

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
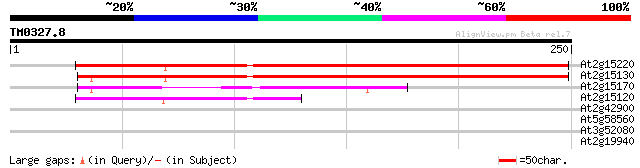
Query= TM0327.8
(250 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g15220 hypothetical protein 213 8e-56
At2g15130 unknown protein 187 5e-48
At2g15170 hypothetical protein 74 7e-14
At2g15120 unknown protein 52 2e-07
At2g42900 unknown protein 36 0.017
At5g58560 unknown protein 32 0.25
At3g52080 putative protein 28 4.6
At2g19940 putative N-acetyl-gamma-glutamyl-phosphate reductase 28 4.6
>At2g15220 hypothetical protein
Length = 225
Score = 213 bits (542), Expect = 8e-56
Identities = 103/221 (46%), Positives = 144/221 (64%), Gaps = 3/221 (1%)
Query: 30 LMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEIG-IPFTQQTIQLASQFILQTFK 88
+ F++ + S ++AV Y V + + + GG RF EIG I + QT++ A+ F+ + F+
Sbjct: 6 IFFVISLMLVVSLVNAVDYSVVDNSGDSTGGRRFRGEIGGISYGTQTLRSATDFVWRLFQ 65
Query: 89 QIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEAT 148
Q + K++ +++ + NG+ A S N I + Y+ G SGDV+ E G++YHE
Sbjct: 66 QTNPSDRKSVTKITLFME--NGDGVAYNSANEIHFNVGYLAGVSGDVKREFTGVVYHEVV 123
Query: 149 HIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGL 208
H WQW G AP GL EG+AD++RLKAG+AP HWV G G WD+GY VTA FLDYCNGL
Sbjct: 124 HSWQWNGAGRAPGGLIEGIADYVRLKAGYAPSHWVGPGRGDRWDQGYDVTARFLDYCNGL 183
Query: 209 RDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFG 249
R+GFVA LN M++ YSD FFV LLGK V+QLW +YK+ +G
Sbjct: 184 RNGFVAELNKKMRNGYSDGFFVDLLGKDVNQLWREYKAKYG 224
>At2g15130 unknown protein
Length = 225
Score = 187 bits (475), Expect = 5e-48
Identities = 95/221 (42%), Positives = 138/221 (61%), Gaps = 4/221 (1%)
Query: 31 MFLVI-IASSGSPIHAVQYQVTNRATGTPGGTRFENEIG-IPFTQQTIQLASQFILQTFK 88
+FLVI + + S + AV + V + +PGG RF NEIG + + +Q+++ A+ F + F+
Sbjct: 6 IFLVISLMLAVSLVSAVDFSVVDNTGDSPGGRRFRNEIGGVSYGEQSLRDATDFTWRLFQ 65
Query: 89 QIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEAT 148
Q + K++ +++ + N N A +S + I ++ + G VR G++YHE
Sbjct: 66 QTNPSDRKDVTKITLFME--NSNGIAYSSQDEIHYNAGSLVDDKGYVRRGFTGVVYHEVV 123
Query: 149 HIWQWFGNKEAPRGLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGL 208
H WQW G AP GL EG+AD++RLKAG+ HWV G G WD+GY VTA FL+YCN L
Sbjct: 124 HSWQWNGAGRAPGGLIEGIADYVRLKAGYVASHWVRPGGGDRWDQGYDVTARFLEYCNDL 183
Query: 209 RDGFVAMLNAMMKDHYSDDFFVKLLGKSVDQLWSDYKSVFG 249
R+GFVA LN M+ Y+D FFV LLGK V+QLW +YK+ +G
Sbjct: 184 RNGFVAELNKKMRSDYNDGFFVDLLGKDVNQLWREYKANYG 224
>At2g15170 hypothetical protein
Length = 134
Score = 73.9 bits (180), Expect = 7e-14
Identities = 50/149 (33%), Positives = 72/149 (47%), Gaps = 31/149 (20%)
Query: 31 MFLVI-IASSGSPIHAVQYQVTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTFKQ 89
+FLVI + + S ++AV + V + +PGG +F +EIG
Sbjct: 6 IFLVICLMLAVSMVNAVDFFVVDNTGDSPGGRKFRDEIG--------------------- 44
Query: 90 IDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEATH 149
G + S IVTA T + ++ + G SGDVR G++YHE H
Sbjct: 45 -----GVSYGKQSRIVTAW---PTTRTWERRYIFNARSLAGNSGDVRVGFTGVIYHEVAH 96
Query: 150 IWQWFGNKE-APRGLTEGVADFMRLKAGF 177
WQWFG + P GL EG+AD++RLKAG+
Sbjct: 97 SWQWFGAADRVPGGLIEGIADYVRLKAGY 125
>At2g15120 unknown protein
Length = 109
Score = 52.4 bits (124), Expect = 2e-07
Identities = 29/102 (28%), Positives = 55/102 (53%), Gaps = 3/102 (2%)
Query: 30 LMFLVIIASSGSPIHAVQYQVTNRATGTPGGTRFENEI-GIPFTQQTIQLASQFILQTFK 88
+ F++ + S ++AV Y V + + + GG RF EI GI + QT++ A+ F+ + F+
Sbjct: 6 IFFVISLMLVVSLVNAVDYSVVDNSGDSTGGRRFRGEIGGISYGTQTLRSATDFVWRLFQ 65
Query: 89 QIDQFGGKNIEHVSVIVTALNGNSAAITSDNNIFLDSNYIEG 130
Q + K++ +++ + NG+ A S N I + Y+ G
Sbjct: 66 QTNPSDRKSVTKITLFME--NGDGVAYNSGNEIHFNVGYLAG 105
>At2g42900 unknown protein
Length = 276
Score = 36.2 bits (82), Expect = 0.017
Identities = 41/185 (22%), Positives = 67/185 (36%), Gaps = 9/185 (4%)
Query: 50 VTNRATGTPGGTRFENEIGIPFTQQTIQLASQFILQTF--KQIDQFGGKNIEHVSVIVTA 107
+ N A +P G RF T I L + F ++ F + + K + HV+V
Sbjct: 59 IINDAKDSPSGKRFALFFESDDTAVRILLDASFFVERFLYEGVPHRLRKPVNHVTVQFCG 118
Query: 108 LNGNSAAITSDNNIFLDSNYIEGFSGDVR------NEVIGILYHEATHIWQWFGNKEAPR 161
+ + S + Y+ S + N V L IW W A
Sbjct: 119 NSSDRVDRFSVTSGASHGEYVIRLSPSLTERKKFSNAVESALRRSMVRIWLWGDESGASP 178
Query: 162 GLTEGVADFMRLKAGFAPPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDGFVAMLNAMMK 221
L G+ +++ +++ H+ G E V LDYC +GF+ LN M+
Sbjct: 179 ELVAGMVEYLAVESR-KRRHFEKFGGNWKDKEKSVYVVSLLDYCERRSEGFIRRLNHGMR 237
Query: 222 DHYSD 226
+ D
Sbjct: 238 LRWDD 242
>At5g58560 unknown protein
Length = 307
Score = 32.3 bits (72), Expect = 0.25
Identities = 29/121 (23%), Positives = 48/121 (38%), Gaps = 12/121 (9%)
Query: 3 LKLPICCILSSCAALIYHWTKMSPIIYLMFLVIIASSGSPIHAVQYQVTNRATGTP---- 58
LK P+ +LS +A IY+W K SPI + + A G + R GT
Sbjct: 175 LKGPLYYVLSITSACIYYW-KSSPIAIAVICNLCAGDG------MADIVGRRFGTEKLPY 227
Query: 59 -GGTRFENEIGIPFTQQTIQLASQFILQTFKQIDQFGGKNIEHVSVIVTALNGNSAAITS 117
F IG+ +A + +F I+ GG + + + + + S I++
Sbjct: 228 NKNKSFAGSIGMATAGFLASVAYMYYFASFGYIEDSGGMILRFLVISIASALVESLPIST 287
Query: 118 D 118
D
Sbjct: 288 D 288
>At3g52080 putative protein
Length = 732
Score = 28.1 bits (61), Expect = 4.6
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query: 10 ILSSCAALIYHWTKMSPIIYLMFLVIIASS-GSPI 43
+L+ H+TK SP I+ + L ++ASS GSPI
Sbjct: 124 VLAFVTTPFLHYTKTSPYIFSLALSLMASSTGSPI 158
>At2g19940 putative N-acetyl-gamma-glutamyl-phosphate reductase
Length = 359
Score = 28.1 bits (61), Expect = 4.6
Identities = 33/141 (23%), Positives = 59/141 (41%), Gaps = 29/141 (20%)
Query: 98 IEHVSVIVTALNGNSAAITSDNNIFLDSNYIEGFSGDVRNEVIGILYHEATHIWQWFGNK 157
I+H ++I+ A +G S A L S EG S G+ H H+ +
Sbjct: 181 IKHENIIIDAKSGVSGAGRGAKEANLYSEIAEGISS------YGVTRHR--HVPE----- 227
Query: 158 EAPRGLTEGVADFMRLKAGFA-PPHWVPRGTGSGWDEGYVVTAYFLDYCNGLRDGFVAML 216
+ +G++D + K + PH +P G + + +++ G+R L
Sbjct: 228 -----IEQGLSDVAQSKVTVSFTPHLMPMIRG-------MQSTIYVEMAPGVR---TEDL 272
Query: 217 NAMMKDHYSDDFFVKLLGKSV 237
+ +K Y D+ FVK+L + V
Sbjct: 273 HQQLKTSYEDEEFVKVLDEGV 293
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,795,801
Number of Sequences: 26719
Number of extensions: 247628
Number of successful extensions: 503
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 492
Number of HSP's gapped (non-prelim): 10
length of query: 250
length of database: 11,318,596
effective HSP length: 97
effective length of query: 153
effective length of database: 8,726,853
effective search space: 1335208509
effective search space used: 1335208509
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0327.8


