRGK-----GDVFKTSFSTKDTWNQ 452
Query: 493 YLEEPDPV--FNWIWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCL 550
++ + V + +W + W L R+ T + Q+ +N + C C
Sbjct: 453 VRKKSNEVAWYKGVWFSHSTPKYQFCTWLALRNRLSTGYRM---QLWNNGSDVKCTFCST 509
Query: 551 AEESGSHLLFSCSNSMLI 568
+ E+ HL FSCS + I
Sbjct: 510 SIETRDHLFFSCSYASAI 527
>At2g02650 putative reverse transcriptase
Length = 365
Score = 51.2 bits (121), Expect = 2e-06
Identities = 28/123 (22%), Positives = 57/123 (45%), Gaps = 3/123 (2%)
Query: 504 IWMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSCS 563
IW + V IK F+WR + G + T L + + + IC CC+ EE+ H++F+C
Sbjct: 37 IWKLHVAPKIKHFLWRCVTGALATNTRLRSRNI---DADPICQRCCIEEETIHHIMFNCP 93
Query: 564 NSMLI*YECHAWLGVSTAQVSDPKVHLLQFSSIGWSKAQKLGESAIWMSVLWSIWCLRNM 623
+ + + +G S + +L + + ++ + + ++W +W RN+
Sbjct: 94 YTQSVWRSANIIIGNQWGPPSSFEDNLNRLIQLSKTQTTNSLDRFLPFWIMWRLWKSRNV 153
Query: 624 VVF 626
+F
Sbjct: 154 FLF 156
>At4g11710 putative protein
Length = 473
Score = 50.4 bits (119), Expect = 3e-06
Identities = 49/238 (20%), Positives = 94/238 (38%), Gaps = 23/238 (9%)
Query: 334 WRFLTEPDSLWCRVVKANCNSCGESSWWKEIQSITVDNGWPWFQ----ESVQKTVFA--- 386
WR ++ DSLW + ++++ + S+W ++ T W W + + +T+
Sbjct: 141 WRIISHADSLWVKWIQSSLLK--KVSFWA-VRENTSLGSWMWRKILKFRDIARTLCKVEI 197
Query: 387 --GNATQFWAENWTGSGSLELRFRRLYNLSCQKREQVSNMGKWVNGVWQWEFKWRRNLQG 444
G T FW ++W+ G RL + + + + K V W + RR +
Sbjct: 198 NNGARTSFWYDDWSDLG-------RLIDSAGDRGAIDLGINKHATVVEAWGNRRRRRHRT 250
Query: 445 REVGWFEELMVILQGIRLVENKKDTWRWKLEGEGGFSFNSSFVFLQDQYLEEPDPVFNWI 504
+ EE +++ R + W+ K E F++ + + + + +
Sbjct: 251 NFLNRVEERLILSWNSRNQAEDRALWKGK-ENRFRSIFSTKDTWNHIRTVSNKVAWYKGV 309
Query: 505 WMVPVPSNIKAFVWRVLLGRIQTRDNLLN*QVLHNALEAICPLCCLAEESGSHLLFSC 562
W +W + R+ T D + + + ++A C LC A ES HL FSC
Sbjct: 310 WFAQAIPKHAFCMWLAVHNRLSTGDRMT---LWNMGVDATCILCNKALESRDHLFFSC 364
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.345 0.151 0.582
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,512,130
Number of Sequences: 26719
Number of extensions: 632254
Number of successful extensions: 2505
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 54
Number of HSP's that attempted gapping in prelim test: 2301
Number of HSP's gapped (non-prelim): 157
length of query: 706
length of database: 11,318,596
effective HSP length: 106
effective length of query: 600
effective length of database: 8,486,382
effective search space: 5091829200
effective search space used: 5091829200
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0330.6


