

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0330.12
(413 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
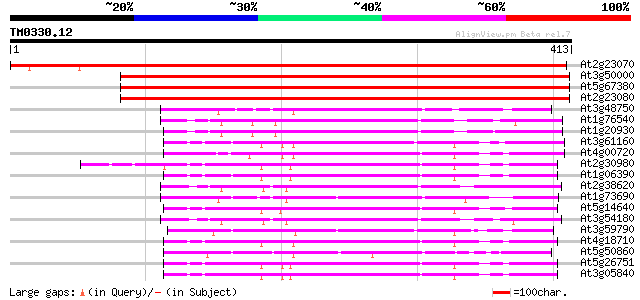
Score E
Sequences producing significant alignments: (bits) Value
At2g23070 putative casein kinase II catalytic (alpha) subunit 674 0.0
At3g50000 CASEIN KINASE II, ALPHA CHAIN 2 (CK II) 615 e-176
At5g67380 casein kinase II catalytic subunit (ATPK12D) 611 e-175
At2g23080 putative casein kinase II catalytic (alpha) subunit 595 e-170
At3g48750 protein kinase (cdc2) 148 6e-36
At1g76540 putative cell division control protein cdc2 146 2e-35
At1g20930 putative cell division control protein, cdc2 kinase 145 3e-35
At3g61160 shaggy-like kinase beta 144 7e-35
At4g00720 Shaggy related protein kinase tetha 144 9e-35
At2g30980 putative shaggy-like protein kinase dzeta 144 9e-35
At1g06390 putative shaggy kinase iota protein 142 3e-34
At2g38620 putative cell division control protein kinase 140 2e-33
At1g73690 cell division protein kinase 139 2e-33
At5g14640 protein kinase MSK-3 - like 139 3e-33
At3g54180 protein kinase cdc2 homolog B 139 3e-33
At3g59790 mitogen-activated protein kinase-like protein 138 6e-33
At4g18710 shaggy-like protein kinase etha (EC 2.7.1.-) 136 2e-32
At5g50860 cyclin-dependent protein kinase-like 134 9e-32
At5g26751 shaggy-like kinase alpha 134 9e-32
At3g05840 shaggy related protein kinase, ASK-GAMMA 132 3e-31
>At2g23070 putative casein kinase II catalytic (alpha) subunit
Length = 432
Score = 674 bits (1739), Expect = 0.0
Identities = 336/428 (78%), Positives = 362/428 (84%), Gaps = 18/428 (4%)
Query: 1 MAIRPFHHFNLLS-------RDRLLYLFRTAQQFHSPPSYLLRSPSFSLRHFAVSAT--- 50
MA+RP F + S + L L + + + LL S +LR FA SA+
Sbjct: 1 MALRPCTGFTISSLRNASAANNNLFSLLSFSSSSPAKRNLLLSSLQDNLRRFASSASLYR 60
Query: 51 --------PAPPQPRRHLPPPPDSMAQKIGKSTRRPGASSKARVYADVNVVRPKEYWDYE 102
Q + + +++AQKIGKS RR GA SKARVYADVNVVRPK+YWDYE
Sbjct: 61 QHLRNQQQQHQQQQQSRVKEKSETLAQKIGKSIRRAGAPSKARVYADVNVVRPKDYWDYE 120
Query: 103 TLTVQWGEQDDYEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIKREIKILQN 162
+L VQWG QDDYEVVRKVGRGKYSEVFEG+H DNEKCVIKILKPVKKKKIKREIKILQN
Sbjct: 121 SLAVQWGVQDDYEVVRKVGRGKYSEVFEGIHATDNEKCVIKILKPVKKKKIKREIKILQN 180
Query: 163 LCGGPNIVKLLDIVRDQQSKTPSLIFEYVNNTDFKVLYPTLSDYDIRYYIYELLKALDYC 222
LCGGPNIVKLLDIVRDQQSKTPSLIFE+VNN DFKVLYPTLSDYD+RYYI+ELLKALD+C
Sbjct: 181 LCGGPNIVKLLDIVRDQQSKTPSLIFEHVNNKDFKVLYPTLSDYDVRYYIFELLKALDFC 240
Query: 223 HSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQ 282
HS+GIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQ
Sbjct: 241 HSRGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQ 300
Query: 283 DYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDELSAYLNKYRIELDP 342
DYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDEL+AYLNKYRIELDP
Sbjct: 301 DYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDELNAYLNKYRIELDP 360
Query: 343 NLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFNPV 402
NL +L+GRHSRKPW KFIN ENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYF P+
Sbjct: 361 NLTSLVGRHSRKPWTKFINSENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFYPI 420
Query: 403 RSAESSRT 410
R+AESSRT
Sbjct: 421 RNAESSRT 428
>At3g50000 CASEIN KINASE II, ALPHA CHAIN 2 (CK II)
Length = 333
Score = 615 bits (1587), Expect = e-176
Identities = 289/331 (87%), Positives = 314/331 (94%)
Query: 82 SKARVYADVNVVRPKEYWDYETLTVQWGEQDDYEVVRKVGRGKYSEVFEGVHCIDNEKCV 141
SKARVY DVNV+RPK+YWDYE+L VQWGEQDDYEVVRKVGRGKYSEVFEG++ +NEKC+
Sbjct: 2 SKARVYTDVNVIRPKDYWDYESLNVQWGEQDDYEVVRKVGRGKYSEVFEGINMNNNEKCI 61
Query: 142 IKILKPVKKKKIKREIKILQNLCGGPNIVKLLDIVRDQQSKTPSLIFEYVNNTDFKVLYP 201
IKILKPVKKKKI+REIKILQNLCGGPNIVKLLD+VRDQ SKTPSLIFEYVN+TDFKVLYP
Sbjct: 62 IKILKPVKKKKIRREIKILQNLCGGPNIVKLLDVVRDQHSKTPSLIFEYVNSTDFKVLYP 121
Query: 202 TLSDYDIRYYIYELLKALDYCHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGK 261
TL+DYDIRYYIYELLKALD+CHSQGIMHRDVKPHNVMIDHE RKLRLIDWGLAEFYHPGK
Sbjct: 122 TLTDYDIRYYIYELLKALDFCHSQGIMHRDVKPHNVMIDHELRKLRLIDWGLAEFYHPGK 181
Query: 262 EYNVRVASRYFKGPELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIA 321
EYNVRVASRYFKGPELLVDLQDYDYSLD+WSLGCMFAGMIFRKEPFFYGHDN DQLVKIA
Sbjct: 182 EYNVRVASRYFKGPELLVDLQDYDYSLDMWSLGCMFAGMIFRKEPFFYGHDNQDQLVKIA 241
Query: 322 KVLGTDELSAYLNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLR 381
KVLGTDEL+AYLNKY++ELD L AL+GRHSRKPW+KFIN +N+HL PEA+D++DKLLR
Sbjct: 242 KVLGTDELNAYLNKYQLELDTQLEALVGRHSRKPWSKFINADNRHLVSPEAIDYLDKLLR 301
Query: 382 YDHQERPTAKEAMAHPYFNPVRSAESSRTRT 412
YDHQ+R TAKEAMAHPYF VR+AESSR RT
Sbjct: 302 YDHQDRLTAKEAMAHPYFAQVRAAESSRMRT 332
>At5g67380 casein kinase II catalytic subunit (ATPK12D)
Length = 333
Score = 611 bits (1575), Expect = e-175
Identities = 287/331 (86%), Positives = 312/331 (93%)
Query: 82 SKARVYADVNVVRPKEYWDYETLTVQWGEQDDYEVVRKVGRGKYSEVFEGVHCIDNEKCV 141
SKARVY +VNV+RPK+YWDYE+L VQWGEQDDYEVVRKVGRGKYSEVFEG++ EKC+
Sbjct: 2 SKARVYTEVNVIRPKDYWDYESLIVQWGEQDDYEVVRKVGRGKYSEVFEGINVNSKEKCI 61
Query: 142 IKILKPVKKKKIKREIKILQNLCGGPNIVKLLDIVRDQQSKTPSLIFEYVNNTDFKVLYP 201
IKILKPVKKKKI+REIKILQNLCGGPNIVKLLD+VRDQ SKTPSLIFEYVN+TDFKVLYP
Sbjct: 62 IKILKPVKKKKIRREIKILQNLCGGPNIVKLLDVVRDQHSKTPSLIFEYVNSTDFKVLYP 121
Query: 202 TLSDYDIRYYIYELLKALDYCHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGK 261
TL+DYDIRYYIYELLKALD+CHSQGIMHRDVKPHNVMIDHE RKLRLIDWGLAEFYHPGK
Sbjct: 122 TLTDYDIRYYIYELLKALDFCHSQGIMHRDVKPHNVMIDHELRKLRLIDWGLAEFYHPGK 181
Query: 262 EYNVRVASRYFKGPELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIA 321
EYNVRVASRYFKGPELLVDLQDYDYSLD+WSLGCMFAGMIFRKEPFFYGHDN DQLVKIA
Sbjct: 182 EYNVRVASRYFKGPELLVDLQDYDYSLDMWSLGCMFAGMIFRKEPFFYGHDNQDQLVKIA 241
Query: 322 KVLGTDELSAYLNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLR 381
KVLGTDEL+AYLNKY++ELDP L AL+GRHSRKPW+KFIN +NQHL PEA+DF+DKLLR
Sbjct: 242 KVLGTDELNAYLNKYQLELDPQLEALVGRHSRKPWSKFINADNQHLVSPEAIDFLDKLLR 301
Query: 382 YDHQERPTAKEAMAHPYFNPVRSAESSRTRT 412
YDHQ+R TAKEAMAH YF VR+AE+SR R+
Sbjct: 302 YDHQDRLTAKEAMAHAYFAQVRAAETSRMRS 332
>At2g23080 putative casein kinase II catalytic (alpha) subunit
Length = 333
Score = 595 bits (1534), Expect = e-170
Identities = 280/331 (84%), Positives = 306/331 (91%)
Query: 82 SKARVYADVNVVRPKEYWDYETLTVQWGEQDDYEVVRKVGRGKYSEVFEGVHCIDNEKCV 141
SKARVY DVNVVRPKEYWDYE+L VQWG QDDYEVVRKVGRGKYSEVFEG + NE+CV
Sbjct: 2 SKARVYTDVNVVRPKEYWDYESLVVQWGHQDDYEVVRKVGRGKYSEVFEGKNVNTNERCV 61
Query: 142 IKILKPVKKKKIKREIKILQNLCGGPNIVKLLDIVRDQQSKTPSLIFEYVNNTDFKVLYP 201
IKILKPVKKKKIKREIKILQNLCGGPNIVKL DIVRD+ SKTPSL+FE+VN+ DFKVLYP
Sbjct: 62 IKILKPVKKKKIKREIKILQNLCGGPNIVKLYDIVRDEHSKTPSLVFEFVNSVDFKVLYP 121
Query: 202 TLSDYDIRYYIYELLKALDYCHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGK 261
TL+DYDIRYYIYELLKALD+CHSQGIMHRDVKPHNVMIDH+ RKLRLIDWGLAEFYHPGK
Sbjct: 122 TLTDYDIRYYIYELLKALDFCHSQGIMHRDVKPHNVMIDHQLRKLRLIDWGLAEFYHPGK 181
Query: 262 EYNVRVASRYFKGPELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIA 321
EYNVRVASRYFKGPELLVDLQDYDYSLD+WSLGCMFAGMIFRKEPFFYGHDN+DQLVKIA
Sbjct: 182 EYNVRVASRYFKGPELLVDLQDYDYSLDMWSLGCMFAGMIFRKEPFFYGHDNHDQLVKIA 241
Query: 322 KVLGTDELSAYLNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLR 381
KVLGT+EL YLNKY+++LDP L AL+GRH KPW+KFIN +NQHL PEA+DF+DKLL+
Sbjct: 242 KVLGTNELDHYLNKYQLDLDPQLEALVGRHVPKPWSKFINADNQHLVSPEAIDFLDKLLQ 301
Query: 382 YDHQERPTAKEAMAHPYFNPVRSAESSRTRT 412
YDHQ+R TA+EAM HPYF V++AESSR RT
Sbjct: 302 YDHQDRLTAREAMDHPYFAQVKAAESSRLRT 332
>At3g48750 protein kinase (cdc2)
Length = 294
Score = 148 bits (373), Expect = 6e-36
Identities = 97/301 (32%), Positives = 161/301 (53%), Gaps = 28/301 (9%)
Query: 112 DDYEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKK-----IKREIKILQNLCGG 166
D YE V K+G G Y V++ + NE +K ++ ++ + REI +L+ +
Sbjct: 2 DQYEKVEKIGEGTYGVVYKARDKVTNETIALKKIRLEQEDEGVPSTAIREISLLKEMQHS 61
Query: 167 PNIVKLLDIVRDQQSKTPSLIFEYVNNTDFKVLYPTLSDYD-----IRYYIYELLKALDY 221
NIVKL D+V + K L+FEY++ D K + D+ I+ Y+Y++L+ + Y
Sbjct: 62 -NIVKLQDVVHSE--KRLYLVFEYLD-LDLKKHMDSTPDFSKDLHMIKTYLYQILRGIAY 117
Query: 222 CHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAE-FYHPGKEYNVRVASRYFKGPELLVD 280
CHS ++HRD+KP N++ID L+L D+GLA F P + + V + +++ PE+L+
Sbjct: 118 CHSHRVLHRDLKPQNLLIDRRTNSLKLADFGLARAFGIPVRTFTHEVVTLWYRAPEILLG 177
Query: 281 LQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDELSAYLNKYR-IE 339
Y +D+WS+GC+FA MI +K P F G DQL KI +++GT Y + +R +
Sbjct: 178 SHHYSTPVDIWSVGCIFAEMISQK-PLFPGDSEIDQLFKIFRIMGT----PYEDTWRGVT 232
Query: 340 LDPNLAALIGRHSRKPWAKFI-NVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPY 398
P+ + + F+ N++ P+ VD + K+L D +R A+ A+ H Y
Sbjct: 233 SLPDYKSAFPKWKPTDLETFVPNLD------PDGVDLLSKMLLMDPTKRINARAALEHEY 286
Query: 399 F 399
F
Sbjct: 287 F 287
>At1g76540 putative cell division control protein cdc2
Length = 313
Score = 146 bits (369), Expect = 2e-35
Identities = 98/321 (30%), Positives = 163/321 (50%), Gaps = 45/321 (14%)
Query: 112 DDYEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIK-----------REIKIL 160
D +E + KVG G Y +V+ EK KI+ +KK ++ REI IL
Sbjct: 12 DAFEKLEKVGEGTYGKVYRA-----REKATGKIVA-LKKTRLHEDEEGVPSTTLREISIL 65
Query: 161 QNLCGGPNIVKLLDIVR--DQQSKTPS-LIFEYVNNT------DFKVLYPTLSDYDIRYY 211
+ L P++V+L+D+ + ++ KT L+FEY++ F+ + I+
Sbjct: 66 RMLARDPHVVRLMDVKQGLSKEGKTVLYLVFEYMDTDVKKFIRSFRSTGKNIPTQTIKSL 125
Query: 212 IYELLKALDYCHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAE-FYHPGKEYNVRVASR 270
+Y+L K + +CH GI+HRD+KPHN+++D + +L++ D GLA F P K+Y + +
Sbjct: 126 MYQLCKGMAFCHGHGILHRDLKPHNLLMDPKTMRLKIADLGLARAFTLPMKKYTHEILTL 185
Query: 271 YFKGPELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDELS 330
+++ PE+L+ Y ++D+WS+GC+FA ++ + F G QL+ I K+ GT
Sbjct: 186 WYRAPEVLLGATHYSTAVDMWSVGCIFAELV-TNQAIFQGDSELQQLLHIFKLFGTP--- 241
Query: 331 AYLNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPE----AVDFVDKLLRYDHQE 386
E+ P ++ L H W AVP VD + K+L+Y+ +
Sbjct: 242 ------NEEMWPGVSTLKNWHEYPQW----KPSTLSSAVPNLDEAGVDLLSKMLQYEPAK 291
Query: 387 RPTAKEAMAHPYFNPVRSAES 407
R +AK AM HPYF+ + S
Sbjct: 292 RISAKMAMEHPYFDDLPEKSS 312
>At1g20930 putative cell division control protein, cdc2 kinase
Length = 315
Score = 145 bits (367), Expect = 3e-35
Identities = 96/316 (30%), Positives = 167/316 (52%), Gaps = 39/316 (12%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIK-----------REIKILQN 162
+E + KVG G Y +V+ EK I+ +KK ++ REI IL+
Sbjct: 16 FEKLEKVGEGTYGKVYRA-----REKATGMIVA-LKKTRLHEDEEGVPPTTLREISILRM 69
Query: 163 LCGGPNIVKLLDIVR--DQQSKTPS-LIFEYVNNT------DFKVLYPTLSDYDIRYYIY 213
L P+IV+L+D+ + +++ KT L+FEYV+ F+ + ++ +Y
Sbjct: 70 LARDPHIVRLMDVKQGINKEGKTVLYLVFEYVDTDLKKFIRSFRQAGQNIPQNTVKCLMY 129
Query: 214 ELLKALDYCHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAE-FYHPGKEYNVRVASRYF 272
+L K + +CH G++HRD+KPHN+++D + L++ D GLA F P K+Y + + ++
Sbjct: 130 QLCKGMAFCHGHGVLHRDLKPHNLLMDRKTMTLKIADLGLARAFTLPMKKYTHEILTLWY 189
Query: 273 KGPELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDELSAY 332
+ PE+L+ Y +D+WS+GC+FA ++ K+ F G QL++I ++LGT
Sbjct: 190 RAPEVLLGATHYSTGVDMWSVGCIFAELV-TKQAIFAGDSELQQLLRIFRLLGTP----- 243
Query: 333 LNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPEA-VDFVDKLLRYDHQERPTAK 391
E+ P ++ L H W K +++ + EA +D + K+L Y+ +R +AK
Sbjct: 244 ----NEEVWPGVSKLKDWHEYPQW-KPLSLSTAVPNLDEAGLDLLSKMLEYEPAKRISAK 298
Query: 392 EAMAHPYFNPVRSAES 407
+AM HPYF+ + S
Sbjct: 299 KAMEHPYFDDLPDKSS 314
>At3g61160 shaggy-like kinase beta
Length = 438
Score = 144 bits (364), Expect = 7e-35
Identities = 96/309 (31%), Positives = 165/309 (53%), Gaps = 29/309 (9%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIK-REIKILQNLCGGPNIVKL 172
Y +G G + VF+ C++ E+ V I K ++ K+ K RE++I++ L PN+V+L
Sbjct: 109 YRAEHVIGTGSFGVVFQA-KCLETEEKVA-IKKVLQDKRYKNRELQIMRML-DHPNVVEL 165
Query: 173 LDIVRDQQSKTP---SLIFEYVNNTDFKVL--YPTLSDYD----IRYYIYELLKALDYCH 223
K +L+ EYV T ++ Y ++ + I+ Y Y++ +A++Y H
Sbjct: 166 KHSFFSTTEKDELYLNLVLEYVPETIYRASRSYTKMNQHMPLIYIQLYTYQICRAMNYLH 225
Query: 224 SQ-GIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQ 282
G+ HRD+KP N+++++ ++++ D+G A+ PG+ + SRY++ PEL+
Sbjct: 226 QVVGVCHRDIKPQNLLVNNVTHEVKICDFGSAKMLIPGEPNISYICSRYYRAPELIFGAT 285
Query: 283 DYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAYLNKYRIE 339
+Y ++D+WS+GC+ A +F P F G + DQLV+I K+LGT +E+ +Y
Sbjct: 286 EYTSAIDMWSVGCVMA-ELFLGHPLFPGETSVDQLVEIIKILGTPAREEIKNMNPRYNDF 344
Query: 340 LDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYF 399
P + A +PW K + PEA+D +LL+Y R TA EA AHP+F
Sbjct: 345 KFPQIKA-------QPWHKIF----RRQVSPEAMDLASRLLQYSPNLRCTALEACAHPFF 393
Query: 400 NPVRSAESS 408
+ +R +S
Sbjct: 394 DDLRDPRAS 402
>At4g00720 Shaggy related protein kinase tetha
Length = 472
Score = 144 bits (363), Expect = 9e-35
Identities = 98/308 (31%), Positives = 159/308 (50%), Gaps = 27/308 (8%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIKREIKILQNLCGGPNIVKLL 173
Y R VG G + VF+ E+ IK + K+ K RE++I++ L PN+V+L
Sbjct: 138 YMAQRVVGTGSFGVVFQAKCLETGEQVAIKKVLQDKRYK-NRELQIMR-LQDHPNVVRLR 195
Query: 174 DI---VRDQQSKTPSLIFEYVNNTDFKVL--YPTLSDYD----IRYYIYELLKALDYCHS 224
D+ +L+ EYV T ++ Y ++ + ++ Y Y++ +AL+Y H
Sbjct: 196 HSFFSTTDKDELYLNLVLEYVPETVYRASKHYTKMNQHMPIIFVQLYTYQICRALNYLHR 255
Query: 225 Q-GIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQD 283
G+ HRD+KP N++++ + +L++ D+G A+ PG+ + SRY++ PEL+ +
Sbjct: 256 VVGVCHRDIKPQNLLVNPQTHQLKICDFGSAKMLVPGEPNISYICSRYYRAPELIFGATE 315
Query: 284 YDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAYLNKYRIEL 340
Y ++D+WS GC+ A ++ +P F G DQLV+I K+LGT +E+ Y
Sbjct: 316 YTNAIDMWSGGCVMAELLL-GQPLFPGESGIDQLVEIIKILGTPTREEIRCMNPNYTEFK 374
Query: 341 DPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFN 400
P + A PW K + PEAVD V +LL+Y R TA EA AHP+F+
Sbjct: 375 FPQIKA-------HPWHKIFHKR----MPPEAVDLVSRLLQYSPNLRCTALEACAHPFFD 423
Query: 401 PVRSAESS 408
+R S
Sbjct: 424 DLRDPNVS 431
>At2g30980 putative shaggy-like protein kinase dzeta
Length = 412
Score = 144 bits (363), Expect = 9e-35
Identities = 111/371 (29%), Positives = 174/371 (45%), Gaps = 38/371 (10%)
Query: 53 PPQPRRHLPPPPDSMAQKIGKSTRRPGASSKARVYADVNVVRPKEYWDYETLTVQWGEQD 112
PPQP P PP K RRP + + A V + + ++ G ++
Sbjct: 8 PPQPPSLAPQPPHLHGGDSLK--RRPDIDNDKEMSAAV--IEGNDAVTGHIISTTIGGKN 63
Query: 113 -------DYEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIKREIKILQNLCG 165
Y R VG G + VF+ C++ + V I K ++ ++ K L L
Sbjct: 64 GEPKQTISYMAERVVGTGSFGIVFQA-KCLETGESVA-IKKVLQDRRYKNRELQLMRLMD 121
Query: 166 GPNIVKLLDIVRDQQSKTP---SLIFEYVNNTDFKVLYPTLSD------YDIRYYIYELL 216
PN+V L ++ +L+ EYV T ++VL S + ++ Y Y++
Sbjct: 122 HPNVVSLKHCFFSTTTRDELFLNLVMEYVPETLYRVLKHYTSSNQRMPIFYVKLYTYQIF 181
Query: 217 KALDYCHSQ-GIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGP 275
+ L Y H+ G+ HRDVKP N+++D + +L D+G A+ G+ + SRY++ P
Sbjct: 182 RGLAYIHTAPGVCHRDVKPQNLLVDPLTHQCKLCDFGSAKVLVKGEANISYICSRYYRAP 241
Query: 276 ELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAY 332
EL+ +Y S+D+WS GC+ A ++ +P F G ++ DQLV+I KVLGT +E+
Sbjct: 242 ELIFGATEYTSSIDIWSAGCVLAELLL-GQPLFPGENSVDQLVEIIKVLGTPTREEIRCM 300
Query: 333 LNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKE 392
Y P + A PW K + PEA+D +LL+Y R TA E
Sbjct: 301 NPNYTDFRFPQIKA-------HPWHKVFHKR----MPPEAIDLASRLLQYSPSLRCTALE 349
Query: 393 AMAHPYFNPVR 403
A AHP+FN +R
Sbjct: 350 ACAHPFFNELR 360
>At1g06390 putative shaggy kinase iota protein
Length = 407
Score = 142 bits (359), Expect = 3e-34
Identities = 97/304 (31%), Positives = 159/304 (51%), Gaps = 29/304 (9%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIK-REIKILQNLCGGPNIVKL 172
Y R VG G + VF+ C++ + V I K ++ ++ K RE+++++ + PN++ L
Sbjct: 70 YMAERVVGTGSFGIVFQA-KCLETGESVA-IKKVLQDRRYKNRELQLMRPM-DHPNVISL 126
Query: 173 LDIVRDQQSKTP---SLIFEYVNNTDFKVLYPTLSD------YDIRYYIYELLKALDYCH 223
S+ +L+ EYV T ++VL S + ++ Y Y++ + L Y H
Sbjct: 127 KHCFFSTTSRDELFLNLVMEYVPETLYRVLRHYTSSNQRMPIFYVKLYTYQIFRGLAYIH 186
Query: 224 S-QGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQ 282
+ G+ HRDVKP N+++D +++L D+G A+ G+ + SRY++ PEL+
Sbjct: 187 TVPGVCHRDVKPQNLLVDPLTHQVKLCDFGSAKVLVKGEPNISYICSRYYRAPELIFGAT 246
Query: 283 DYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAYLNKYRIE 339
+Y S+D+WS GC+ A ++ +P F G ++ DQLV+I KVLGT +E+ Y
Sbjct: 247 EYTASIDIWSAGCVLAELLL-GQPLFPGENSVDQLVEIIKVLGTPTREEIRCMNPNYTDF 305
Query: 340 LDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYF 399
P + A PW K + PEA+D +LL+Y R TA EA AHP+F
Sbjct: 306 RFPQIKA-------HPWHKVFHKR----MPPEAIDLASRLLQYSPSLRCTALEACAHPFF 354
Query: 400 NPVR 403
N +R
Sbjct: 355 NELR 358
>At2g38620 putative cell division control protein kinase
Length = 311
Score = 140 bits (352), Expect = 2e-33
Identities = 96/326 (29%), Positives = 167/326 (50%), Gaps = 48/326 (14%)
Query: 112 DDYEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIK-----------REIKIL 160
+ YE + KVG G Y +V++ + EK K++ +KK +++ REI +L
Sbjct: 2 EKYEKLEKVGEGTYGKVYKAM-----EKTTGKLVA-LKKTRLEMDEEGIPPTALREISLL 55
Query: 161 QNLCGGPNIVKLLDIVRDQQSKTPS----------LIFEYVNNTDFKVLYPT-------- 202
Q L IV+LL + QSK + L+FEY++ TD K +
Sbjct: 56 QMLSQSIYIVRLLCVEHVIQSKDSTVSHSPKSNLYLVFEYLD-TDLKKFIDSHRKGSNPR 114
Query: 203 -LSDYDIRYYIYELLKALDYCHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAE-FYHPG 260
L ++ ++++L K + +CHS G++HRD+KP N+++D ++ L++ D GL+ F P
Sbjct: 115 PLEASLVQRFMFQLFKGVAHCHSHGVLHRDLKPQNLLLDKDKGILKIADLGLSRAFTVPL 174
Query: 261 KEYNVRVASRYFKGPELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKI 320
K Y + + +++ PE+L+ Y ++D+WS+GC+FA MI R++ F G + QL+ I
Sbjct: 175 KAYTHEIVTLWYRAPEVLLGSTHYSTAVDIWSVGCIFAEMI-RRQALFPGDSEFQQLLHI 233
Query: 321 AKVLGTDELSAYLNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLL 380
++LGT + P + AL H W PE +D + ++L
Sbjct: 234 FRLLGTPTEQQW---------PGVMALRDWHVYPKWEPQDLSRAVPSLSPEGIDLLTQML 284
Query: 381 RYDHQERPTAKEAMAHPYFNPVRSAE 406
+Y+ ER +AK A+ HPYF+ + ++
Sbjct: 285 KYNPAERISAKAALDHPYFDSLDKSQ 310
>At1g73690 cell division protein kinase
Length = 398
Score = 139 bits (351), Expect = 2e-33
Identities = 92/311 (29%), Positives = 167/311 (53%), Gaps = 41/311 (13%)
Query: 112 DDYEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKK-----IKREIKILQNLCGG 166
D Y +G+G Y VF+ + E IK ++ K+K+ REIK+L+ L
Sbjct: 9 DRYLKREVLGQGTYGVVFKATDTKNGETVAIKKIRLGKEKEGVNVTALREIKLLKEL-KH 67
Query: 167 PNIVKLLDIVRDQQSKTPSLIFEYVNNTDFKVLYPT----LSDYDIRYYIYELLKALDYC 222
P+I++L+D +++ ++FE++ TD + + LS D++ Y+ +LK L+YC
Sbjct: 68 PHIIELIDAFPHKENL--HIVFEFME-TDLEAVIRDRNLYLSPGDVKSYLQMILKGLEYC 124
Query: 223 HSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFY-HPGKEYNVRVASRYFKGPELLVDL 281
H + ++HRD+KP+N++I +L+L D+GLA + PG+++ +V +R+++ PELL
Sbjct: 125 HGKWVLHRDMKPNNLLIG-PNGQLKLADFGLARIFGSPGRKFTHQVFARWYRAPELLFGA 183
Query: 282 QDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDELSAYLN------- 334
+ YD ++D+W+ GC+FA ++ R+ PF G+ + DQL KI GT + + +
Sbjct: 184 KQYDGAVDVWAAGCIFAELLLRR-PFLQGNSDIDQLSKIFAAFGTPKADQWPDMICLPDY 242
Query: 335 -KYRIELDPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEA 393
+Y+ P+L +L+ S +A+D + K+ YD + R + ++A
Sbjct: 243 VEYQFVPAPSLRSLLPTVSE-----------------DALDLLSKMFTYDPKSRISIQQA 285
Query: 394 MAHPYFNPVRS 404
+ H YF S
Sbjct: 286 LKHRYFTSAPS 296
>At5g14640 protein kinase MSK-3 - like
Length = 410
Score = 139 bits (350), Expect = 3e-33
Identities = 94/303 (31%), Positives = 149/303 (49%), Gaps = 27/303 (8%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIKREIKILQNLCGGPNIVKLL 173
Y R VG+G + VF+ C++ + V I K ++ K+ K L PN+V L
Sbjct: 74 YMAERIVGQGSFGIVFQA-KCLETGETVA-IKKVLQDKRYKNRELQTMRLLDHPNVVSLK 131
Query: 174 DIVRDQQSKTP---SLIFEYVNNTDFKV------LYPTLSDYDIRYYIYELLKALDYCHS 224
K +L+ EYV T ++V + ++ Y Y++ +AL Y H
Sbjct: 132 HCFFSTTEKDELYLNLVLEYVPETVYRVSKHYSRANQRMPIIYVKLYTYQICRALAYIHG 191
Query: 225 Q-GIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQD 283
G+ HRD+KP N++++ +++L D+G A+ G+ + SRY++ PEL+ +
Sbjct: 192 GVGVCHRDIKPQNLLVNPHTHQVKLCDFGSAKVLVKGEPNISYICSRYYRAPELIFGATE 251
Query: 284 YDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAYLNKYRIEL 340
Y ++D+WS GC+ A ++ +P F G DQLV+I KVLGT +E+ Y
Sbjct: 252 YTTTIDIWSAGCVLAELLL-GQPLFPGESGVDQLVEIIKVLGTPTREEIKCMNPNYTEFK 310
Query: 341 DPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFN 400
P + A PW K + PEAVD V +LL+Y R TA EA+ HP+F+
Sbjct: 311 FPQIKA-------HPWHKIFHKRTP----PEAVDLVSRLLQYSPNLRSTAMEAIVHPFFD 359
Query: 401 PVR 403
+R
Sbjct: 360 ELR 362
>At3g54180 protein kinase cdc2 homolog B
Length = 309
Score = 139 bits (350), Expect = 3e-33
Identities = 97/328 (29%), Positives = 168/328 (50%), Gaps = 54/328 (16%)
Query: 112 DDYEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIK-----------REIKIL 160
+ YE + KVG G Y +V++ + EK K++ +KK +++ REI +L
Sbjct: 2 EKYEKLEKVGEGTYGKVYKAM-----EKGTGKLVA-LKKTRLEMDEEGIPPTALREISLL 55
Query: 161 QNLCGGPNIVKLLDIVRDQQSKTPS--------LIFEYVNNTDFKVLYPT---------L 203
Q L +V+LL + Q T S L+FEY++ TD K + L
Sbjct: 56 QMLSTSIYVVRLLCVEHVHQPSTKSQSTKSNLYLVFEYLD-TDLKKFIDSYRKGPNPKPL 114
Query: 204 SDYDIRYYIYELLKALDYCHSQGIMHRDVKPHNVMIDHEQRKLRLIDWGLAE-FYHPGKE 262
+ I+ +++L K + +CHS G++HRD+KP N+++ ++ L++ D GL F P K
Sbjct: 115 EPFLIQKLMFQLCKGVAHCHSHGVLHRDLKPQNLLLVKDKELLKIADLGLGRAFTVPLKS 174
Query: 263 YNVRVASRYFKGPELLVDLQDYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAK 322
Y + + +++ PE+L+ Y +D+WS+GC+FA M+ R++ F G + QL+ I +
Sbjct: 175 YTHEIVTLWYRAPEVLLGSTHYSTGVDMWSVGCIFAEMV-RRQALFPGDSEFQQLLHIFR 233
Query: 323 VLGTDELSAYLNKYRIELDPNLAALIGRHSRKPWAKFINVENQHLAV----PEAVDFVDK 378
+LGT + P ++ L H W ++ LAV P+ VD + K
Sbjct: 234 LLGTPTEQQW---------PGVSTLRDWHVYPKW----EPQDLTLAVPSLSPQGVDLLTK 280
Query: 379 LLRYDHQERPTAKEAMAHPYFNPVRSAE 406
+L+Y+ ER +AK A+ HPYF+ + ++
Sbjct: 281 MLKYNPAERISAKTALDHPYFDSLDKSQ 308
>At3g59790 mitogen-activated protein kinase-like protein
Length = 393
Score = 138 bits (347), Expect = 6e-33
Identities = 95/300 (31%), Positives = 155/300 (51%), Gaps = 32/300 (10%)
Query: 117 VRKVGRGKYSEVFEGVHCIDNEKCVIKILKPV-----KKKKIKREIKILQNLCGGPNIVK 171
+R +GRG V V NEK IK + V + K+ REIK+L++ NIV
Sbjct: 63 IRPIGRGACGIVCSAVDSETNEKVAIKKITQVFDNTIEAKRTLREIKLLRHF-DHENIVA 121
Query: 172 LLDIVRDQQSKTPSLIFEYVNNTDFKVLYPTLSDYDIR-----YYIYELLKALDYCHSQG 226
+ D++ Q + ++ +F + SD ++ Y++Y++L+ L Y HS
Sbjct: 122 IRDVILPPQRDSFEDVYIVNELMEFDLYRTLKSDQELTKDHGMYFMYQILRGLKYIHSAN 181
Query: 227 IMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQDYDY 286
++HRD+KP N+++ Q L++ D+GLA V +R+++ PELL+ DY
Sbjct: 182 VLHRDLKPSNLLLS-TQCDLKICDFGLARATPESNLMTEYVVTRWYRAPELLLGSSDYTA 240
Query: 287 SLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT------DELSAYLNKYRIEL 340
++D+WS+GC+F I +EP F G D +QL + +++GT LS Y +Y +L
Sbjct: 241 AIDVWSVGCIFM-EIMNREPLFPGKDQVNQLRLLLELIGTPSEEELGSLSEYAKRYIRQL 299
Query: 341 DPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFN 400
P L + KF NV P A+D V+K+L +D ++R + KEA+AHPY +
Sbjct: 300 -PTLP------RQSFTEKFPNVP------PLAIDLVEKMLTFDPKQRISVKEALAHPYLS 346
>At4g18710 shaggy-like protein kinase etha (EC 2.7.1.-)
Length = 380
Score = 136 bits (342), Expect = 2e-32
Identities = 94/303 (31%), Positives = 153/303 (50%), Gaps = 27/303 (8%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIKREIKILQNLCGGPNIVKLL 173
Y R VG G + VF+ C++ + V I K ++ ++ K L + PN+V L
Sbjct: 40 YMAERVVGTGSFGIVFQA-KCLETGETVA-IKKVLQDRRYKNRELQLMRVMDHPNVVCLK 97
Query: 174 DIVRDQQSKTP---SLIFEYVNNTDFKVLYPTLSDYD------IRYYIYELLKALDYCHS 224
SK +L+ EYV + ++VL S ++ Y+Y++ + L Y H+
Sbjct: 98 HCFFSTTSKDELFLNLVMEYVPESLYRVLKHYSSANQRMPLVYVKLYMYQIFRGLAYIHN 157
Query: 225 -QGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQD 283
G+ HRD+KP N+++D ++++ D+G A+ G+ + SR+++ PEL+ +
Sbjct: 158 VAGVCHRDLKPQNLLVDPLTHQVKICDFGSAKQLVKGEANISYICSRFYRAPELIFGATE 217
Query: 284 YDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAYLNKYRIEL 340
Y S+D+WS GC+ A ++ +P F G + DQLV+I KVLGT +E+ Y
Sbjct: 218 YTTSIDIWSAGCVLAELLL-GQPLFPGENAVDQLVEIIKVLGTPTREEIRCMNPHYTDFR 276
Query: 341 DPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFN 400
P + A PW K + PEA+DF +LL+Y R TA EA AHP+F+
Sbjct: 277 FPQIKA-------HPWHKIFHKR----MPPEAIDFASRLLQYSPSLRCTALEACAHPFFD 325
Query: 401 PVR 403
+R
Sbjct: 326 ELR 328
>At5g50860 cyclin-dependent protein kinase-like
Length = 580
Score = 134 bits (337), Expect = 9e-32
Identities = 94/297 (31%), Positives = 161/297 (53%), Gaps = 23/297 (7%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKI-----LKPVKKKKIKREIKILQNLCGGPN 168
YE + K+G+G YS V++ + + +K L+ K + REI +L+ L PN
Sbjct: 114 YEKLEKIGQGTYSNVYKAKDLLSGKIVALKKVRFDNLEAESVKFMAREILVLRRL-NHPN 172
Query: 169 IVKLLDIVRDQQSKTPSLIFEYVNNTDFKVLYPTLS-DYD---IRYYIYELLKALDYCHS 224
++KL +V + S + L+FEY+ + D L T +D ++ ++ +LL L++CHS
Sbjct: 173 VIKLQGLVTSRVSCSLYLVFEYMEH-DLSGLAATQGLKFDLPQVKCFMKQLLSGLEHCHS 231
Query: 225 QGIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNV--RVASRYFKGPELLVDLQ 282
+G++HRD+K N++ID++ L++ D+GLA FY P ++ + RV + +++ PELL+
Sbjct: 232 RGVLHRDIKGSNLLIDND-GILKIADFGLATFYDPKQKQTMTSRVVTLWYRPPELLLGAT 290
Query: 283 DYDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGTDELSAYLNKYRIELDP 342
Y +DLWS GC+ A ++ K P G +QL KI K+ G+ +Y KYR+ P
Sbjct: 291 SYGTGVDLWSAGCIMAELLAGK-PVMPGRTEVEQLHKIFKLCGSPS-DSYWKKYRL---P 345
Query: 343 NLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYF 399
N +H P+ + + E + P +V V+ LL D +R T+ A+ +F
Sbjct: 346 NATLFKPQH---PYKRCV-AEAFNGFTPSSVHLVETLLTIDPADRGTSTSALNSEFF 398
>At5g26751 shaggy-like kinase alpha
Length = 405
Score = 134 bits (337), Expect = 9e-32
Identities = 92/303 (30%), Positives = 148/303 (48%), Gaps = 27/303 (8%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIKREIKILQNLCGGPNIVKLL 173
Y R VG G + VF+ C++ + V I K ++ ++ K L PN+V L
Sbjct: 69 YMAERVVGHGSFGVVFQA-KCLETGETVA-IKKVLQDRRYKNRELQTMRLLDHPNVVSLK 126
Query: 174 DIVRDQQSKTP---SLIFEYVNNTDFKVL--YPTLSD----YDIRYYIYELLKALDYCHS 224
K +L+ EYV T +V+ Y L+ ++ Y Y++ +AL Y H
Sbjct: 127 HCFFSTTEKDELYLNLVLEYVPETVHRVIKHYNKLNQRMPLIYVKLYTYQIFRALSYIHR 186
Query: 225 Q-GIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQD 283
G+ HRD+KP N++++ +++L D+G A+ G+ + SRY++ PEL+ +
Sbjct: 187 CIGVCHRDIKPQNLLVNPHTHQVKLCDFGSAKVLVKGEPNISYICSRYYRAPELIFGATE 246
Query: 284 YDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAYLNKYRIEL 340
Y ++D+WS GC+ A ++ +P F G DQLV+I KVLGT +E+ Y
Sbjct: 247 YTTAIDVWSAGCVLAELLL-GQPLFPGESGVDQLVEIIKVLGTPTREEIKCMNPNYTEFK 305
Query: 341 DPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFN 400
P + A PW K + PEAVD V +LL+Y R A + + HP+F+
Sbjct: 306 FPQIKA-------HPWHKIFHKR----MPPEAVDLVSRLLQYSPNLRSAALDTLVHPFFD 354
Query: 401 PVR 403
+R
Sbjct: 355 ELR 357
>At3g05840 shaggy related protein kinase, ASK-GAMMA
Length = 409
Score = 132 bits (333), Expect = 3e-31
Identities = 91/303 (30%), Positives = 149/303 (49%), Gaps = 27/303 (8%)
Query: 114 YEVVRKVGRGKYSEVFEGVHCIDNEKCVIKILKPVKKKKIKREIKILQNLCGGPNIVKLL 173
Y R VG G + VF+ C++ + V I K ++ ++ K L PN+V L
Sbjct: 73 YMAERVVGHGSFGVVFQA-KCLETGETVA-IKKVLQDRRYKNRELQTMRLLDHPNVVSLK 130
Query: 174 DIVRDQQSKTP---SLIFEYVNNTDFKVL--YPTLSD----YDIRYYIYELLKALDYCHS 224
K +L+ EYV T +V+ Y L+ ++ Y Y++ ++L Y H
Sbjct: 131 HCFFSTTEKDELYLNLVLEYVPETVHRVIKHYNKLNQRMPLVYVKLYTYQIFRSLSYIHR 190
Query: 225 Q-GIMHRDVKPHNVMIDHEQRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQD 283
G+ HRD+KP N++++ +++L D+G A+ G+ + SRY++ PEL+ +
Sbjct: 191 CIGVCHRDIKPQNLLVNPHTHQVKLCDFGSAKVLVKGEPNISYICSRYYRAPELIFGATE 250
Query: 284 YDYSLDLWSLGCMFAGMIFRKEPFFYGHDNYDQLVKIAKVLGT---DELSAYLNKYRIEL 340
Y ++D+WS GC+ A ++ +P F G DQLV+I KVLGT +E+ Y
Sbjct: 251 YTTAIDVWSAGCVLAELLL-GQPLFPGESGVDQLVEIIKVLGTPTREEIKCMNPNYTEFK 309
Query: 341 DPNLAALIGRHSRKPWAKFINVENQHLAVPEAVDFVDKLLRYDHQERPTAKEAMAHPYFN 400
P + A PW K + PEAVD V +LL+Y R A +++ HP+F+
Sbjct: 310 FPQIKA-------HPWHKIFHKR----MPPEAVDLVSRLLQYSPNLRCAALDSLVHPFFD 358
Query: 401 PVR 403
+R
Sbjct: 359 ELR 361
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,883,131
Number of Sequences: 26719
Number of extensions: 463408
Number of successful extensions: 3819
Number of sequences better than 10.0: 891
Number of HSP's better than 10.0 without gapping: 366
Number of HSP's successfully gapped in prelim test: 525
Number of HSP's that attempted gapping in prelim test: 2448
Number of HSP's gapped (non-prelim): 1256
length of query: 413
length of database: 11,318,596
effective HSP length: 102
effective length of query: 311
effective length of database: 8,593,258
effective search space: 2672503238
effective search space used: 2672503238
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0330.12


