

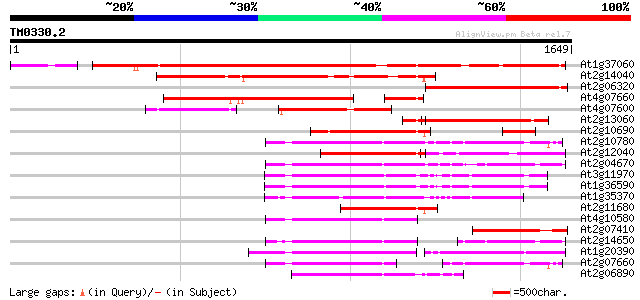
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0330.2
(1649 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g37060 Athila retroelment ORF 1, putative 1316 0.0
At2g14040 putative retroelement pol polyprotein 723 0.0
At2g06320 putative retroelement pol polyprotein 541 e-153
At4g07660 putative athila transposon protein 535 e-152
At4g07600 449 e-126
At2g13060 pseudogene 444 e-124
At2g10690 putative retroelement pol polyprotein 442 e-123
At2g10780 pseudogene 390 e-108
At2g12040 T10J7.2 389 e-108
At2g04670 putative retroelement pol polyprotein 370 e-102
At3g11970 hypothetical protein 354 3e-97
At1g36590 hypothetical protein 348 1e-95
At1g35370 hypothetical protein 340 4e-93
At2g11680 putative retroelement pol polyprotein 332 1e-90
At4g10580 putative reverse-transcriptase -like protein 304 2e-82
At2g07410 putative retroelement pol polyprotein 291 2e-78
At2g14650 putative retroelement pol polyprotein 281 2e-75
At1g20390 hypothetical protein 273 6e-73
At2g07660 putative retroelement pol polyprotein 263 8e-70
At2g06890 putative retroelement integrase 260 5e-69
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 1316 bits (3405), Expect = 0.0
Identities = 696/1428 (48%), Positives = 935/1428 (64%), Gaps = 101/1428 (7%)
Query: 244 TPTSEQDPLDQPQIVVKTVGKKSLEELIERFIDHTSSNYKSHDMAIKSLESKLGHLAKQM 303
TP S+ + Q + + G L + + + +Y ++ +++L SK+ ++ Q
Sbjct: 350 TPDSDLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQT 409
Query: 304 SDNHQGKFSSETLTLIE--QENTAVVSTRSGRVL---HEPEEKIEGDKNEEAEVERDEGV 358
KF+ + + +E ++ RSG+ L P + E +++ E G
Sbjct: 410 GSTAAPKFTGPSGKSMSNSKEYAHAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGN 469
Query: 359 MEERARE------PTRTRVV----------------NTHTPELRK--IPFPKALVKKNLD 394
E+A E PTR N P K +PFP K +
Sbjct: 470 SAEKAIEEPILHQPTRPLAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQ 529
Query: 395 K*FSKFLEVFKKLHINIPFSEALEQMPIYAKFMKDILSKRRKLSEVDETILMTEECSAIL 454
K + + K L + +P + L +P K++KD++++R K EV ++++ ECSAI+
Sbjct: 530 KYKALLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMITERIK--EVQGMVVLSHECSAII 587
Query: 455 QRKM-PKKRRDPGSFTIPVEIEGLTVVEALCDLGASINLMPLSMFKRLNLGEVTPTMISL 513
Q+K+ PKK DPGSFT+P + L + LCDLGAS++LMPL + K+L + P ISL
Sbjct: 588 QQKIIPKKLGDPGSFTLPCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISL 647
Query: 514 QMADRSLKTPYGIIEDVVVKVDKFVFPVDFVILDMEEDSKVPLILGRPFLATGEAEIKVA 573
+ADRS++ +G++ED+ V + P DFV+L+M+E+ K PLILGRPFLA A I V
Sbjct: 648 ILADRSVRISHGLLEDLPVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVK 707
Query: 574 KGTLTLKVGED-EVLFNIFDSLKHRADE-EVFRCEVVDELVCEEFVRISIKDPLETTIME 631
KG + L +G D ++ F+I +++K E +F E +D L + + D L++ + +
Sbjct: 708 KGKIDLNLGRDLKMTFDITNTMKKPTIEGNIFWIEEMDMLADKMLEELGETDHLQSALTK 767
Query: 632 GLEIEDQNLERELDSLY---HEVNATLSQLECVSSLATKSIWKEELTRDEEIPIEE-KSE 687
+ D +LE L Y H T + +SL S + L D+ I+ K +
Sbjct: 768 DSKEGDLHLET-LGGEYILDHITRPTAHSVYSTTSLDHNSSSEANLVSDDWSEIKAPKVD 826
Query: 688 LKPLPSSLKYAYLEEGENKPVILNSALTPLEEEKLLRVLRDHKSALGWTIDDIKGISPAI 747
LKPLP L+Y +L PVI+N LT + L+ L ++ A+G+++DDIKGISP +
Sbjct: 827 LKPLPKGLRYVFLGLNSTYPVIVNDELTADQVNLLITELMKYRKAIGYSLDDIKGISPTL 886
Query: 748 CMHKILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKK 807
C H+I LE ++PQRRLNP++K+VV+KEI+KLLDAGVIYPISDS WVSPV VPKK
Sbjct: 887 CTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKK 946
Query: 808 GGITVVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCF 867
GG+TVV N +ELIPTR +T R+CI+YR+LN +RK+HFPLPFID ML++LA H YYCF
Sbjct: 947 GGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCF 1006
Query: 868 LDGYSGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETC 927
LD YSG+ QI + P DQ KT FTCPY FAYKRMPFGLCNAPATFQRCM +IFSDLIE
Sbjct: 1007 LDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEM 1066
Query: 928 IEIFMDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGI 987
+E+FMDDFSV+G +F +CL NL VLKRC+ETNLVLNWEKCHFMVR+GIVLG K+SE GI
Sbjct: 1067 VEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGI 1126
Query: 988 EVDRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDE 1047
EVD+AKI+V+ +L PP +K IRSFLGHAGFYR FIKDFSKLA+P+T LL KE F FD+
Sbjct: 1127 EVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDD 1186
Query: 1048 NCLKAFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTV 1107
CL AF+ IK++L+TAP++ AP+W PFEI+
Sbjct: 1187 ECLTAFKLIKEALITAPIVQAPNWDFPFEII----------------------------T 1218
Query: 1108 LNEAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWV 1167
+++AQ Y TTEKELL VVFA EKFR Y++G KV ++TDHAALRH++AK+D+KPRL+RW+
Sbjct: 1219 MDDAQVRYATTEKELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWI 1278
Query: 1168 LLLQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPDEKLLAVS--TEEPLPWY 1225
LLLQEFD+EI+D++G +NGVADHLSR+ + I + P+E+L+A+ E+ LPWY
Sbjct: 1279 LLLQEFDMEIVDKKGIENGVADHLSRMR--IEDEVLIDDSMPEEQLMAIQQLNEKKLPWY 1336
Query: 1226 VHFANFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDGVIRRCIPEVNFEKIL 1285
N+ V+G P +L+ +KKKF D + WD+P+L+ +C D + R C+ E E IL
Sbjct: 1337 ADHVNYLVSGEEPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKIYRTCVSEDEIEGIL 1396
Query: 1286 WYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRRNEMPLK 1345
+CHG +YGGHF+ +T +K+LQ+GF+WP++ +D++ F+ CD
Sbjct: 1397 LHCHGFAYGGHFATFKTMSKILQAGFWWPSMFKDAQEFISKCD----------------- 1439
Query: 1346 NILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVVVAFLKKNI 1405
E FDVWGIDFMGPFP S+G +YIL+A+DYVSKWVEA A TND++VV+ K I
Sbjct: 1440 ---SFENFDVWGIDFMGPFPSSYGNKYILVAIDYVSKWVEAIASHTNDARVVLKLFKTII 1496
Query: 1406 FTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILE 1465
F RFGVPR +ISDGG HF N+ FE+LL+K+GVKHK TSGQVEISNRE+K ILE
Sbjct: 1497 FPRFGVPRIVISDGGKHFINKGFENLLKKHGVKHK--------TSGQVEISNREIKAILE 1548
Query: 1466 KVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELEHKAYWAIRKLN 1525
K V S+RKDWS KL+D LWAYRTAFKTPIGT+PF+L++GK+CHLPVELE+KA WA++ LN
Sbjct: 1549 KTVGSTRKDWSAKLNDTLWAYRTAFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLN 1608
Query: 1526 FDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFVSGQLVLLFNSR 1585
FD K A EKRL+QLN+L++ RL AYES+ IYKE+TK +HD+KI++R+F G VLLFNSR
Sbjct: 1609 FDIKTAEEKRLIQLNDLNKIRLEAYESSKIYKERTKSFHDKKIVSRDFKVGDQVLLFNSR 1668
Query: 1586 LRLFPGKLKSRWSGPFVVKRVFPHGAVEVENPETKNTFTVNGQRLKVY 1633
LRLFPGKLKSRWSGPF V V P+GA+ + FTVNGQRLK Y
Sbjct: 1669 LRLFPGKLKSRWSGPFSVTAVRPYGAITLAG--KNGDFTVNGQRLKKY 1714
Score = 120 bits (302), Expect = 5e-27
Identities = 69/200 (34%), Positives = 101/200 (50%), Gaps = 22/200 (11%)
Query: 3 EDPHAHMERFIRNCNTYRVMNVPSEAIRLSLFPFSLKDAAEDWLNSQPQGSLTTWEDLAE 62
EDP H++ F R C+ ++ V + +L LFPFSL D A W S PQGS+T+W D +
Sbjct: 60 EDPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKK 119
Query: 63 KFTTRFFPRSLLRKLKNDIMTFAQSTDENLYEAWEHFKKLLRKCPQHNLTQAECVAKFYD 122
F +FF S +L+NDI F Q+ +E YEAWE FK +CP H +
Sbjct: 120 AFLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERFKGYQTQCPHHEML---------- 169
Query: 123 GLLYSSRFGLDAASSGEFDALSPQVGYNLIEKMAMR--AMNSENERQIRRSSFEVETYDK 180
LD AS+G F + G+ ++E +A N + +R IR SS E + +
Sbjct: 170 ---------LDTASNGNFLNKDVEDGWEVVENLAQSDGNYNEDYDRSIRTSSDSDEKHRR 220
Query: 181 -LIASNKQLSEEVAEMRKHI 199
+ A N +L + + +KHI
Sbjct: 221 EMKAMNDKLDKLLLMQQKHI 240
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 723 bits (1867), Expect = 0.0
Identities = 410/868 (47%), Positives = 535/868 (61%), Gaps = 116/868 (13%)
Query: 431 LSKRRKLSEVDETILMTE----ECSAILQRKMPKKRRDPGSFTIPVEIEGLTVVEALCDL 486
+ + ++ S DE + + ECSAILQ +P KR DPGSF +P I T LCDL
Sbjct: 33 IQREKEYSLFDEIMRQLQRYSPECSAILQNVIPVKREDPGSFVLPSRIGEYTFDRCLCDL 92
Query: 487 GASINLMPLSMFKRLNLGEVTPTMISLQMADRSLKTPYGIIEDVVVKVDKFVFPVDFVIL 546
GA ++LMP S+ KRL TPT +SL + DRS+ P G+ EDV V+V F P DFVI+
Sbjct: 93 GAGVSLMPFSVAKRLGDTNFTPTKMSLVLGDRSISFPVGVAEDVQVRVGNFYIPTDFVII 152
Query: 547 DMEEDSKVPLILGRPFLATGEAEIKVAKGTLTLKVGEDEVLFNIFDSLKHRADE-EVFRC 605
+++E+ + LILGRPFL A I V K + L++G+ FN+ + E + F
Sbjct: 153 ELDEEPRHRLILGRPFLNIVAALIDVRKSKINLRIGDIVQEFNMERIMSKPTTECQTFWV 212
Query: 606 EVVDELVCEEFVRISIKDPLETTIMEGLEIEDQNLERELDSLYHEVNATLSQLECVSSLA 665
+++DELV E ++ +DPL+T + + E E L E +++ SS
Sbjct: 213 DIMDELVNELLAELNTEDPLQTVLTKE--------ESEFGYL-GEATTRFARILDSSSPM 263
Query: 666 TKSIWKEELTRDEEIPIEE-------------------KSELKPLPSSLKYAYLEEGENK 706
TK + EL +E +E+ K ELKPLP+ L+ A+L
Sbjct: 264 TKVVAFAELGDNE---VEKALVVSSPKDCDDWSELNAPKMELKPLPARLRVAFLGPNSTY 320
Query: 707 PVILNSALTPLEEEKLLRVLRDHKSALGWTIDDIKGISPAICMHKILLEENYKPIVQPQR 766
VI+N L +E LL LR ++ ALG+++DDI GISP +CMH I LE V+ QR
Sbjct: 321 LVIINPELNNVESALLLCELRKYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQR 380
Query: 767 RLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGITVVANENNELIPTRQV 826
RLN +++DVV+KEI+KLLDAG+IYPISDS WVSPV VVPKKGG+ V+ NE NELIPTR V
Sbjct: 381 RLNSNLRDVVKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTV 440
Query: 827 TKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEK 886
T R+CIDYR+LNS TRKD+FPL FIDQML++L+ YYCFLDGY G+ QI + P+DQEK
Sbjct: 441 TGHRMCIDYRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEK 500
Query: 887 TAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACL 946
T FTCPY FAY+RMPFGLCNAPATFQ CM IFSD+IE +E+F+DDF
Sbjct: 501 TTFTCPYGTFAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFMEVFIDDF----------- 549
Query: 947 GNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNI 1006
LVL+RC++ +LVLNWEK HFMVRDGIVLGHK+SE+G+EVDRAKIE++
Sbjct: 550 ----LVLQRCEDKHLVLNWEKSHFMVRDGIVLGHKISEKGVEVDRAKIEIMR-------- 597
Query: 1007 KGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSLVTAPVI 1066
FLGHAGFYRRFIKDFSK+A+P T LL KE FD+ CL+AF +K+SLV+A ++
Sbjct: 598 -----FLGHAGFYRRFIKDFSKIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIV 652
Query: 1067 VAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEKELLGVV 1126
P+W LPFE++CDASD +GAVL Q+K++ L+ IYYAS L+ AQ
Sbjct: 653 QPPEWELPFEVICDASDYVVGAVLGQRKDKKLHAIYYASRTLDGAQ-------------- 698
Query: 1127 FACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQEFDLEIIDRRGKDNG 1186
V+VHT HAALR+L +K+D+KPRL+RW+LLLQ+FDLEI D++G +N
Sbjct: 699 --------------VIVHTVHAALRYLLSKKDAKPRLLRWILLLQDFDLEIKDKKGIENE 744
Query: 1187 VADHLSRLEGGACSPIPIQEEFPDEKL-------------LAVST---------EEPLPW 1224
VAD+LSRL + +++ P E L L VST E LPW
Sbjct: 745 VADNLSRLR--VQEEVLMRDILPGENLASIEDCYMDEVGRLRVSTLELMTLHTGESNLPW 802
Query: 1225 YVHFANFRVAGLIPHDLTWQQKKKFLHD 1252
Y FAN+ + P D T KKK L +
Sbjct: 803 YADFANYLSCEVPPPDFTGYFKKKLLKE 830
>At2g06320 putative retroelement pol polyprotein
Length = 466
Score = 541 bits (1394), Expect = e-153
Identities = 254/417 (60%), Positives = 317/417 (75%), Gaps = 2/417 (0%)
Query: 1223 PWYVHFANFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDGVIRRCIPEVNFE 1282
PWY N+ G+ P +LT ++K F D Y WD+P+L+ +C D + R + E E
Sbjct: 38 PWYADHVNYLACGIEPPNLTSYERKNFFRDIHHYYWDEPYLYTLCKDKIYRSYVSEDEVE 97
Query: 1283 KILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRRNEM 1342
IL +CH S+YGGHF+ +T +K+LQ+GF+WPT+ +D++ FV CD CQR GNISRRNEM
Sbjct: 98 GILLHCHDSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEM 157
Query: 1343 PLKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVVVAFLK 1402
P ILE+++FDVWGIDFMGPFP S+G +YIL+ VDYVSKWVEA A TND+KVV+ K
Sbjct: 158 PQNPILEVDIFDVWGIDFMGPFPSSYGNKYILVVVDYVSKWVEAIASPTNDAKVVLKLFK 217
Query: 1403 KNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKR 1462
IF RFGV +ISDGG HF N+ FE+LL+K+GVKHKV+TPYHPQTSGQVEISNRE+K
Sbjct: 218 TIIFPRFGVSWVVISDGGKHFINKVFENLLKKHGVKHKVATPYHPQTSGQVEISNREIKT 277
Query: 1463 ILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELEHKAYWAIR 1522
ILEK V +RKDWS KLDDALWAY+TAFKTPIGT+PF+L+ K+CHL VELE+KA WA++
Sbjct: 278 ILEKTVGITRKDWSTKLDDALWAYKTAFKTPIGTTPFNLLCVKSCHLHVELEYKAMWAVK 337
Query: 1523 KLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFVSGQLVLLF 1582
LNFD K A EKRL+QL+ELDE RL AYES+ IYKE+TK +HD+KI+ ++F G VLLF
Sbjct: 338 LLNFDIKTAEEKRLIQLSELDEIRLEAYESSKIYKERTKLFHDKKIITKDFQVGDQVLLF 397
Query: 1583 NSRLRLFPGKLKSRWSGPFVVKRVFPHGAVEVENPETKNTFTVNGQRLKVYHGGEVL 1639
NSRL++FPGKLKSRWSGPF + V P+GAV + FTVNGQRLK Y ++L
Sbjct: 398 NSRLKIFPGKLKSRWSGPFCITEVRPYGAVTLAG--KSGVFTVNGQRLKKYLANQIL 452
>At4g07660 putative athila transposon protein
Length = 724
Score = 535 bits (1378), Expect = e-152
Identities = 288/596 (48%), Positives = 389/596 (64%), Gaps = 38/596 (6%)
Query: 452 AILQRKM-PKKRRDPGSFTIPVEIEGLTVVEALCDLGASINLMPLSMFKRLNLGEVTPTM 510
AI Q+K+ PKK DPGSFT+P + L LCDLGA ++LMPLS+ KRL +
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLGFTQYKSCN 62
Query: 511 ISLQMADRSLKTPYGIIEDVVVKVDKFVFPVDFVILDMEEDSKVPLILGRPFLATGEAEI 570
ISL +ADRS++ P+ + E++ +++ P DFV+L+M+E+ K PLILGRPFLAT A
Sbjct: 63 ISLILADRSVRIPHSLFENLPIRIGAVDIPTDFVVLEMDEEPKDPLILGRPFLATAGAMN 122
Query: 571 KVAKGTLTLKVGE-DEVLFNIFDSLKHRADE-EVFRCEVVDELVCEEFVRISIKDPLETT 628
V KG + L +G+ + F++ D++K + ++F E +D+L E + +D L +
Sbjct: 123 DVKKGKIDLNLGKYCRMTFDVKDAMKKPTIKGQLFWIEEIDQLADELLEERAEEDHLYSA 182
Query: 629 IMEG-----LEIEDQNLERELDS--------LYHEVNATLSQLECVSSLATKSIW----- 670
+ + L +E ++ LDS + E+N +++ +S + +
Sbjct: 183 LTKRGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPETEVMVMSEEGSTQVQPAHSR 242
Query: 671 -------KEELTRDEEIPI----------EEKSELKPLPSSLKYAYLEEGENKPVILNSA 713
++ E+ I K +LK LP L+Y + PVI+N
Sbjct: 243 TYSTNNSTSTISNSGELIIPTSDDWSELKAPKVDLKSLPKGLRYVFFGPNSTYPVIINVE 302
Query: 714 LTPLEEEKLLRVLRDHKSALGWTIDDIKGISPAICMHKILLEENYKPIVQPQRRLNPSMK 773
L E LL LR +K A+G+++ DIK ISP++C H+I LE ++PQRRLN ++K
Sbjct: 303 LNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLNLNLK 362
Query: 774 DVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGITVVANENNELIPTRQVTKWRVCI 833
+VV+KEI+KLLDAGVIYPISDS WV PV VPKKGG+TVV NE +ELIPTR +T RVCI
Sbjct: 363 EVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCI 422
Query: 834 DYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTAFTCPY 893
DYR+LN+ +RKDHFPLPF +QML+ LA H Y CFLDGYSG+ QI + P DQEKT FTCPY
Sbjct: 423 DYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPY 482
Query: 894 DVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGNLALVL 953
FAYKRMPFGLCNAP TFQRCM +IFSDLIE +E+FMDDFSV+GP+F +CL NL VL
Sbjct: 483 GTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRVL 542
Query: 954 KRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKGI 1009
+ +ETNLVLNWEKC+FMV++GIVLGHK+SE+GIEVD+ KI+V+ +L PP +K I
Sbjct: 543 TKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
Score = 140 bits (352), Expect = 8e-33
Identities = 69/116 (59%), Positives = 91/116 (77%), Gaps = 2/116 (1%)
Query: 1101 IYYASTVLNEAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSK 1160
IYYAS L+EAQ Y TTEKELL VVFA +KFR Y++ KV V+TDHAALRH++AK+D+K
Sbjct: 598 IYYASRTLDEAQGRYATTEKELLVVVFAFKKFRSYLVESKVTVYTDHAALRHMYAKKDTK 657
Query: 1161 PRLIRWVLLLQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPDEKLLAV 1216
PRL+R +LLLQEFD+EI++++G +NG ADHLSR+ PI I + P+E+L+ V
Sbjct: 658 PRLLRGILLLQEFDMEIVEKKGIENGAADHLSRMR--IEEPILIDDSMPEEQLMVV 711
>At4g07600
Length = 630
Score = 449 bits (1154), Expect = e-126
Identities = 218/341 (63%), Positives = 261/341 (75%), Gaps = 26/341 (7%)
Query: 789 IYPISDSEW-------VSPVQVVPKKGGITVVANENNELIPTRQVTKWRVCIDYRRLNSV 841
I P SD +W VSPVQ +PKKGGITV+ NE +ELIP R +T R+CIDYR LN+
Sbjct: 309 IIPTSD-DWSKLKAPKVSPVQCIPKKGGITVIKNEKDELIPIRTITGLRMCIDYRNLNAA 367
Query: 842 TRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTAFTCPYDVFAYKRM 901
+R DHFPLPF DQML++LA H YYCFLDGYSG+ QI + P D EKT FTCPY FAY+RM
Sbjct: 368 SRNDHFPLPFTDQMLERLANHPYYCFLDGYSGFFQIPIHPNDHEKTTFTCPYGTFAYERM 427
Query: 902 PFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGNLALVLKRCQETNL 961
PFGLCNAPATFQRCM +IFSD+IE +E+FMDDFSV+GP+F +CL NL VL RC+ETNL
Sbjct: 428 PFGLCNAPATFQRCMTSIFSDIIEEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNL 487
Query: 962 VLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRR 1021
VLNWEKCHFMV++GI+LGHK+SE+GIEVD+ FLGHAGFYRR
Sbjct: 488 VLNWEKCHFMVKEGIMLGHKISEKGIEVDKG------------------CFLGHAGFYRR 529
Query: 1022 FIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSLVTAPVIVAPDWSLPFEIMCDA 1081
FIKDFSK+A+P+T LL KE F FDE+C K+F +IK++LV+APV+ A +W PFEIMCDA
Sbjct: 530 FIKDFSKIARPLTRLLCKETEFEFDEDCPKSFHTIKEALVSAPVVRATNWDYPFEIMCDA 589
Query: 1082 SDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEKEL 1122
D A+GAVL QK ++ L+VIYYAS L+EAQ Y TTEKEL
Sbjct: 590 LDYAVGAVLGQKIDKKLHVIYYASRTLDEAQGRYATTEKEL 630
Score = 152 bits (385), Expect = 1e-36
Identities = 97/276 (35%), Positives = 162/276 (58%), Gaps = 12/276 (4%)
Query: 400 FLEVFKKLHINIPFSEALEQMPIYAKFMKDILSKRRKLSEVDETILMTEECSAILQRKM- 458
F + K++ + IP +AL + KF+KD++ +R + EV ++++ ECSAI+Q+K+
Sbjct: 2 FAKNIKEVELRIPLVDALALILDTHKFLKDLIVER--IQEVQGMVVLSHECSAIIQKKIV 59
Query: 459 PKKRRDPGSFTIPVEIEGLTVVEALCDLGASINLMPLSMFKRLNLGEVTPTMISLQMADR 518
PKK DPGSFT+P + + LCDLGA ++ MPLS+ KRL + ISL +ADR
Sbjct: 60 PKKLSDPGSFTLPCFLGTVAFNRCLCDLGALVSPMPLSIAKRLGFTQYKSCNISLILADR 119
Query: 519 SLKTPYGIIEDVVVKVDKFVFPVDFVILDMEEDSKVPLILGRPFLATGEAEIKVAKGTLT 578
S++ +G+++++ +++ DFVIL+M+E+ K PLIL RPFLAT A I V KG +
Sbjct: 120 SVRISHGLLKNLPIRIGAAEISTDFVILEMDEEPKDPLILRRPFLATAGAMIDVKKGKID 179
Query: 579 LKVGED-EVLFNIFDSLKH-RADEEVFRCEVVDELVCEEFVRISIKDPLETTIMEG---- 632
L +G+D + F+I D++K + ++F E +D+L E ++ +D L + + +
Sbjct: 180 LNLGKDFRMTFDIKDAMKKPNIEGQLFWIEEMDQLADELLEELAQEDHLNSALTKTGQDG 239
Query: 633 -LEIEDQNLERELDSLYHEVNATLSQLECVSSLATK 667
L ++ ++ LDS H+ E ++ ATK
Sbjct: 240 FLHLKVLGYQKLLDS--HKAMEESKPFEELNGPATK 273
>At2g13060 pseudogene
Length = 930
Score = 444 bits (1142), Expect = e-124
Identities = 208/372 (55%), Positives = 265/372 (70%), Gaps = 6/372 (1%)
Query: 1211 EKLLAVSTEEPLPWYVHFANFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDG 1270
E + E+ PWY N+ A + P + T KK+FL + + Y WD+P+L+K DG
Sbjct: 125 ENCAVTAVEKDYPWYADIVNYLAADVEPDNFTDYNKKRFLREIRRYQWDEPYLYKHSYDG 184
Query: 1271 VIRRCIPEVNFEKILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRC 1330
+ RRCI IL +CH SSYGGHF+ +T +KVLQ+ F+WPT+ RD++ F+ CD C
Sbjct: 185 IYRRCIAATEVPSILSHCHSSSYGGHFATFKTVSKVLQADFWWPTMFRDTQKFISQCDPC 244
Query: 1331 QRTGNISRRNEMPLKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALS 1390
QR G IS+RNEMP LE+E+FD WGIDFMGPFPPS YIL+ VDYVSKWVEA A
Sbjct: 245 QRRGKISKRNEMPPNFRLEVEVFDRWGIDFMGPFPPSNKNLYILVDVDYVSKWVEAIASL 304
Query: 1391 TNDSKVVVAFLKKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTS 1450
NDS VV+ K IF RFGVPR +ISDG HF N+ E LL +YGV+H+V+TPYHPQTS
Sbjct: 305 KNDSAVVMKLFKSIIFPRFGVPRIVISDGDKHFINKILEKLLLQYGVQHRVATPYHPQTS 364
Query: 1451 GQVEISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLP 1510
GQVE+SNR++K ILEK V ++K+WS KL DALWAY+TAFKTP+GT+PFHL++GKACHLP
Sbjct: 365 GQVEVSNRQIKEILEKTVGKAKKEWSYKLYDALWAYKTAFKTPLGTTPFHLLYGKACHLP 424
Query: 1511 VELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILN 1570
VELEHKA WA++ +NFD K A E +ELDE R+ AY+++ +YKE+TK +HD+KIL
Sbjct: 425 VELEHKAAWAVKMMNFDIKSAGE------SELDEIRIHAYDNSKLYKERTKAYHDKKILT 478
Query: 1571 REFVSGQLVLLF 1582
R F +L F
Sbjct: 479 RTFEPNDQILRF 490
Score = 71.2 bits (173), Expect = 5e-12
Identities = 32/65 (49%), Positives = 47/65 (72%), Gaps = 2/65 (3%)
Query: 1156 KQDSKPRLIRWVLLLQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPDEKLLA 1215
K+D+KPRL+ W+LLLQEFD+E+ D++G +NGVADHLSR+ +PI + P+E +
Sbjct: 3 KKDAKPRLLIWILLLQEFDIEVRDKKGVENGVADHLSRIR--IDDDVPINDFLPEENIYM 60
Query: 1216 VSTEE 1220
+ T E
Sbjct: 61 IDTVE 65
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 442 bits (1136), Expect = e-123
Identities = 220/372 (59%), Positives = 273/372 (73%), Gaps = 24/372 (6%)
Query: 884 QEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFD 943
++KT FTCP FAYKRM FGLCNAP TFQR M +IFSD IE +E+FMDDFS F
Sbjct: 163 KKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDDFS----GFS 218
Query: 944 ACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPP 1003
+CL NL VL+RC+ETNLVLNWEKCHFMV +GIVLGHK+S +GIEVD+AKI+V+ +L PP
Sbjct: 219 SCLLNLCRVLERCEETNLVLNWEKCHFMVHEGIVLGHKISGKGIEVDKAKIDVMIQLQPP 278
Query: 1004 TNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSLVTA 1063
+K IRSFLGHA FYRRFIKDFSK+A+P+T LL KE F FDE+CLKAF IK++LV+A
Sbjct: 279 KTVKDIRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCLKAFHLIKETLVSA 338
Query: 1064 PVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEKELL 1123
P+ AP+W PFEIMCDA D A+GAVL QK + L+VIYYAS ++EAQ Y T EKELL
Sbjct: 339 PIFQAPNWDHPFEIMCDAFDYAVGAVLDQKIDDKLHVIYYASRTMDEAQTRYATIEKELL 398
Query: 1124 GVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQEFDLEIIDRRGK 1183
VVFA EK R Y++GFKV V+ DHAALRH++AK+++K RL+RW+LLLQEFD+EIID++G
Sbjct: 399 AVVFAFEKIRSYLVGFKVKVYIDHAALRHIYAKKETKSRLLRWILLLQEFDMEIIDKKGV 458
Query: 1184 DNGVADHLSRLEGGACSPIPIQEEFPDEKLLAVS------------------TEEPLPWY 1225
+NGVADHLSR+ +PI + P+E+L+ EE LPWY
Sbjct: 459 ENGVADHLSRMR--IEDSVPIDDTMPEEQLMFYDLVNKSFDTKDMPEEADAVEEEKLPWY 516
Query: 1226 VHFANFRVAGLI 1237
N+ +G +
Sbjct: 517 ADLVNYLTSGQV 528
Score = 135 bits (339), Expect = 3e-31
Identities = 61/98 (62%), Positives = 80/98 (81%)
Query: 1449 TSGQVEISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACH 1508
TSGQVE+SNR++K IL KVV S++DWS KLD+ LWAYRTA+KTPIG +PF +++GK+CH
Sbjct: 524 TSGQVEVSNRQMKDILTKVVGVSKRDWSTKLDETLWAYRTAYKTPIGRTPFQMLYGKSCH 583
Query: 1509 LPVELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFR 1546
LPVE+E+KA WA + LN + K A EKR + L+EL+E R
Sbjct: 584 LPVEVEYKAIWATKLLNLEIKGAQEKRPIDLHELEEIR 621
Score = 42.7 bits (99), Expect = 0.002
Identities = 20/49 (40%), Positives = 30/49 (60%)
Query: 685 KSELKPLPSSLKYAYLEEGENKPVILNSALTPLEEEKLLRVLRDHKSAL 733
K +LKPLP L+YA+L + PVI+N L+ + +LL LR ++ L
Sbjct: 102 KLDLKPLPKGLRYAFLGPNDTYPVIINDGLSDEQVNQLLNELRKYRGQL 150
>At2g10780 pseudogene
Length = 1611
Score = 390 bits (1001), Expect = e-108
Identities = 288/905 (31%), Positives = 454/905 (49%), Gaps = 90/905 (9%)
Query: 752 ILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGIT 811
I LE PI + R+ P+ ++K++ +LLD G I P S S W +PV V KK G
Sbjct: 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRP-SSSPWGAPVLFVKKKDG-- 707
Query: 812 VVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGY 871
+R+CIDYR LN VT K+ +PLP ID+++D+L G Q++ +D
Sbjct: 708 ----------------SFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLA 751
Query: 872 SGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIF 931
SGY+QI + P D KTAF YD F + MPFGL NAPA F + M +F D ++ + IF
Sbjct: 752 SGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIF 811
Query: 932 MDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDR 991
++D V+ +++A +L VL+R +E L KC F R LGH +S++G+ VD
Sbjct: 812 INDILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDP 871
Query: 992 AKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLK 1051
KI I++ P P N IRSFLG AG+YRRF+ F+ +A+P+T L K+ F + + C K
Sbjct: 872 EKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEK 931
Query: 1052 AFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEA 1111
+F +K L APV+V P+ P+ + DAS + LG VL QK VI YAS L +
Sbjct: 932 SFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKH 987
Query: 1112 QRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQ 1171
++NY T + E+ VVF + +R Y+ G KV ++TDH +L+++F + + R RW+ L+
Sbjct: 988 EKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVA 1047
Query: 1172 EFDLEIIDRRGKDNGVADHLSRLEG---GACSPIPIQEEFPDEKLLAVSTE-EPLPWYVH 1227
+++L+I GK N VAD LSR S + + + A+S E EPL
Sbjct: 1048 DYNLDIAYHPGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPL----G 1103
Query: 1228 FANFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDGVI----RRCIPEVNF-- 1281
A L+ Q++ + + K + ++ ++ ++G I R C+P
Sbjct: 1104 LGAADQADLLSRIRLAQERDE---EIKGWAQNNKTEYQTSNNGTIVVNGRVCVPNDRALK 1160
Query: 1282 EKILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRRNE 1341
E+IL H S + H G + L+ ++W + +D +V C CQ + ++
Sbjct: 1161 EEILREAHQSKFSIH-PGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQL---VKAEHQ 1216
Query: 1342 MP---LKNILEIE-LFDVWGIDFMGPFPPSFGCQY--ILLAVDYVSKWVEAAALSTNDSK 1395
+P L+N+ E +D +DF+ P ++ + + VD ++K A+S D
Sbjct: 1217 VPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGA 1276
Query: 1396 VVVAFLKKNIFTRF-GVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVE 1454
++A + R G+P +I+SD T F ++ +++ + G + +ST YHPQT Q E
Sbjct: 1277 EIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSE 1336
Query: 1455 ISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELE 1514
+ + L+ +L V +W + L +AY +F+ IG SP+ ++G+AC P+
Sbjct: 1337 RTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPL--- 1393
Query: 1515 HKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKE---KTKKWHDRKILNR 1571
W E+RL +DE R KE + K + +++
Sbjct: 1394 ------------CWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKEL 1441
Query: 1572 EFVSGQLVLL----------FNSRLRLFPGKLKSRWSGPF-VVKRVFPHGAV--EVENPE 1618
EF G LV L F SR +L P R+ GP+ V++RV GAV +++ P
Sbjct: 1442 EFQVGDLVYLKAMTYKGAGRFTSRKKLSP-----RYVGPYKVIERV---GAVAYKLDLPP 1493
Query: 1619 TKNTF 1623
N F
Sbjct: 1494 KLNAF 1498
>At2g12040 T10J7.2
Length = 976
Score = 389 bits (999), Expect = e-108
Identities = 183/308 (59%), Positives = 240/308 (77%), Gaps = 2/308 (0%)
Query: 913 QRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMV 972
+R M +IF+D+IE +E+FMDDFSV+G F+ CL NL VL RC+E +LVLNWEKCHF V
Sbjct: 79 ERGMMSIFTDMIEDIMEVFMDDFSVYGSLFEDCLENLYKVLARCEEKHLVLNWEKCHFRV 138
Query: 973 RDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKP 1032
+DGIVLGH++SE GIE DRAKIEV+ L N+K +RSFLGHAGFYRRFIKDFSK+A+P
Sbjct: 139 QDGIVLGHRISEYGIEADRAKIEVMTSLQALDNLKAVRSFLGHAGFYRRFIKDFSKIARP 198
Query: 1033 MTNLLEKEAPFTFDENCLKAFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQ 1092
+T+LL KE F F + C AF+ IK++L++AP++ PDW LPFE+MCDASD A+GAVL Q
Sbjct: 199 LTSLLCKEVKFEFTQECHDAFQQIKQALISAPIVQPPDWDLPFEVMCDASDFAVGAVLGQ 258
Query: 1093 KKERVLYVIYYASTVLNEAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRH 1152
+K++ L+ IYYAS L++A+RNY TTEKE L VVFA EKFR Y++G KV+VHTDHAAL++
Sbjct: 259 RKDKKLHAIYYASRTLDDAKRNYATTEKEFLAVVFAFEKFRSYLVGLKVIVHTDHAALKY 318
Query: 1153 LFAKQDSKPRLIRWVLLLQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPDEK 1212
L +D+KPRL+RW+LLLQEFD+E+ D++G + VADHLSR+ +PI + P+E
Sbjct: 319 LMQNKDAKPRLLRWILLLQEFDIEVRDKKGVEKCVADHLSRIR--IDDDVPINDFLPEEN 376
Query: 1213 LLAVSTEE 1220
+ + T E
Sbjct: 377 IYMIDTAE 384
Score = 320 bits (819), Expect = 6e-87
Identities = 186/427 (43%), Positives = 239/427 (55%), Gaps = 41/427 (9%)
Query: 1209 PDEKLLAVSTEEPLPWYVHFANFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICS 1268
P E + E+ PWY + A + + KK+FL + + Y WD P+L
Sbjct: 445 PTENCAVTALEKDYPWYADIVIYLGADVELDNFIDYNKKRFLREIRRYYWDKPYL----- 499
Query: 1269 DGVIRRCIPEVNFEKILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCD 1328
I + F + C+ S S A V SG S + S D
Sbjct: 500 ----PISIVLMEFIGEMHCCNRDSIQSSSSRLLVAYNV--SGC--------SEIHIAS-D 544
Query: 1329 RCQRTGNISRRNEMPLKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAA 1388
CQR G IS NEMP K ILE+++FD WGI+FMGP PPS YIL+ VDYVSKWVEA A
Sbjct: 545 PCQRRGKISNCNEMPQKFILEVKVFDCWGINFMGPIPPSNKNLYILVVVDYVSKWVEAIA 604
Query: 1389 LSTNDSKVVVAFLKKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQ 1448
NDS VV+ K IF FGVPR +ISDGG HF N+ LL +YGV+H+V+TPYHPQ
Sbjct: 605 SPKNDSAVVMKLFKCIIFPHFGVPRIVISDGGKHFINKILTKLLLQYGVQHRVATPYHPQ 664
Query: 1449 TSGQVEISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACH 1508
TSGQVE+SNR++K ILEK V AFKTP+GT+PFHL++GKACH
Sbjct: 665 TSGQVEVSNRQIKEILEKTV--------------------AFKTPLGTTPFHLLYGKACH 704
Query: 1509 LPVELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKI 1568
LPVELEHKA W ++ LNFD K A E+RL+QLNELDE ++ AY+++ +YKE+TK +HD+KI
Sbjct: 705 LPVELEHKAAWTVKMLNFDIKSAGERRLIQLNELDEIQIHAYDNSKLYKERTKAYHDKKI 764
Query: 1569 LNREFVSGQLVLLFNSRLRLFPGKLKSRWSGPFVVKRVFPHGAVEVENPETKNT-FTVNG 1627
L R F + F G+ K ++ +F H + E E T FT++
Sbjct: 765 LTRTFEPKDQKKKKTRETKPFKGRGKKSERVGSLLIPIFEHFRIRFEGEEVNTTRFTMDE 824
Query: 1628 QRLKVYH 1634
LK H
Sbjct: 825 SYLKNSH 831
Score = 30.4 bits (67), Expect = 8.9
Identities = 15/33 (45%), Positives = 21/33 (63%)
Query: 749 MHKILLEENYKPIVQPQRRLNPSMKDVVRKEII 781
MH+I LE+ K V+ QRRLN K +RK ++
Sbjct: 1 MHRIHLEDESKSSVEHQRRLNLIQKKRLRKRLL 33
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 370 bits (949), Expect = e-102
Identities = 283/900 (31%), Positives = 429/900 (47%), Gaps = 99/900 (11%)
Query: 752 ILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGIT 811
I LE P+ + R+ P+ ++K++ LL G I P S S W +PV V KK G
Sbjct: 509 IELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRP-STSPWGAPVLFVKKKDG-- 565
Query: 812 VVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGY 871
+R+CIDYR LN VT K+ +PLP ID++LD+L G + +D
Sbjct: 566 ----------------SFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLT 609
Query: 872 SGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIF 931
SGY+QI +A D KTAF Y F + MPF L NAPA F R M ++F + ++ + IF
Sbjct: 610 SGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIF 669
Query: 932 MDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDR 991
+DD V+ + + +L V+++ +E L KC F R+ LGH VS G+ VD
Sbjct: 670 IDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDP 729
Query: 992 AKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLK 1051
KIE I P PTN IRSFL G+YRRF+K F+ +A+PMT L K+ PF + C +
Sbjct: 730 EKIEAIRDWPRPTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEE 789
Query: 1052 AFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEA 1111
F S+K+ L + PV+ P+ P+ + DAS + LG VL Q+ + VI YAS L +
Sbjct: 790 GFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQRGK----VIAYASRQLRKH 845
Query: 1112 QRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQ 1171
+ NY T + E+ V+FA + +R Y+ G KV V TDH +L+++F + + R +RW+ L+
Sbjct: 846 EGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVA 905
Query: 1172 EFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPD--EKLLAVSTEEPLPWYVHFA 1229
++DLEI GK N VAD LSR GA ++ + L V EPL
Sbjct: 906 DYDLEIAYHPGKANVVADALSRKRVGAAPGQSVEALVSEIGALRLCVVAREPL----GLE 961
Query: 1230 NFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDGVI----RRCIP--EVNFEK 1283
A L+ Q+K + L A + ++ ++G I R C+P E +
Sbjct: 962 AVDRADLLTRARLAQEKDEGLIAASKAEGSE---YQFAANGTIFVYGRVCVPKDEELRRE 1018
Query: 1284 ILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRRNEMP 1343
IL H S + H G + L+ + W + RD +V CD CQ + +++P
Sbjct: 1019 ILSEAHASMFSIH-PGATKMYRDLKRYYQWVGMKRDVANWVAECDVCQL---VKAEHQVP 1074
Query: 1344 LKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVVVAFLKK 1403
I + +D ++K A+ D V+A KK
Sbjct: 1075 ---------------------------DAIWVIMDRLTKSAHFLAIRKTDGAAVLA--KK 1105
Query: 1404 ---NIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNREL 1460
I GVP +I+SD + F + + K G K ++ST YHPQT GQ E + + L
Sbjct: 1106 YVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTL 1165
Query: 1461 KRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELEHKAYWA 1520
+ +L V W+ L +AY +++ IG +PF ++G+ C P+ +
Sbjct: 1166 EDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPLRWTQVEERS 1225
Query: 1521 IRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFVSGQLVL 1580
I ++ + R+L+LN + + + + + D++ EF G V
Sbjct: 1226 IYGADYVQETTERIRVLKLNMKEA------------QARQRSYADKRRRELEFEVGDRVY 1273
Query: 1581 LFNSRLR-----LFPGKLKSRWSGPF-VVKRVFPHGAVEVENPETKNTFTVNGQRLKVYH 1634
L + LR + KL R+ GPF +V+RV P A +E P+ F KV+H
Sbjct: 1274 LKMAMLRGPNRSILETKLSPRYMGPFRIVERVGP-VAYRLELPDVMRAFH------KVFH 1326
>At3g11970 hypothetical protein
Length = 1499
Score = 354 bits (908), Expect = 3e-97
Identities = 269/845 (31%), Positives = 408/845 (47%), Gaps = 82/845 (9%)
Query: 750 HKILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGG 809
HKI L E P+ Q R + K+ + K + LL G + S S + SPV +V KK G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQA-SSSPYASPVVLVKKKDG 649
Query: 810 ITVVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLD 869
WR+C+DYR LN +T KD FP+P I+ ++D+L G + +D
Sbjct: 650 T------------------WRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKID 691
Query: 870 GYSGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIE 929
+GY+Q+ + P+D +KTAF F Y MPFGL NAPATFQ M IF + +
Sbjct: 692 LRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVL 751
Query: 930 IFMDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEV 989
+F DD V+ + + +L V + + L KC F V LGH +S +GIE
Sbjct: 752 VFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIET 811
Query: 990 DRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENC 1049
D AKI+ +++ P PT +K +R FLG AG+YRRF++ F +A P+ L + +A F +
Sbjct: 812 DPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDA-FEWTAVA 870
Query: 1050 LKAFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLN 1109
+AFE +K +L APV+ P + F + DA +GAVL Q+ + Y+ S L
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYI----SRQLK 926
Query: 1110 EAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLL 1169
Q + + EKELL V+FA K+R Y+L ++ TD +L++L ++ + P +W+
Sbjct: 927 GKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPK 986
Query: 1170 LQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPDEKLLAVSTEEPLPWYVHFA 1229
L EFD EI R+GK+N VAD LSR+EG E + V + +A
Sbjct: 987 LLEFDYEIQYRQGKENVVADALSRVEG---------SEVLHMAMTVVECDLLKDIQAGYA 1037
Query: 1230 NFRVAGLIPHDLTWQQKKKFLHDAKSYL-WDDPFLFKICSDGVIRR----CIPEVNFEK- 1283
N + +T Q+ D+K Y W S ++RR +P + K
Sbjct: 1038 N---DSQLQDIITALQRDP---DSKKYFSW---------SQNILRRKSKIVVPANDNIKN 1082
Query: 1284 -ILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQR-----TGNIS 1337
IL + HGS GGH SG + ++ FYW + +D +A++ SC CQ+ +
Sbjct: 1083 TILLWLHGSGVGGH-SGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPG 1141
Query: 1338 RRNEMPLKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVV 1397
+P+ + + E+ +DF+ P S G I++ VD +SK ALS S +
Sbjct: 1142 LLQPLPIPDTIWSEV----SMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALT 1197
Query: 1398 VAF-LKKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEIS 1456
VA N+F G P +I+SD F + + GV K+++ YHPQ+ GQ E+
Sbjct: 1198 VAHAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVV 1257
Query: 1457 NRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKA--CHLPVELE 1514
NR L+ L + + WS+ L A + Y T + + +PF +V+G+ HLP
Sbjct: 1258 NRCLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPY--- 1314
Query: 1515 HKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFV 1574
L + KVA R LQ E ++ L + + K++ D+ REF
Sbjct: 1315 ---------LPGESKVAVVARSLQ--EREDMLLFLKFHLMRAQHRMKQFADQHRTEREFE 1363
Query: 1575 SGQLV 1579
G V
Sbjct: 1364 IGDYV 1368
>At1g36590 hypothetical protein
Length = 1499
Score = 348 bits (893), Expect = 1e-95
Identities = 268/845 (31%), Positives = 407/845 (47%), Gaps = 82/845 (9%)
Query: 750 HKILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGG 809
HKI L E P+ Q R + K+ + K + LL G + S S + SPV +V KK G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQA-SSSPYASPVVLVKKKDG 649
Query: 810 ITVVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLD 869
WR+C+DYR LN +T KD FP+P I+ ++D+L G + +D
Sbjct: 650 T------------------WRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKID 691
Query: 870 GYSGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIE 929
+GY+Q+ + P+D +KTAF F Y MPFGL NAPATFQ M IF + +
Sbjct: 692 LRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVL 751
Query: 930 IFMDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEV 989
+F DD V+ + + +L V + + L KC F V LGH +S +GIE
Sbjct: 752 VFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIET 811
Query: 990 DRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENC 1049
D AKI+ +++ P PT +K +R FLG AG+YRRF++ F +A P+ L + +A F +
Sbjct: 812 DPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDA-FEWTAVA 870
Query: 1050 LKAFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLN 1109
+AFE +K +L APV+ P + F + DA +GAVL Q+ + Y+ S L
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYI----SRQLK 926
Query: 1110 EAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLL 1169
Q + + EKELL V+FA K+R Y+L ++ TD +L++L ++ + P +W+
Sbjct: 927 GKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPK 986
Query: 1170 LQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPDEKLLAVSTEEPLPWYVHFA 1229
L EFD EI R+GK+N VAD LSR+EG E + V + +A
Sbjct: 987 LLEFDYEIQYRQGKENVVADALSRVEG---------SEVLHMAMTVVECDLLKDIQAGYA 1037
Query: 1230 NFRVAGLIPHDLTWQQKKKFLHDAKSYL-WDDPFLFKICSDGVIRR----CIPEVNFEK- 1283
N + +T Q+ D+K Y W S ++RR +P + K
Sbjct: 1038 N---DSQLQDIITALQRDP---DSKKYFSW---------SQNILRRKSKIVVPANDNIKN 1082
Query: 1284 -ILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQR-----TGNIS 1337
IL + HGS GGH SG + ++ FY + +D +A++ SC CQ+ +
Sbjct: 1083 TILLWLHGSGVGGH-SGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPG 1141
Query: 1338 RRNEMPLKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVV 1397
+P+ + + E+ +DF+ P S G I++ VD +SK ALS S +
Sbjct: 1142 LLQPLPIPDTIWSEV----SMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALT 1197
Query: 1398 VA-FLKKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEIS 1456
VA N+F G P +I+SD F + + GV K+++ YHPQ+ GQ E+
Sbjct: 1198 VAQAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVV 1257
Query: 1457 NRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKA--CHLPVELE 1514
NR L+ L + + WS+ L A + Y T + + +PF +V+G+ HLP
Sbjct: 1258 NRCLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPY--- 1314
Query: 1515 HKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFV 1574
L + KVA R LQ E ++ L + + K++ D+ REF
Sbjct: 1315 ---------LPGESKVAVVARSLQ--EREDMLLFLKFHLMRAQHRMKQFADQHRTEREFE 1363
Query: 1575 SGQLV 1579
G V
Sbjct: 1364 IGDYV 1368
>At1g35370 hypothetical protein
Length = 1447
Score = 340 bits (872), Expect = 4e-93
Identities = 248/771 (32%), Positives = 389/771 (50%), Gaps = 66/771 (8%)
Query: 750 HKILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGG 809
HKI L E P+ Q R KD + K + ++ +G I +S S + SPV +V KK G
Sbjct: 570 HKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQ-VSSSPFASPVVLVKKKDG 628
Query: 810 ITVVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLD 869
WR+C+DY LN +T KD F +P I+ ++D+L G + +D
Sbjct: 629 T------------------WRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKID 670
Query: 870 GYSGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIE 929
+GY+Q+ + P+D +KTAF F Y M FGL NAPATFQ M ++F D + +
Sbjct: 671 LRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVL 730
Query: 930 IFMDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEV 989
+F DD ++ + + +L LV + + L K H LGH +S R IE
Sbjct: 731 VFFDDILIYSSSIEEHKEHLRLVFEVMRLHKLFAKGSKEH--------LGHFISAREIET 782
Query: 990 DRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENC 1049
D AKI+ +++ P PT +K +R FLG AG+YRRF+++F +A P+ + L K F +
Sbjct: 783 DPAKIQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIAGPL-HALTKTDGFCWSLEA 841
Query: 1050 LKAFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLN 1109
AF+++K L APV+ P + F + DA + AVL QK + Y+ S L
Sbjct: 842 QSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQKGHPLAYI----SRQLK 897
Query: 1110 EAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLL 1169
Q + + EKELL +FA K+R Y+L ++ TD +L++L ++ + P +W+
Sbjct: 898 GKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPK 957
Query: 1170 LQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIP---IQEEFPDEKLLAVSTEEPLPWYV 1226
L EFD EI R+GK+N VAD LSR+EG + ++ +F E +A ++
Sbjct: 958 LLEFDYEIQYRQGKENLVADALSRVEGSEVLHMALSIVECDFLKEIQVAYESD------- 1010
Query: 1227 HFANFRVAGLIPHDLTWQQKKKFLHDAKS-YLWDDPFLFKICSDGVIRRCIPEVNFEKIL 1285
G++ ++ Q+ DAK Y W L + S V+ + N K+L
Sbjct: 1011 --------GVLKDIISALQQHP---DAKKHYSWSQDIL-RRKSKIVVPNDVEITN--KLL 1056
Query: 1286 WYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQ--RTGNISRRNEMP 1343
+ H S GG SG + + ++S FYW + +D +AF+ SC CQ ++ N + +
Sbjct: 1057 QWLHCSGMGGR-SGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQ 1115
Query: 1344 LKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVVV--AFL 1401
I + DV +DF+ P S G I++ VD +SK AL+ S + V AFL
Sbjct: 1116 PLPIPDKIWCDV-SMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFL 1174
Query: 1402 KKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELK 1461
N++ G P +I+SD F + ++ + GV+ ++S+ YHPQ+ GQ E+ NR L+
Sbjct: 1175 -DNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLE 1233
Query: 1462 RILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKA--CHLP 1510
L + + W++ L A + Y T + + +PF LV+G+A HLP
Sbjct: 1234 NYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLP 1284
>At2g11680 putative retroelement pol polyprotein
Length = 301
Score = 332 bits (850), Expect = 1e-90
Identities = 165/303 (54%), Positives = 221/303 (72%), Gaps = 19/303 (6%)
Query: 971 MVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLA 1030
MVR+GIVLGHK+S++G++V++AK++V+ +L PP K IRSFLGHAGFYRRFIKDFSKLA
Sbjct: 1 MVREGIVLGHKISKKGVQVNKAKVDVMMQLQPPKTGKDIRSFLGHAGFYRRFIKDFSKLA 60
Query: 1031 KPMTNLLEKEAPFTFDENCLKAFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVL 1090
+P+T LL KEA F F+E CL AF+S K++LV+AP++ AP+ PFEIMCD SD A+GAVL
Sbjct: 61 RPLTRLLCKEAEFIFEEECLTAFKSTKEALVSAPIVQAPNRDYPFEIMCDDSDYAVGAVL 120
Query: 1091 CQKKERVLYVIYYASTVLNEAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAAL 1150
QK +R L+VIYYAS +++ Q Y TTEKELL VVFA EKFR Y++G KV V+TDHAAL
Sbjct: 121 GQKIDRKLHVIYYASGTMDDTQVRYATTEKELLAVVFAFEKFRSYLVGSKVTVYTDHAAL 180
Query: 1151 RHLFAKQDSKPRLIRWVLLLQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPD 1210
RH++AK+D+KPRL+RW+LLLQEFD+EI+D++G +NGVADHLSR+ +PI + P+
Sbjct: 181 RHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGVADHLSRMR--IEDEVPIDDSMPE 238
Query: 1211 EKLLAV-----------------STEEPLPWYVHFANFRVAGLIPHDLTWQQKKKFLHDA 1253
E+L+A+ + E+ L WY + G P +L+ +KKKF D
Sbjct: 239 EQLMAIQQLNESAQINKSLDQVCAVEDKLMWYADHVKYLFGGEEPPNLSSYEKKKFFKDI 298
Query: 1254 KSY 1256
+
Sbjct: 299 NHF 301
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 304 bits (779), Expect = 2e-82
Identities = 178/447 (39%), Positives = 252/447 (55%), Gaps = 23/447 (5%)
Query: 752 ILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGIT 811
I LE P+ + R+ P+ ++K++ LL G I P S S W +PV V KK G
Sbjct: 483 IELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRP-STSPWGAPVLFVKKKDG-- 539
Query: 812 VVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGY 871
+R+CIDYR LN VT K+ +PLP ID++LD+L G + +D
Sbjct: 540 ----------------SFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLT 583
Query: 872 SGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIF 931
SGY+QI +A D KTAF Y F + MPFGL NAPA F R M ++F + ++ + IF
Sbjct: 584 SGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIF 643
Query: 932 MDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDR 991
+DD V+ + + +L V+++ +E L KC F R+ LGH VS G+ VD
Sbjct: 644 IDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDP 703
Query: 992 AKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLK 1051
KIE I P PTN IRSFLG AG+YRRF+K F+ +A+PMT L K+ PF + + C +
Sbjct: 704 EKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEE 763
Query: 1052 AFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEA 1111
F S+K+ L + PV+ P+ P+ + DAS + LG VL Q + VI YAS L +
Sbjct: 764 GFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHGK----VIAYASRQLMKH 819
Query: 1112 QRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQ 1171
+ NY T + E+ V+FA + +R Y+ G KV V TDH +L+++F + + R RW+ L+
Sbjct: 820 EGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVA 879
Query: 1172 EFDLEIIDRRGKDNGVADHLSRLEGGA 1198
++DLEI GK N V D LSR GA
Sbjct: 880 DYDLEIAYHPGKANVVVDALSRKRVGA 906
>At2g07410 putative retroelement pol polyprotein
Length = 411
Score = 291 bits (746), Expect = 2e-78
Identities = 158/283 (55%), Positives = 190/283 (66%), Gaps = 37/283 (13%)
Query: 1361 MGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVVVAFLKKNIFTRFGVPRAIISDGG 1420
MG FP S+G +YIL+AVDYVSKWVEA TND+KVV+ K IFTRFGV +ISDG
Sbjct: 1 MGSFPSSYGNKYILVAVDYVSKWVEAIVSPTNDAKVVLKLFKTIIFTRFGVHMVVISDGR 60
Query: 1421 THFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILEKVVDSSRKDWSRKLD 1480
HF N+AFE+LL+K+GVKHKV+T YHPQTSGQVEISNRE+K ILEK V +RKD S KLD
Sbjct: 61 KHFINKAFENLLKKHGVKHKVATSYHPQTSGQVEISNREIKAILEKTVGVTRKDLSAKLD 120
Query: 1481 DALWAY----RTAFKTPIGTSPFHLVFGKACHLPVELEHKAYWAIRKLNFDWKVASEKRL 1536
DALWAY +TAFKTPIGT+PF+L++ K+ HLP+ELE KA WA++ +NFD K A E+RL
Sbjct: 121 DALWAYKTAFKTAFKTPIGTTPFNLLYAKSGHLPIELEFKAMWAVKLMNFDIKTAEEERL 180
Query: 1537 LQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFVSGQLVLLFNSRLRLFPGKLKSR 1596
+QLN+LDE RL AYES+ YK K KLKSR
Sbjct: 181 IQLNDLDEIRLEAYESSKNYKGKR-------------------------------KLKSR 209
Query: 1597 WSGPFVVKRVFPHGAVEVENPETKNTFTVNGQRLKVYHGGEVL 1639
GPF V + P+GAV + FTVNGQRLK Y ++L
Sbjct: 210 LFGPFCVTEIRPYGAVTL--AVKSGDFTVNGQRLKKYLEDQIL 250
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 281 bits (719), Expect = 2e-75
Identities = 169/447 (37%), Positives = 244/447 (53%), Gaps = 35/447 (7%)
Query: 752 ILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGIT 811
I LE P+ + R+ P+ ++K++ LL A PV V KK G
Sbjct: 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGA-------------PVLFVKKKDG-- 514
Query: 812 VVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGY 871
+R+CIDYR LN VT K+ +PLP ID++LD+L G + +D
Sbjct: 515 ----------------SFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLT 558
Query: 872 SGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIF 931
SGY+ I +A D KTAF Y F + MPFGL NAPA F R M ++F ++++ + IF
Sbjct: 559 SGYHLIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIF 618
Query: 932 MDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDR 991
+DD V+ + + +L V+++ +E L KC F R+ LGH VS G+ VD
Sbjct: 619 IDDILVYSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDP 678
Query: 992 AKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLK 1051
KIE I PTN IRSFLG AG+YRRF+K F+ +A+PMT L K+ PF + C +
Sbjct: 679 EKIEAIRDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEE 738
Query: 1052 AFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEA 1111
F S+K+ L + PV+ P+ P+ + DAS + LG VL Q+ + VI YAS L +
Sbjct: 739 GFVSLKEMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGK----VIAYASRQLRKH 794
Query: 1112 QRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQ 1171
+ NY T + E+ V+FA + +R Y+ G V V TDH +L+++F + + R +W+ L+
Sbjct: 795 EGNYPTHDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVA 854
Query: 1172 EFDLEIIDRRGKDNGVADHLSRLEGGA 1198
++DLEI GK N VAD LS GA
Sbjct: 855 DYDLEIAYHPGKANVVADALSHKRVGA 881
Score = 90.5 bits (223), Expect = 7e-18
Identities = 87/331 (26%), Positives = 147/331 (44%), Gaps = 37/331 (11%)
Query: 1317 NRDSRAFVESCDRCQRTGNISRRNEMPLKNILEIEL----FDVWGIDFMGPFPPSFGCQY 1372
+ D +V CD CQ + +++P + + + +D IDF+ P S
Sbjct: 940 SEDVANWVAECDVCQL---VKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTKDA 996
Query: 1373 ILLAVDYVSKWVEAAALSTNDSKVVVAFLKK---NIFTRFGVPRAIISDGGTHFCNRAFE 1429
I + VD ++K A+ D V+A KK I GVP +I+SD + F + +
Sbjct: 997 IWVIVDRLTKSAHFLAIRKTDGAAVLA--KKYVSEIVKLHGVPVSIVSDRDSKFTSAFWR 1054
Query: 1430 SLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTA 1489
+ + G K ++ST YHPQT GQ E + + L+ +L V W+ L +AY +
Sbjct: 1055 AFQAEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNS 1114
Query: 1490 FKTPIGTSPFHLVFGKACHLPVELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRA 1549
+ IG +PF ++ + C P+ L +I ++ + R+L+LN +
Sbjct: 1115 YPASIGMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEA----- 1169
Query: 1550 YESASIYKEKTKKWHDRKILNREFVSGQLVLLFNSRLR-----LFPGKLKSRWSGPF-VV 1603
+++ + + D++ EF G V L + LR + KL R+ GPF +V
Sbjct: 1170 -------QDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIV 1222
Query: 1604 KRVFPHGAVEVENPETKNTFTVNGQRLKVYH 1634
+RV P A +E P+ F KV+H
Sbjct: 1223 ERVGP-VAYRLELPDVMRAFH------KVFH 1246
>At1g20390 hypothetical protein
Length = 1791
Score = 273 bits (698), Expect = 6e-73
Identities = 163/495 (32%), Positives = 252/495 (49%), Gaps = 20/495 (4%)
Query: 701 EEGENKPVILNSALTPLEEEKLLRVLRDHKSALGWTIDDIKGISPAICMHKILLEENYKP 760
E + V + + ++P +L+ +L+ + W+I+D+KGI PAI H++ ++ +KP
Sbjct: 718 ESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKP 777
Query: 761 IVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGITVVANENNEL 820
+ Q +R+L P V +E+ KLL AG I + EW++ VV KK G
Sbjct: 778 VKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNG----------- 826
Query: 821 IPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVA 880
KWRVC+DY LN KD +PLP ID++++ +G+ F+D +SGYNQI +
Sbjct: 827 -------KWRVCVDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMH 879
Query: 881 PEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGP 940
+DQEKT+F + YK M FGL NA AT+QR + + +D I +E+++DD V
Sbjct: 880 KDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSL 939
Query: 941 NFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKL 1000
+ + +L+ + LN KC F V G LG+ V++RGIE + +I I +L
Sbjct: 940 KPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILEL 999
Query: 1001 PPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSL 1060
P P N + ++ G RFI + P NLL++ A F +D++ +AFE +K L
Sbjct: 1000 PSPRNAREVQRLTGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYL 1059
Query: 1061 VTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEK 1120
T P++V P+ + SD A+ +VL ++ I+Y S L EA+ Y EK
Sbjct: 1060 STPPILVKPEVGETLYLYIAVSDHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEK 1119
Query: 1121 ELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQEFDLEIIDR 1180
L VV + K RPY + V TD LR R+ +W + L E+D++ R
Sbjct: 1120 AALAVVTSARKLRPYFQSHTIAVLTDQ-PLRVALHSPSQSGRMTKWAVELSEYDIDFRPR 1178
Query: 1181 RG-KDNGVADHLSRL 1194
K +AD L L
Sbjct: 1179 PAMKSQVLADFLIEL 1193
Score = 193 bits (491), Expect = 6e-49
Identities = 123/420 (29%), Positives = 205/420 (48%), Gaps = 10/420 (2%)
Query: 1220 EPLPWYVHFANFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDGVIRRCIPEV 1279
+P W V ++ G +P D W ++ + AK L + L K+ + G + C+
Sbjct: 1376 DPTDWRVEIRDYLSDGTLPSD-KWTARRLRIKAAKYTLMKE-HLLKVSAFGAMLNCLHGT 1433
Query: 1280 NFEKILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRR 1339
+I+ H + G H G A K+ + GFYWPT+ D + F C++CQR +
Sbjct: 1434 EINEIMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQ 1493
Query: 1340 NEMPLKNILEIELFDVWGIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVVVA 1399
L+ + F W +D +GP P S ++IL+ DY +KWVEA + +T + V
Sbjct: 1494 PTELLRAGVAPYPFMRWAMDIVGPMPASRQKRFILVMTDYFTKWVEAESYATIRANDVQN 1553
Query: 1400 FLKKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRE 1459
F+ K I R G+P II+D G+ F + +FE+ + ++ STP +PQ +GQ E +N+
Sbjct: 1554 FVWKFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNKT 1613
Query: 1460 LKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELEHKAYW 1519
+ L+K +D + W+ +LD LW+YRT ++ +PF +G P E+ Y
Sbjct: 1614 ILSGLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEV---GYS 1670
Query: 1520 AIRKLNFDWKVASEKRLL--QLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFVSGQ 1577
++R+ R++ +L++L+E R A Y+ K +++K+ NR F G
Sbjct: 1671 SLRRSMMVKNPELNDRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGD 1730
Query: 1578 LVL--LFNSRLRLFPGKLKSRWSGPFVVKRVFPHGAVEVENPE-TKNTFTVNGQRLKVYH 1634
LVL +F + + GKL + W G + V ++ G E+ T T N LK Y+
Sbjct: 1731 LVLRKVFENTAEINAGKLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTWNSMHLKRYY 1790
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 263 bits (671), Expect = 8e-70
Identities = 152/386 (39%), Positives = 219/386 (56%), Gaps = 23/386 (5%)
Query: 752 ILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGIT 811
I LE PI + R+ P+ ++K++ +LL G I P S S W +PV V KK G
Sbjct: 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRP-SSSPWGAPVLFVKKKDG-- 202
Query: 812 VVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGY 871
+R+CIDYR LN VT K+ +PLP ID+++D+L G Q++ +D
Sbjct: 203 ----------------SFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLA 246
Query: 872 SGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIF 931
SGY+QI + P D KTAF Y F + MPFGL NAPA F + M +F D ++ + IF
Sbjct: 247 SGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIF 306
Query: 932 MDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDR 991
+DD V +++A +L VL+R +E L K F R LGH +S++G+ VD
Sbjct: 307 IDDILVHSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDP 366
Query: 992 AKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLK 1051
KI I++ P P N IRSFLG AG+YRRF+ F+ +A+P+T L K+ F + + C K
Sbjct: 367 EKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEK 426
Query: 1052 AFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEA 1111
+F +K L+ APV+V P+ P+ + DAS + LG VL QK VI YAS L +
Sbjct: 427 SFLELKAMLINAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKH 482
Query: 1112 QRNYTTTEKELLGVVFACEKFRPYIL 1137
++NY T + E+ VVFA + +R Y++
Sbjct: 483 EKNYPTHDLEMAAVVFALKIWRSYLI 508
Score = 100 bits (249), Expect = 7e-21
Identities = 100/376 (26%), Positives = 168/376 (44%), Gaps = 52/376 (13%)
Query: 1273 RRCIPEVNF--EKILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRC 1330
R C+P E+IL H S + H G + L+ ++W + +D +V C C
Sbjct: 542 RVCVPNDRALKEEILREAHQSKFSIH-PGSNKMYRDLKRYYHWVGMRKDVARWVAKCPTC 600
Query: 1331 QRTGNISRRNEMP---LKNILEIEL-FDVWGIDFMGPFPPSFGCQY--ILLAVDYVSKWV 1384
Q + +++P L+N+ E +D +DF+ P ++ + + VD ++K
Sbjct: 601 QL---VKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSA 657
Query: 1385 EAAALSTNDSKVVVAFLKKNIFTRF-GVPRAIISDGGTHFCNRAFESLLEKYGVKHKVST 1443
A+S D ++A + R G+P +I+SD T F ++ + + + G + +ST
Sbjct: 658 HFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLST 717
Query: 1444 PYHPQTSGQVEISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVF 1503
YHPQT GQ E + + L+ +L V +W + L +AY +F+ IG SP+ ++
Sbjct: 718 AYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALY 777
Query: 1504 GKACHLPVELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKE---KT 1560
G+AC P+ W E+RL +DE R KE +
Sbjct: 778 GRACRTPL---------------CWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQ 822
Query: 1561 KKWHDRKILNREFVSGQLVLL----------FNSRLRLFPGKLKSRWSGPF-VVKRVFPH 1609
K + +++ EF G LV L F SR +L P R+ GP+ V++RV
Sbjct: 823 KSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSP-----RYVGPYKVIERV--- 874
Query: 1610 GAV--EVENPETKNTF 1623
GAV +++ P N F
Sbjct: 875 GAVAYKLDLPPKLNVF 890
>At2g06890 putative retroelement integrase
Length = 1215
Score = 260 bits (664), Expect = 5e-69
Identities = 165/505 (32%), Positives = 256/505 (50%), Gaps = 31/505 (6%)
Query: 829 WRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTA 888
WR+C D R +N+VT K P+P +D MLD+L G + +D SGY+QI + D+ KTA
Sbjct: 469 WRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWKTA 528
Query: 889 FTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGN 948
F + ++ + MPFGL +AP+TF R M + I + ++ DD V+ + + +
Sbjct: 529 FKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDILVYSESLREHIEH 588
Query: 949 LALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKG 1008
L VL ++ L N +KC F + + LG VS G++VD K++ I P P +
Sbjct: 589 LDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKVDEEKVKAIRDWPSPKTVGE 648
Query: 1009 IRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSLVTAPVIVA 1068
+RSF G AGFYRRF KDFS + P+T +++K+ F +++ +AF+S+K L APV++
Sbjct: 649 VRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQEEAFQSLKDKLTNAPVLIL 708
Query: 1069 PDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEKELLGVVFA 1128
++ FEI CDAS + +GAVL Q ++ +I + S L A NY T +KEL +V A
Sbjct: 709 SEFLKTFEIECDASGIGIGAVLMQDQK----LIAFFSEKLGGATLNYPTYDKELYALVRA 764
Query: 1129 CEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLLLQEFDLEIIDRRGKDNGVA 1188
++++ Y+ V+HTDH +L+HL +Q R RWV ++ F I ++GKDN VA
Sbjct: 765 LQRWQHYLWPKVFVIHTDHESLKHLKGQQKLNKRHARWVEFIETFAYVIKYKKGKDNVVA 824
Query: 1189 DHLSRLEGGACSPIPIQEEFPDEKLLAVSTEEPLPWYVHFANFRVAGLIPHDLTWQQKKK 1248
D LS ++ +ST + F + HD K
Sbjct: 825 DALS------------------QRYTLLSTLNVK--LMGFEQIKEVYETDHDFQEVYKAC 864
Query: 1249 FLHDAKSYLWDDPFLFKICSDGVIRRCIPEVNFEKI-LWYCHGSSYGGHFSGERTAAKVL 1307
+ Y D FLF R C+P + + + HG GHF +T +V+
Sbjct: 865 EKFASGRYFRQDKFLFY-----ENRLCVPNCSLRDLFVREAHGGGLMGHFGIAKT-LEVM 918
Query: 1308 QSGFYWPTLNRDSRAFVESCDRCQR 1332
F WP + D + C+ C++
Sbjct: 919 TEHFRWPHMKCDVKRICGRCNTCKQ 943
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,207,698
Number of Sequences: 26719
Number of extensions: 1728101
Number of successful extensions: 5362
Number of sequences better than 10.0: 208
Number of HSP's better than 10.0 without gapping: 152
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 4825
Number of HSP's gapped (non-prelim): 436
length of query: 1649
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1536
effective length of database: 8,299,349
effective search space: 12747800064
effective search space used: 12747800064
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0330.2


