

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0327.19
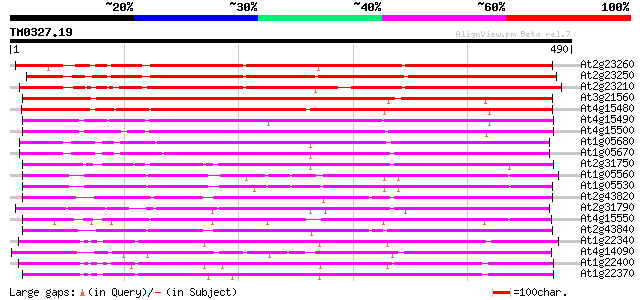
(490 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g23260 putative glucosyltransferase 394 e-110
At2g23250 putative glucosyltransferase 384 e-107
At2g23210 putative glucosyltransferase 373 e-103
At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferas... 364 e-101
At4g15480 indole-3-acetate beta-glucosyltransferase like protein 359 2e-99
At4g15490 indole-3-acetate beta-glucosyltransferase like protein 352 2e-97
At4g15500 indole-3-acetate beta-glucosyltransferase like protein 333 1e-91
At1g05680 indole-3-acetate beta-glucosyltransferase like protein 307 8e-84
At1g05670 UDP glycosyltransferase UGT74E1 306 2e-83
At2g31750 putative glucosyltransferase 296 1e-80
At1g05560 UDP-Glucose Transferase (UGT75B2) 290 9e-79
At1g05530 hypothetical protein 290 9e-79
At2g43820 putative glucosyltransferase 290 1e-78
At2g31790 putative glucosyltransferase 290 2e-78
At4g15550 UDP-glucose:indole-3-acetate beta-D-glucosyltransferas... 286 2e-77
At2g43840 putative glucosyltransferase 285 5e-77
At1g22340 hypothetical protein 268 7e-72
At4g14090 glucosyltransferase like protein 263 2e-70
At1g22400 Putative UDP-glucose glucosyltransferase 254 8e-68
At1g22370 putative UDP-glucose glucosyltransferase 254 1e-67
>At2g23260 putative glucosyltransferase
Length = 456
Score = 394 bits (1013), Expect = e-110
Identities = 210/476 (44%), Positives = 301/476 (63%), Gaps = 33/476 (6%)
Query: 6 SRAEELHVMLVAFSAQGHINPLLRLGK--SLLARGLQVTLATTELVYHRVFKSSSTTAAT 63
S +E HV++V QGHINP+L+L K SL ++ L + LAT E S+ +
Sbjct: 4 SEGQETHVLMVTLPFQGHINPMLKLAKHLSLSSKNLHINLATIE---------SARDLLS 54
Query: 64 TDTIPTNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNS 123
T P + ++FFSDGL P + P+ + + +G +NLS +I+
Sbjct: 55 TVEKPRY----PVDLVFFSDGL---PKEDPKAPETLLKSLNKVGAMNLSKIIE------E 101
Query: 124 KKLACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNV 183
K+ +CII++PF PWV VAA I CA LWIQ C Y++YY +Y TN FP LE+ V
Sbjct: 102 KRYSCIISSPFTPWVPAVAASHNISCAILWIQACGAYSVYYRYYMKTNSFPDLEDLNQTV 161
Query: 184 QLPGLPLLKPQDLPSFILPTNPFGA-LSKVLADMLKDMKKLKWVLANSFYELEKEVIDSM 242
+LP LPLL+ +DLPSF+LP+ GA ++A+ ++ +KWVL NSFYELE E+I+SM
Sbjct: 162 ELPALPLLEVRDLPSFMLPSG--GAHFYNLMAEFADCLRYVKWVLVNSFYELESEIIESM 219
Query: 243 AETYPVIPVGPLLPLSLLGVDENEDV---GIEMWKPQDSCLEWLNDQPPSSVIYISFGSL 299
A+ PVIP+GPL+ LLG E E + ++ K D C+EWL+ Q SSV+YISFGS+
Sbjct: 220 ADLKPVIPIGPLVSPFLLGDGEEETLDGKNLDFCKSDDCCMEWLDKQARSSVVYISFGSM 279
Query: 300 IVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKV 359
+ ++E+IA ALKN FLWVI+ ++ ++ + Q +E +G+V+ W PQ K+
Sbjct: 280 LETLENQVETIAKALKNRGLPFLWVIRPKEKAQNVAVLQEMVKE--GQGVVLEWSPQEKI 337
Query: 360 LVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDS-D 418
L H AI+CF+THCGWNS +E + AG P++AYP WTDQP +A+L+ DVF +G+R+ DS D
Sbjct: 338 LSHEAISCFVTHCGWNSTMETVVAGVPVVAYPSWTDQPIDARLLVDVFGIGVRMRNDSVD 397
Query: 419 GFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
G ++ E+ER IE + GP + +++R AAELKR AR A+A GGSS RN+ F+ +I
Sbjct: 398 GELKVEEVERCIEAVTEGPAAVDIRRRAAELKRVARLALAPGGSSTRNLDLFISDI 453
>At2g23250 putative glucosyltransferase
Length = 438
Score = 384 bits (985), Expect = e-107
Identities = 202/465 (43%), Positives = 293/465 (62%), Gaps = 29/465 (6%)
Query: 15 LVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNFTTN 74
+VA + QGH+NP+L+ K L L TLATTE ++ ++T D
Sbjct: 1 MVALAFQGHLNPMLKFAKHLARTNLHFTLATTE-------QARDLLSSTADE-----PHR 48
Query: 75 GIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIINNPF 134
+ + FFSDGL P PD ++ G NLS +I+ K+ CII+ PF
Sbjct: 49 PVDLAFFSDGL---PKDDPRDPDTLAKSLKKDGAKNLSKIIE------EKRFDCIISVPF 99
Query: 135 VPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPLLKPQ 194
PWV VAA IPCA LWIQ C +++YY +Y TN FP LE+ V+LP LPLL+ +
Sbjct: 100 TPWVPAVAAAHNIPCAILWIQACGAFSVYYRYYMKTNPFPDLEDLNQTVELPALPLLEVR 159
Query: 195 DLPSFILPTNPFGA-LSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIPVGP 253
DLPS +LP+ GA ++ ++A+ +K +KWVL NSFYELE E+I+SM++ P+IP+GP
Sbjct: 160 DLPSLMLPSQ--GANVNTLMAEFADCLKDVKWVLVNSFYELESEIIESMSDLKPIIPIGP 217
Query: 254 LLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESIATA 313
L+ LLG DE + ++MWK D C+EWL+ Q SSV+YISFGS++ ++E+IATA
Sbjct: 218 LVSPFLLGNDEEKT--LDMWKVDDYCMEWLDKQARSSVVYISFGSILKSLENQVETIATA 275
Query: 314 LKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIACFLTHCG 373
LKN FLWVI+ ++ +++ + Q +E K G+V W Q K+L H AI+CF+THCG
Sbjct: 276 LKNRGVPFLWVIRPKEKGENVQVLQEMVKEGK--GVVTEWGQQEKILSHMAISCFITHCG 333
Query: 374 WNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDS-DGFVETGELERAIEE 432
WNS +E + G P++AYP W DQP +A+L+ DVF +G+R++ D+ DG ++ E+ER IE
Sbjct: 334 WNSTIETVVTGVPVVAYPTWIDQPLDARLLVDVFGIGVRMKNDAIDGELKVAEVERCIEA 393
Query: 433 IVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEILAT 477
+ GP + +++R A ELK AAR A++ GGSS +N+ +F+ +I T
Sbjct: 394 VTEGPAAADMRRRATELKHAARSAMSPGGSSAQNLDSFISDIPIT 438
>At2g23210 putative glucosyltransferase
Length = 453
Score = 373 bits (957), Expect = e-103
Identities = 200/479 (41%), Positives = 300/479 (61%), Gaps = 41/479 (8%)
Query: 9 EELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIP 68
+E HV++VA QGH+NP+L+ K L L TLAT E S+ ++TD P
Sbjct: 7 QETHVLMVALPFQGHLNPMLKFAKHLARTNLHFTLATIE--------SARDLLSSTDE-P 57
Query: 69 TNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLAC 128
+ + ++FFSDGL P D P D+ L E+L + + K I K+ C
Sbjct: 58 HSL----VDLVFFSDGL-PKDD-----PRDHEPLTESLRKVGANNFSK---IIEGKRFDC 104
Query: 129 IINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGL 188
II+ PF PWV VAA IPCA LWI+ CA +++YY +Y TN FP LE+P V+LPGL
Sbjct: 105 IISVPFTPWVPAVAAAHNIPCAILWIEACAGFSVYYRYYMKTNSFPDLEDPNQKVELPGL 164
Query: 189 PLLKPQDLPSFILPTNPFGAL-SKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYP 247
P L+ +DLP+ +LP++ GA+ + ++A+ ++ +K +KWVLANSFYELE +I+SM + P
Sbjct: 165 PFLEVRDLPTLMLPSH--GAIFNTLMAEFVECLKDVKWVLANSFYELESVIIESMFDLKP 222
Query: 248 VIPVGPLLPLSLLGVDEN---EDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSA 304
+IP+GPL+ LLG DE+ + ++MWK D C+EWL+ Q S++ S
Sbjct: 223 IIPIGPLVSPFLLGADEDKILDGKSLDMWKADDYCMEWLDKQV----------SILKSSE 272
Query: 305 KKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPA 364
++E+IATALKN FLWVI+ ++ +++ + + EE +G+V+ W Q K+L H A
Sbjct: 273 NQVETIATALKNRGVPFLWVIRPKEKAENVDVLEDMVEE--GQGVVIEWGQQEKILCHMA 330
Query: 365 IACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQD-SDGFVET 423
I+CF+THCGWNS +E + +G PM+AYP W DQP +A+L+ DVF +G+R++ D DG ++
Sbjct: 331 ISCFVTHCGWNSTIETVVSGVPMVAYPTWFDQPLDARLLVDVFGIGVRMKNDVVDGELKV 390
Query: 424 GELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEILATRLDVM 482
E+ER I+ + G + +++R AAELK+A R A+A GGS RN+ F+++I + DV+
Sbjct: 391 AEVERCIDAVTKGTDAADMRRRAAELKQATRSAMAPGGSLARNLDLFINDIKIVKNDVI 449
>At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferase,
putative
Length = 496
Score = 364 bits (934), Expect = e-101
Identities = 188/469 (40%), Positives = 286/469 (60%), Gaps = 14/469 (2%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
HVMLV+F QGH+NPLLRLGK L ++GL +T TTE ++ S+ + +
Sbjct: 12 HVMLVSFPGQGHVNPLLRLGKLLASKGLLITFVTTESWGKKMRISNKIQDRVLKPVGKGY 71
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIIN 131
+ FF DGL + + ++ +E +G + L+K + + + C+IN
Sbjct: 72 ----LRYDFFDDGLPEDDEASRTNLTILRPHLELVGKREIKNLVKRYKEVTKQPVTCLIN 127
Query: 132 NPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPLL 191
NPFV WV DVA + +IPCA LW+Q CA A YY++++N FPT PE++VQ+ G+PLL
Sbjct: 128 NPFVSWVCDVAEDLQIPCAVLWVQSCACLAAYYYYHHNLVDFPTKTEPEIDVQISGMPLL 187
Query: 192 KPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIPV 251
K ++PSFI P++P AL +V+ D +K + K + ++F LEK++ID M+ +
Sbjct: 188 KHDEIPSFIHPSSPHSALREVIIDQIKRLHKTFSIFIDTFNSLEKDIIDHMSTLSLPGVI 247
Query: 252 GPLLPLSLLGVDENED-VGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESI 310
PL PL + D V + + +P D C+EWL+ QP SSV+YISFG++ L ++++ I
Sbjct: 248 RPLGPLYKMAKTVAYDVVKVNISEPTDPCMEWLDSQPVSSVVYISFGTVAYLKQEQIDEI 307
Query: 311 ATALKNSNCKFLWVIKKQD---GKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIAC 367
A + N++ FLWVI++Q+ K+ LP EE K +G +V WC Q KVL HP++AC
Sbjct: 308 AYGVLNADVTFLWVIRQQELGFNKEKHVLP----EEVKGKGKIVEWCSQEKVLSHPSVAC 363
Query: 368 FLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLE--QDSDGFVETGE 425
F+THCGWNS +EA+++G P + +PQW DQ T+A + DV++ G+RL + + V E
Sbjct: 364 FVTHCGWNSTMEAVSSGVPTVCFPQWGDQVTDAVYMIDVWKTGVRLSRGEAEERLVPREE 423
Query: 426 LERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
+ + E+ G K+ ELK+NA + K A AVA GGSSDRN++ FV+++
Sbjct: 424 VAERLREVTKGEKAIELKKNALKWKEEAEAAVARGGSSDRNLEKFVEKL 472
>At4g15480 indole-3-acetate beta-glucosyltransferase like protein
Length = 490
Score = 359 bits (921), Expect = 2e-99
Identities = 193/470 (41%), Positives = 293/470 (62%), Gaps = 15/470 (3%)
Query: 11 LHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTN 70
+HVMLV+F QGH+NPLLRLGK + ++GL VT TTEL ++ +++ + +
Sbjct: 18 IHVMLVSFQGQGHVNPLLRLGKLIASKGLLVTFVTTELWGKKMRQANKIVDGELKPVGSG 77
Query: 71 FTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
I FF + + D+ + Y+A +E++G +S L++ + N + ++C+I
Sbjct: 78 ----SIRFEFFDEEWAEDDDR-RADFSLYIAHLESVGIREVSKLVRRYEEAN-EPVSCLI 131
Query: 131 NNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPL 190
NNPF+PWV VA EF IPCA LW+Q CA ++ YYH+ + + FPT PEL+V+LP +P+
Sbjct: 132 NNPFIPWVCHVAEEFNIPCAVLWVQSCACFSAYYHYQDGSVSFPTETEPELDVKLPCVPV 191
Query: 191 LKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIP 250
LK ++PSF+ P++ F + + K++ K VL +SF LE+EVID M+ PV
Sbjct: 192 LKNDEIPSFLHPSSRFTGFRQAILGQFKNLSKSFCVLIDSFDSLEQEVIDYMSSLCPVKT 251
Query: 251 VGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESI 310
VGPL ++ DV ++ K D CLEWL+ +P SSV+YISFG++ L +++E I
Sbjct: 252 VGPLFKVAR---TVTSDVSGDICKSTDKCLEWLDSRPKSSVVYISFGTVAYLKQEQIEEI 308
Query: 311 ATALKNSNCKFLWVIK--KQDGK-DSLPLPQGFKEET-KNRGMVVPWCPQTKVLVHPAIA 366
A + S FLWVI+ D K ++ LPQ KE + K +GM+V WCPQ +VL HP++A
Sbjct: 309 AHGVLKSGLSFLWVIRPPPHDLKVETHVLPQELKESSAKGKGMIVDWCPQEQVLSHPSVA 368
Query: 367 CFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDS--DGFVETG 424
CF+THCGWNS +E++++G P++ PQW DQ T+A + DVF+ G+RL + + + V
Sbjct: 369 CFVTHCGWNSTMESLSSGVPVVCCPQWGDQVTDAVYLIDVFKTGVRLGRGATEERVVPRE 428
Query: 425 ELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
E+ + E G K+EEL++NA + K A AVA GGSSD+N + FV+++
Sbjct: 429 EVAEKLLEATVGEKAEELRKNALKWKAEAEAAVAPGGSSDKNFREFVEKL 478
>At4g15490 indole-3-acetate beta-glucosyltransferase like protein
Length = 479
Score = 352 bits (904), Expect = 2e-97
Identities = 182/468 (38%), Positives = 282/468 (59%), Gaps = 12/468 (2%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
HVMLV+F QGH+NPLLRLGK + ++GL VT TTE + + + ++ D +
Sbjct: 8 HVMLVSFPGQGHVNPLLRLGKLIASKGLLVTFVTTEKPWGKKMRQANKIQ---DGVLKPV 64
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIIN 131
I FFSDG + D+ + D + +E +G + L+K + N + + C+IN
Sbjct: 65 GLGFIRFEFFSDGF-ADDDEKRFDFDAFRPHLEAVGKQEIKNLVKRY---NKEPVTCLIN 120
Query: 132 NPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPLL 191
N FVPWV DVA E IP A LW+Q CA YY++++ +FPT P+++V++P LPLL
Sbjct: 121 NAFVPWVCDVAEELHIPSAVLWVQSCACLTAYYYYHHRLVKFPTKTEPDISVEIPCLPLL 180
Query: 192 KPQDLPSFILPTNPFGALSKVLADMLKDMKKLK--WVLANSFYELEKEVIDSMAETYPVI 249
K ++PSF+ P++P+ A ++ D LK + K ++ ++F ELEK+++D M++ P
Sbjct: 181 KHDEIPSFLHPSSPYTAFGDIILDQLKRFENHKSFYLFIDTFRELEKDIMDHMSQLCPQA 240
Query: 250 PVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLES 309
+ P+ PL + + DV ++ +P C+EWL+ + PSSV+YISFG++ L +++E
Sbjct: 241 IISPVGPLFKMAQTLSSDVKGDISEPASDCMEWLDSREPSSVVYISFGTIANLKQEQMEE 300
Query: 310 IATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIACFL 369
IA + +S LWV+ + + + P E + +G +V WCPQ +VL HPAIACFL
Sbjct: 301 IAHGVLSSGLSVLWVV-RPPMEGTFVEPHVLPRELEEKGKIVEWCPQERVLAHPAIACFL 359
Query: 370 THCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDS--DGFVETGELE 427
+HCGWNS +EA+ AG P++ +PQW DQ T+A ++DVF+ G+RL + + + V +
Sbjct: 360 SHCGWNSTMEALTAGVPVVCFPQWGDQVTDAVYLADVFKTGVRLGRGAAEEMIVSREVVA 419
Query: 428 RAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEIL 475
+ E G K+ EL+ NA K A AVADGGSSD N + FVD+++
Sbjct: 420 EKLLEATVGEKAVELRENARRWKAEAEAAVADGGSSDMNFKEFVDKLV 467
>At4g15500 indole-3-acetate beta-glucosyltransferase like protein
Length = 475
Score = 333 bits (855), Expect = 1e-91
Identities = 181/468 (38%), Positives = 267/468 (56%), Gaps = 17/468 (3%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
HVMLV+F QGHI+PLLRLGK + ++GL VT TTE + + ++ D +
Sbjct: 9 HVMLVSFPGQGHISPLLRLGKIIASKGLIVTFVTTEEPLGKKMRQANNIQ---DGVLKPV 65
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIIN 131
+ FF DG D L +E G + L+K + + + C+IN
Sbjct: 66 GLGFLRFEFFEDGFVYKEDFDLLQKS-----LEVSGKREIKNLVKKY---EKQPVRCLIN 117
Query: 132 NPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPL- 190
N FVPWV D+A E +IP A LW+Q CA A YY++++ +FPT PE+ V +P PL
Sbjct: 118 NAFVPWVCDIAEELQIPSAVLWVQSCACLAAYYYYHHQLVKFPTETEPEITVDVPFKPLT 177
Query: 191 LKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIP 250
LK ++PSF+ P++P ++ + + +K + K VL +F ELEK+ ID M++ P +
Sbjct: 178 LKHDEIPSFLHPSSPLSSIGGTILEQIKRLHKPFSVLIETFQELEKDTIDHMSQLCPQVN 237
Query: 251 VGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESI 310
P+ PL + D+ ++ KP C+EWL+ + PSSV+YISFG+L L +++ I
Sbjct: 238 FNPIGPLFTMAKTIRSDIKGDISKPDSDCIEWLDSREPSSVVYISFGTLAFLKQNQIDEI 297
Query: 311 ATALKNSNCKFLWVIKKQDGKDSLPL-PQGFKEETKNRGMVVPWCPQTKVLVHPAIACFL 369
A + NS LWV++ + L + P E + +G +V WC Q KVL HPA+ACFL
Sbjct: 298 AHGILNSGLSCLWVLRPP--LEGLAIEPHVLPLELEEKGKIVEWCQQEKVLAHPAVACFL 355
Query: 370 THCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQ--DSDGFVETGELE 427
+HCGWNS +EA+ +G P+I +PQW DQ TNA + DVF+ G+RL + + V E+
Sbjct: 356 SHCGWNSTMEALTSGVPVICFPQWGDQVTNAVYMIDVFKTGLRLSRGASDERIVPREEVA 415
Query: 428 RAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEIL 475
+ E G K+ EL+ NA K A AVA GG+S+RN Q FVD+++
Sbjct: 416 ERLLEATVGEKAVELRENARRWKEEAESAVAYGGTSERNFQEFVDKLV 463
>At1g05680 indole-3-acetate beta-glucosyltransferase like protein
Length = 453
Score = 307 bits (787), Expect = 8e-84
Identities = 165/467 (35%), Positives = 274/467 (58%), Gaps = 24/467 (5%)
Query: 9 EELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIP 68
E H++++ F QGHI P+ + K L ++GL++TL K S D+I
Sbjct: 3 EGSHLIVLPFPGQGHITPMSQFCKRLASKGLKLTLVLVS------DKPSPPYKTEHDSIT 56
Query: 69 TNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLAC 128
+NG F +G +P D DDYM VE L L++ ++ + A
Sbjct: 57 VFPISNG-----FQEGEEPLQDL-----DDYMERVETSIKNTLPKLVEDMKLSGNPPRA- 105
Query: 129 IINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNV-QLPG 187
I+ + +PW+ DVA + + A + QP + AIYYH + + P+ + + P
Sbjct: 106 IVYDSTMPWLLDVAHSYGLSGAVFFTQPWLVTAIYYHVFKGSFSVPSTKYGHSTLASFPS 165
Query: 188 LPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYP 247
P+L DLPSF+ ++ + + +++ D L ++ ++ VL N+F +LE++++ + +P
Sbjct: 166 FPMLTANDLPSFLCESSSYPNILRIVVDQLSNIDRVDIVLCNTFDKLEEKLLKWVQSLWP 225
Query: 248 VIPVGPLLPLSLLG--VDENEDVGIEMWKPQDS-CLEWLNDQPPSSVIYISFGSLIVLSA 304
V+ +GP +P L + E+++ G ++ + + C+EWLN + P+SV+Y+SFGSL++L
Sbjct: 226 VLNIGPTVPSMYLDKRLSEDKNYGFSLFNAKVAECMEWLNSKEPNSVVYLSFGSLVILKE 285
Query: 305 KKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPA 364
++ +A LK S FLWV+++ + + LP+ + EE +G++V W PQ VL H +
Sbjct: 286 DQMLELAAGLKQSGRFFLWVVRETE---THKLPRNYVEEIGEKGLIVSWSPQLDVLAHKS 342
Query: 365 IACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETG 424
I CFLTHCGWNS LE ++ G PMI P WTDQPTNAK + DV++VG+R++ + DGFV
Sbjct: 343 IGCFLTHCGWNSTLEGLSLGVPMIGMPHWTDQPTNAKFMQDVWKVGVRVKAEGDGFVRRE 402
Query: 425 ELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFV 471
E+ R++EE++ G K +E+++NA + K A+EAV++GGSSD++I FV
Sbjct: 403 EIMRSVEEVMEGEKGKEIRKNAEKWKVLAQEAVSEGGSSDKSINEFV 449
>At1g05670 UDP glycosyltransferase UGT74E1
Length = 453
Score = 306 bits (784), Expect = 2e-83
Identities = 168/467 (35%), Positives = 272/467 (57%), Gaps = 24/467 (5%)
Query: 9 EELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIP 68
E HV+++ F AQGHI P+ + K L ++ L++TL K S DTI
Sbjct: 3 EGSHVIVLPFPAQGHITPMSQFCKRLASKSLKITLVLVS------DKPSPPYKTEHDTIT 56
Query: 69 TNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLAC 128
+NG F +G + + D D+YM VE+ L LI+ ++ + A
Sbjct: 57 VVPISNG-----FQEGQERSEDL-----DEYMERVESSIKNRLPKLIEDMKLSGNPPRAL 106
Query: 129 IINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNV-QLPG 187
+ ++ +PW+ DVA + + A + QP + AIYYH + + P+ + + P
Sbjct: 107 VYDST-MPWLLDVAHSYGLSGAVFFTQPWLVSAIYYHVFKGSFSVPSTKYGHSTLASFPS 165
Query: 188 LPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYP 247
LP+L DLPSF+ ++ + + + + D L ++ ++ VL N+F +LE++++ + +P
Sbjct: 166 LPILNANDLPSFLCESSSYPYILRTVIDQLSNIDRVDIVLCNTFDKLEEKLLKWIKSVWP 225
Query: 248 VIPVGPLLPLSLLG--VDENEDVGIEMWKPQDS-CLEWLNDQPPSSVIYISFGSLIVLSA 304
V+ +GP +P L + E+++ G ++ + + C+EWLN + PSSV+Y+SFGSL+VL
Sbjct: 226 VLNIGPTVPSMYLDKRLAEDKNYGFSLFGAKIAECMEWLNSKQPSSVVYVSFGSLVVLKK 285
Query: 305 KKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPA 364
+L +A LK S FLWV+++ + + LP+ + EE +G+ V W PQ +VL H +
Sbjct: 286 DQLIELAAGLKQSGHFFLWVVRETERRK---LPENYIEEIGEKGLTVSWSPQLEVLTHKS 342
Query: 365 IACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETG 424
I CF+THCGWNS LE ++ G PMI P W DQPTNAK + DV++VG+R++ DSDGFV
Sbjct: 343 IGCFVTHCGWNSTLEGLSLGVPMIGMPHWADQPTNAKFMEDVWKVGVRVKADSDGFVRRE 402
Query: 425 ELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFV 471
E R +EE++ + +E+++NA + K A+EAV++GGSSD+NI FV
Sbjct: 403 EFVRRVEEVMEAEQGKEIRKNAEKWKVLAQEAVSEGGSSDKNINEFV 449
>At2g31750 putative glucosyltransferase
Length = 456
Score = 296 bits (759), Expect = 1e-80
Identities = 168/470 (35%), Positives = 270/470 (56%), Gaps = 28/470 (5%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
+V++ +F QGHINPLL+ K LL++ + VT TT ++ + + + T AT +P +F
Sbjct: 8 NVLVFSFPIQGHINPLLQFSKRLLSKNVNVTFLTTSSTHNSILRRAITGGATA--LPLSF 65
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIIN 131
DG + + T +PD + EN+ +LS LI + K ++
Sbjct: 66 VP-------IDDGFEEDHPSTDTSPDYFAKFQENVSR-SLSELISSM----DPKPNAVVY 113
Query: 132 NPFVPWVADVAAEFK-IPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPL 190
+ +P+V DV + + A + Q + A Y HF +F +N +V LP +P
Sbjct: 114 DSCLPYVLDVCRKHPGVAAASFFTQSSTVNATYIHFLRG--EFKEFQN---DVVLPAMPP 168
Query: 191 LKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIP 250
LK DLP F+ N L ++++ ++ + + L NSF ELE EV+ M +PV
Sbjct: 169 LKGNDLPVFLYDNNLCRPLFELISSQFVNVDDIDFFLVNSFDELEVEVLQWMKNQWPVKN 228
Query: 251 VGPLLPLSLLG--VDENEDVGIEMWKPQ-DSCLEWLNDQPPSSVIYISFGSLIVLSAKKL 307
+GP++P L + ++D GI ++ Q + CL+WL+ +PP SVIY+SFGSL VL ++
Sbjct: 229 IGPMIPSMYLDKRLAGDKDYGINLFNAQVNECLDWLDSKPPGSVIYVSFGSLAVLKDDQM 288
Query: 308 ESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIAC 367
+A LK + FLWV+++ + K LP + E+ ++G++V W PQ +VL H +I C
Sbjct: 289 IEVAAGLKQTGHNFLWVVRETETKK---LPSNYIEDICDKGLIVNWSPQLQVLAHKSIGC 345
Query: 368 FLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETGELE 427
F+THCGWNS LEA++ G +I P ++DQPTNAK + DV++VG+R++ D +GFV E+
Sbjct: 346 FMTHCGWNSTLEALSLGVALIGMPAYSDQPTNAKFIEDVWKVGVRVKADQNGFVPKEEIV 405
Query: 428 RAIEEIVS--GPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEIL 475
R + E++ K +E+++NA L AREA++DGG+SD+NI FV +I+
Sbjct: 406 RCVGEVMEDMSEKGKEIRKNARRLMEFAREALSDGGNSDKNIDEFVAKIV 455
>At1g05560 UDP-Glucose Transferase (UGT75B2)
Length = 469
Score = 290 bits (743), Expect = 9e-79
Identities = 175/484 (36%), Positives = 261/484 (53%), Gaps = 48/484 (9%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLAR-GLQVTLATTELVYHRVFKSSSTTAATTDTIPTN 70
H +LV F AQGH+NP LR + L+ R G +VT T V+H I +
Sbjct: 5 HFLLVTFPAQGHVNPSLRFARRLIKRTGARVTFVTCVSVFHN------------SMIANH 52
Query: 71 FTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
+ L FSDG D T ++ G LS I+ N + C+I
Sbjct: 53 NKVENLSFLTFSDGFDDGGISTYEDRQKRSVNLKVNGDKALSDFIEATK-NGDSPVTCLI 111
Query: 131 NNPFVPWVADVAAEFKIPCACLWIQPCALYAIYY-HFYNNTNQFPTLENPELNVQLPGLP 189
+ W VA F++P A LWIQP ++ IYY HF N + F +LP L
Sbjct: 112 YTILLNWAPKVARRFQLPSALLWIQPALVFNIYYTHFMGNKSVF----------ELPNLS 161
Query: 190 LLKPQDLPSFILPTNP----FGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAET 245
L+ +DLPSF+ P+N + A +++ ++K+ K +L N+F LE E + +
Sbjct: 162 SLEIRDLPSFLTPSNTNKGAYDAFQEMMEFLIKETKPK--ILINTFDSLEPEALTAFPNI 219
Query: 246 YPVIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLE-WLNDQPPSSVIYISFGSLIVLSA 304
++ VGPLLP + N+ V K Q S WL+ + SSVIY+SFG+++ LS
Sbjct: 220 -DMVAVGPLLPTEIFSGSTNKSV-----KDQSSSYTLWLDSKTESSVIYVSFGTMVELSK 273
Query: 305 KKLESIATALKNSNCKFLWVIK-------KQDGKDSLPLPQ--GFKEETKNRGMVVPWCP 355
K++E +A AL FLWVI K +G++ + + GF+ E + GM+V WC
Sbjct: 274 KQIEELARALIEGKRPFLWVITDKSNRETKTEGEEETEIEKIAGFRHELEEVGMIVSWCS 333
Query: 356 QTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQ 415
Q +VL H A+ CF+THCGW+S LE++ G P++A+P W+DQPTNAKL+ + ++ G+R+ +
Sbjct: 334 QIEVLSHRAVGCFVTHCGWSSTLESLVLGVPVVAFPMWSDQPTNAKLLEESWKTGVRVRE 393
Query: 416 DSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEIL 475
+ DG VE GE+ R +E ++ KS EL+ NA + KR A EA +GGSSD+N++ FV++I
Sbjct: 394 NKDGLVERGEIRRCLEAVME-EKSVELRENAKKWKRLAMEAGREGGSSDKNMEAFVEDIC 452
Query: 476 ATRL 479
L
Sbjct: 453 GESL 456
>At1g05530 hypothetical protein
Length = 455
Score = 290 bits (743), Expect = 9e-79
Identities = 175/477 (36%), Positives = 257/477 (53%), Gaps = 41/477 (8%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLL-ARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTN 70
H +LV F AQGH+NP LR + L+ G +VT AT V HR IP +
Sbjct: 5 HFLLVTFPAQGHVNPSLRFARRLIKTTGARVTFATCLSVIHR------------SMIPNH 52
Query: 71 FTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
+ L FSDG D + + E G LS I+ + N ++C+I
Sbjct: 53 NNVENLSFLTFSDGFDDGVISNTDDVQNRLVHFERNGDKALSDFIEANQ-NGDSPVSCLI 111
Query: 131 NNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPL 190
WV VA F +P LWIQP + IYY++ N + P LP
Sbjct: 112 YTILPNWVPKVARRFHLPSVHLWIQPAFAFDIYYNYSTGNNSV---------FEFPNLPS 162
Query: 191 LKPQDLPSFILPTNPFGALSKV---LADMLKDMKKLKWVLANSFYELEKEVIDSMAETYP 247
L+ +DLPSF+ P+N A V L D LK+ K +L N+F LE E + ++
Sbjct: 163 LEIRDLPSFLSPSNTNKAAQAVYQELMDFLKEESNPK-ILVNTFDSLEPEFLTAIPNI-E 220
Query: 248 VIPVGPLLPLSLL-GVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKK 306
++ VGPLLP + G + +D+ + S WL+ + SSVIY+SFG+++ LS K+
Sbjct: 221 MVAVGPLLPAEIFTGSESGKDLSRD--HQSSSYTLWLDSKTESSVIYVSFGTMVELSKKQ 278
Query: 307 LESIATALKNSNCKFLWVIK-------KQDGKDSLPLPQ--GFKEETKNRGMVVPWCPQT 357
+E +A AL FLWVI K +G++ + + GF+ E + GM+V WC Q
Sbjct: 279 IEELARALIEGGRPFLWVITDKLNREAKIEGEEETEIEKIAGFRHELEEVGMIVSWCSQI 338
Query: 358 KVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDS 417
+VL H AI CFLTHCGW+S LE++ G P++A+P W+DQP NAKL+ ++++ G+R+ ++S
Sbjct: 339 EVLRHRAIGCFLTHCGWSSSLESLVLGVPVVAFPMWSDQPANAKLLEEIWKTGVRVRENS 398
Query: 418 DGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
+G VE GE+ R +E ++ KS EL+ NA + KR A EA +GGSSD+N++ FV +
Sbjct: 399 EGLVERGEIMRCLEAVMEA-KSVELRENAEKWKRLATEAGREGGSSDKNVEAFVKSL 454
>At2g43820 putative glucosyltransferase
Length = 449
Score = 290 bits (742), Expect = 1e-78
Identities = 162/469 (34%), Positives = 260/469 (54%), Gaps = 35/469 (7%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
HV+ V + QGHI P + K L +GL+ TLA T V++ + S
Sbjct: 7 HVLAVPYPTQGHITPFRQFCKRLHFKGLKTTLALTTFVFNSINPDLS------------- 53
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIIN 131
I + SDG D +T + DDY+ + G ++ +I+ H +++ + CI+
Sbjct: 54 --GPISIATISDGYDHGGFETADSIDDYLKDFKTSGSKTIADIIQKHQTSDNP-ITCIVY 110
Query: 132 NPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPLL 191
+ F+PW DVA EF + + QPCA+ +YY Y N N L + + LP L
Sbjct: 111 DAFLPWALDVAREFGLVATPFFTQPCAVNYVYYLSYIN--------NGSLQLPIEELPFL 162
Query: 192 KPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIPV 251
+ QDLPSF + + A +++ + +K +VL NSF ELE + ++ PV+ +
Sbjct: 163 ELQDLPSFFSVSGSYPAYFEMVLQQFINFEKADFVLVNSFQELELHENELWSKACPVLTI 222
Query: 252 GPLLPLSLLGVDENEDVGIEM----WKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKL 307
GP +P L D G ++ K C+ WL+ +P SV+Y++FGS+ L+ ++
Sbjct: 223 GPTIPSIYLDQRIKSDTGYDLNLFESKDDSFCINWLDTRPQGSVVYVAFGSMAQLTNVQM 282
Query: 308 ESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEET-KNRGMVVPWCPQTKVLVHPAIA 366
E +A+A+ SN FLWV++ + + LP GF E K + +V+ W PQ +VL + AI
Sbjct: 283 EELASAV--SNFSFLWVVRSSEEEK---LPSGFLETVNKEKSLVLKWSPQLQVLSNKAIG 337
Query: 367 CFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSD-GFVETGE 425
CFLTHCGWNS +EA+ G PM+A PQWTDQP NAK + DV++ G+R++ + + G + E
Sbjct: 338 CFLTHCGWNSTMEALTFGVPMVAMPQWTDQPMNAKYIQDVWKAGVRVKTEKESGIAKREE 397
Query: 426 LERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
+E +I+E++ G +S+E+K+N + + A +++ +GGS+D NI TFV +
Sbjct: 398 IEFSIKEVMEGERSKEMKKNVKKWRDLAVKSLNEGGSTDTNIDTFVSRV 446
>At2g31790 putative glucosyltransferase
Length = 457
Score = 290 bits (741), Expect = 2e-78
Identities = 165/474 (34%), Positives = 259/474 (53%), Gaps = 30/474 (6%)
Query: 6 SRAEELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTD 65
S A++ HV+ + QGHINP+++L K L +G+ TL HR +S + T
Sbjct: 2 SEAKKGHVLFFPYPLQGHINPMIQLAKRLSKKGITSTLIIASKD-HREPYTSDDYSITVH 60
Query: 66 TIPTNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKK 125
TI F + F D LD + T + D+++ + +N K
Sbjct: 61 TIHDGFFPHEHPHAKFVD-LDRFHNSTSRSLTDFISSAK--------------LSDNPPK 105
Query: 126 LACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPT--LENPELNV 183
+I +PF+P+ D+A + + + QP +YYH T P ENP L
Sbjct: 106 --ALIYDPFMPFALDIAKDLDLYVVAYFTQPWLASLVYYHINEGTYDVPVDRHENPTL-A 162
Query: 184 QLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMA 243
PG PLL DLPSF + L + + ++ + +L N+F +LE +V+ M
Sbjct: 163 SFPGFPLLSQDDLPSFACEKGSYPLLHEFVVRQFSNLLQADCILCNTFDQLEPKVVKWMN 222
Query: 244 ETYPVIPVGPLLPLSLLG--VDENEDVGIEMWK--PQDSCLEWLNDQPPSSVIYISFGSL 299
+ +PV +GP++P L + E++D +E K P +S L+WL ++P SV+Y++FG+L
Sbjct: 223 DQWPVKNIGPVVPSKFLDNRLPEDKDYELENSKTEPDESVLKWLGNRPAKSVVYVAFGTL 282
Query: 300 IVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEET--KNRGMVVPWCPQT 357
+ LS K+++ IA A+ + FLW +++ + LP GF EE K+ G+V W PQ
Sbjct: 283 VALSEKQMKEIAMAISQTGYHFLWSVRESERSK---LPSGFIEEAEEKDSGLVAKWVPQL 339
Query: 358 KVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDS 417
+VL H +I CF++HCGWNS LEA+ G PM+ PQWTDQPTNAK + DV+++G+R+ D
Sbjct: 340 EVLAHESIGCFVSHCGWNSTLEALCLGVPMVGVPQWTDQPTNAKFIEDVWKIGVRVRTDG 399
Query: 418 DGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFV 471
+G E+ R I E++ G + +E+++N +LK AREA+++GGSSD+ I FV
Sbjct: 400 EGLSSKEEIARCIVEVMEGERGKEIRKNVEKLKVLAREAISEGGSSDKKIDEFV 453
>At4g15550 UDP-glucose:indole-3-acetate beta-D-glucosyltransferase
(iaglu)
Length = 474
Score = 286 bits (732), Expect = 2e-77
Identities = 173/488 (35%), Positives = 264/488 (53%), Gaps = 54/488 (11%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLAR--GLQVTLATTELVYHRVFKSSSTTAATTDTIPT 69
H + V F AQGHINP L L K L G +VT A + Y+R S T+ +P
Sbjct: 13 HFLFVTFPAQGHINPSLELAKRLAGTISGARVTFAASISAYNRRMFS-------TENVPE 65
Query: 70 N--FTTNGIHVLFFSDGLDP-------NPDQTQLTPDDYMALVENLGPINLSTLIKTHFI 120
F T +SDG D + Q ++M+ + G L+ LI+ +
Sbjct: 66 TLIFAT-------YSDGHDDGFKSSAYSDKSRQDATGNFMSEMRRRGKETLTELIEDNRK 118
Query: 121 NNSKKLACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPT--LEN 178
N + C++ + WVA++A EF +P A LW+QP +++I+YH++N +
Sbjct: 119 QN-RPFTCVVYTILLTWVAELAREFHLPSALLWVQPVTVFSIFYHYFNGYEDAISEMANT 177
Query: 179 PELNVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKL--KWVLANSFYELEK 236
P +++LP LPLL +D+PSFI+ +N + L + + +K+ +L N+F ELE
Sbjct: 178 PSSSIKLPSLPLLTVRDIPSFIVSSNVYAFLLPAFREQIDSLKEEINPKILINTFQELEP 237
Query: 237 EVIDSMAETYPVIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISF 296
E + S+ + + ++PVGPLL L + + +EWL+ + SSV+Y+SF
Sbjct: 238 EAMSSVPDNFKIVPVGPLLTLRT------------DFSSRGEYIEWLDTKADSSVLYVSF 285
Query: 297 GSLIVLSAKKLESIATALKNSNCKFLWVI-------KKQDGKDSLPLPQGFKEETKNRGM 349
G+L VLS K+L + AL S FLWVI K+ + + F+EE GM
Sbjct: 286 GTLAVLSKKQLVELCKALIQSRRPFLWVITDKSYRNKEDEQEKEEDCISSFREELDEIGM 345
Query: 350 VVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRV 409
VV WC Q +VL H +I CF+THCGWNS LE++ +G P++A+PQW DQ NAKL+ D ++
Sbjct: 346 VVSWCDQFRVLNHRSIGCFVTHCGWNSTLESLVSGVPVVAFPQWNDQMMNAKLLEDCWKT 405
Query: 410 GIRL----EQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDR 465
G+R+ E++ V++ E+ R IEE++ K+EE + NA K A EAV +GGSS
Sbjct: 406 GVRVMEKKEEEGVVVVDSEEIRRCIEEVME-DKAEEFRGNATRWKDLAAEAVREGGSSFN 464
Query: 466 NIQTFVDE 473
+++ FVDE
Sbjct: 465 HLKAFVDE 472
>At2g43840 putative glucosyltransferase
Length = 449
Score = 285 bits (728), Expect = 5e-77
Identities = 157/469 (33%), Positives = 264/469 (55%), Gaps = 35/469 (7%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
HV+ V F +QGHI P+ + K L ++G + T T +++ + S+
Sbjct: 7 HVLAVPFPSQGHITPIRQFCKRLHSKGFKTTHTLTTFIFNTIHLDPSSP----------- 55
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIIN 131
I + SDG D + + +Y+ + G ++ +I+ H + + CI+
Sbjct: 56 ----ISIATISDGYDQGGFSSAGSVPEYLQNFKTFGSKTVADIIRKHQ-STDNPITCIVY 110
Query: 132 NPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPLL 191
+ F+PW D+A +F + A + Q CA+ I Y Y N N L + + LPLL
Sbjct: 111 DSFMPWALDLAMDFGLAAAPFFTQSCAVNYINYLSYIN--------NGSLTLPIKDLPLL 162
Query: 192 KPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIPV 251
+ QDLP+F+ PT A +++ + K +VL NSF++L+ + +++ PV+ +
Sbjct: 163 ELQDLPTFVTPTGSHLAYFEMVLQQFTNFDKADFVLVNSFHDLDLHEEELLSKVCPVLTI 222
Query: 252 GPLLPLSLLGV----DENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKL 307
GP +P L D + D+ + K C +WL+ +P SV+YI+FGS+ LS++++
Sbjct: 223 GPTVPSMYLDQQIKSDNDYDLNLFDLKEAALCTDWLDKRPEGSVVYIAFGSMAKLSSEQM 282
Query: 308 ESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEET-KNRGMVVPWCPQTKVLVHPAIA 366
E IA+A+ SN +LWV++ + LP GF E K++ +V+ W PQ +VL + AI
Sbjct: 283 EEIASAI--SNFSYLWVVR---ASEESKLPPGFLETVDKDKSLVLKWSPQLQVLSNKAIG 337
Query: 367 CFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSD-GFVETGE 425
CF+THCGWNS +E ++ G PM+A PQWTDQP NAK + DV++VG+R++ + + G + E
Sbjct: 338 CFMTHCGWNSTMEGLSLGVPMVAMPQWTDQPMNAKYIQDVWKVGVRVKAEKESGICKREE 397
Query: 426 LERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
+E +I+E++ G KS+E+K NA + + A +++++GGS+D NI FV +I
Sbjct: 398 IEFSIKEVMEGEKSKEMKENAGKWRDLAVKSLSEGGSTDININEFVSKI 446
>At1g22340 hypothetical protein
Length = 487
Score = 268 bits (684), Expect = 7e-72
Identities = 168/491 (34%), Positives = 268/491 (54%), Gaps = 32/491 (6%)
Query: 8 AEELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTI 67
A++ HV+ V + AQGHINP+L++ K L A+G VT T ++R+ +S A D
Sbjct: 9 AQKPHVVCVPYPAQGHINPMLKVAKLLYAKGFHVTFVNTLYNHNRLLRSRGPNAL--DGF 66
Query: 68 PTNFTTNGIHVLFFSDGL-DPNPDQTQLTPDDYMALVEN-LGPINLSTLIKTHFINNSKK 125
P+ F I DGL + + D+TQ TP M++ +N L P L + + ++
Sbjct: 67 PS-FRFESI-----PDGLPETDGDRTQHTPTVCMSIEKNCLAPFK-EILRRINDKDDVPP 119
Query: 126 LACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYN---------NTNQFPTL 176
++CI+++ + + D A E +P W + HFY + +
Sbjct: 120 VSCIVSDGVMSFTLDAAEELGVPEVIFWTNSACGFMTILHFYLFIEKGLSPFKDESYMSK 179
Query: 177 ENPELNVQ-LPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELE 235
E+ + + +P + L+ +D+PS+I TNP + L ++ K+ ++ N+F ELE
Sbjct: 180 EHLDTVIDWIPSMKNLRLKDIPSYIRTTNPDNIMLNFLIREVERSKRASAIILNTFDELE 239
Query: 236 KEVIDSMAETYP-VIPVGPLLPLSLLGVDENEDVG---IEMWKPQDSCLEWLNDQPPSSV 291
+VI SM P V +GPL L ++E ++G + +W+ + CL+WL+ + P+SV
Sbjct: 240 HDVIQSMQSILPPVYSIGPLHLLVKEEINEASEIGQMGLNLWREEMECLDWLDTKTPNSV 299
Query: 292 IYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQ--DGKDSLPLPQGFKEETKNRGM 349
++++FG + V+SAK+LE A L S +FLWVI+ G+ + LPQ F ET +R M
Sbjct: 300 LFVNFGCITVMSAKQLEEFAWGLAASRKEFLWVIRPNLVVGEAMVVLPQEFLAETIDRRM 359
Query: 350 VVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRV 409
+ WCPQ KVL HPAI FLTHCGWNS LE++A G PMI +P +++QPTN K D + V
Sbjct: 360 LASWCPQEKVLSHPAIGGFLTHCGWNSTLESLAGGVPMICWPCFSEQPTNCKFCCDEWGV 419
Query: 410 GIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVA-DGGSSDRNIQ 468
GI + +D V+ E+E + E++ G K ++L+ A E +R A EA GSS N++
Sbjct: 420 GIEIGKD----VKREEVETVVRELMDGEKGKKLREKAEEWRRLAEEATRYKHGSSVMNLE 475
Query: 469 TFVDEILATRL 479
T + ++ L
Sbjct: 476 TLIHKVFLENL 486
>At4g14090 glucosyltransferase like protein
Length = 456
Score = 263 bits (671), Expect = 2e-70
Identities = 166/489 (33%), Positives = 269/489 (54%), Gaps = 54/489 (11%)
Query: 2 TSSESRAEELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTA 61
TS H +LV F AQGHINP L+L L+ G VT +T + R+ + ST
Sbjct: 3 TSVNGSHRRPHYLLVTFPAQGHINPALQLANRLIHHGATVTYSTAVSAHRRMGEPPST-- 60
Query: 62 ATTDTIPTNFTTNGIHVLFFSDGLDPNPDQTQLTPDD--YMALVENLGPINLSTLIKTHF 119
G+ +F+DG D D + D YM+ ++ G L +IK +
Sbjct: 61 ------------KGLSFAWFTDGFD---DGLKSFEDQKIYMSELKRCGSNALRDIIKANL 105
Query: 120 --INNSKKLACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLE 177
++ + +I + VPWV+ VA EF +P LWI+P + IYY+++N + +
Sbjct: 106 DATTETEPITGVIYSVLVPWVSTVAREFHLPTTLLWIEPATVLDIYYYYFNTSYKHLFDV 165
Query: 178 NPELNVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKW-----VLANSFY 232
P ++LP LPL+ DLPSF+ P+ AL L + + ++ L+ +L N+F
Sbjct: 166 EP---IKLPKLPLITTGDLPSFLQPSK---ALPSALVTLREHIEALETESNPKILVNTFS 219
Query: 233 ELEKEVIDSMAETYPVIPVGPLLPLSLLGVDENEDVGIEMWKPQDS-CLEWLNDQPPSSV 291
LE + + S+ E +IP+GPL+ S + +++K D +WL+ + SV
Sbjct: 220 ALEHDALTSV-EKLKMIPIGPLVSSS--------EGKTDLFKSSDEDYTKWLDSKLERSV 270
Query: 292 IYISFGSLIV-LSAKKLESIATALKNSNCKFLWVIKKQDGKDS-----LPLPQGFKEETK 345
IYIS G+ L K +E++ + +N FLW++++++ ++ L L +G
Sbjct: 271 IYISLGTHADDLPEKHMEALTHGVLATNRPFLWIVREKNPEEKKKNRFLELIRG-----S 325
Query: 346 NRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSD 405
+RG+VV WC QT VL H A+ CF+THCGWNS LE++ +G P++A+PQ+ DQ T AKLV D
Sbjct: 326 DRGLVVGWCSQTAVLAHCAVGCFVTHCGWNSTLESLESGVPVVAFPQFADQCTTAKLVED 385
Query: 406 VFRVGIRLEQDSDGFVETGELERAIEEIVS-GPKSEELKRNAAELKRAAREAVADGGSSD 464
+R+G++++ +G V+ E+ R +E+++S G ++EE++ NA + K A +A A+GG SD
Sbjct: 386 TWRIGVKVKVGEEGDVDGEEIRRCLEKVMSGGEEAEEMRENAEKWKAMAVDAAAEGGPSD 445
Query: 465 RNIQTFVDE 473
N++ FVDE
Sbjct: 446 LNLKGFVDE 454
>At1g22400 Putative UDP-glucose glucosyltransferase
Length = 489
Score = 254 bits (649), Expect = 8e-68
Identities = 168/491 (34%), Positives = 259/491 (52%), Gaps = 40/491 (8%)
Query: 8 AEELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTI 67
+++ HV+ V + AQGHINP++R+ K L ARG VT T ++R +S + A D +
Sbjct: 9 SQKPHVVCVPYPAQGHINPMMRVAKLLHARGFYVTFVNTVYNHNRFLRSRGSNAL--DGL 66
Query: 68 PTNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVEN-----LGPINLSTLIKTHFINN 122
P+ F I +DGL P+ D AL E+ L P L + + +N
Sbjct: 67 PS-FRFESI-----ADGL---PETDMDATQDITALCESTMKNCLAPFR-ELLQRINAGDN 116
Query: 123 SKKLACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYN---------NTNQF 173
++CI+++ + + DVA E +P W + Y HFY +
Sbjct: 117 VPPVSCIVSDGCMSFTLDVAEELGVPEVLFWTTSGCAFLAYLHFYLFIEKGLCPLKDESY 176
Query: 174 PTLENPELNVQ--LPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSF 231
T E E V +P + +K +D+PSFI TNP + + K+ ++ N+F
Sbjct: 177 LTKEYLEDTVIDFIPTMKNVKLKDIPSFIRTTNPDDVMISFALRETERAKRASAIILNTF 236
Query: 232 YELEKEVIDSMAETYP-VIPVGPLLPLSLLGVDENEDVGI---EMWKPQDSCLEWLNDQP 287
+LE +V+ +M P V VGPL L+ ++E ++G+ +WK + CL+WL+ +
Sbjct: 237 DDLEHDVVHAMQSILPPVYSVGPLHLLANREIEEGSEIGMMSSNLWKEEMECLDWLDTKT 296
Query: 288 PSSVIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQ--DGKDSLPLPQGFKEETK 345
+SVIYI+FGS+ VLS K+L A L S +FLWVI+ G++++ +P F ETK
Sbjct: 297 QNSVIYINFGSITVLSVKQLVEFAWGLAGSGKEFLWVIRPDLVAGEEAM-VPPDFLMETK 355
Query: 346 NRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSD 405
+R M+ WCPQ KVL HPAI FLTHCGWNS+LE+++ G PM+ +P + DQ N K D
Sbjct: 356 DRSMLASWCPQEKVLSHPAIGGFLTHCGWNSILESLSCGVPMVCWPFFADQQMNCKFCCD 415
Query: 406 VFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADG-GSSD 464
+ VGI + G V+ E+E + E++ G K ++++ A E +R A +A GSS
Sbjct: 416 EWDVGIEI----GGDVKREEVEAVVRELMDGEKGKKMREKAVEWQRLAEKATEHKLGSSV 471
Query: 465 RNIQTFVDEIL 475
N +T V + L
Sbjct: 472 MNFETVVSKFL 482
>At1g22370 putative UDP-glucose glucosyltransferase
Length = 479
Score = 254 bits (648), Expect = 1e-67
Identities = 160/477 (33%), Positives = 248/477 (51%), Gaps = 26/477 (5%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
HV+ + F AQGHINP+L++ K L ARG VT T ++R+ +S + D +P+ F
Sbjct: 13 HVVCIPFPAQGHINPMLKVAKLLYARGFHVTFVNTNYNHNRLIRSRGPNSL--DGLPS-F 69
Query: 72 TTNGIHVLFFSDGL-DPNPDQTQLTPDDYMALVEN-LGPINLSTLIKTHFINNSKKLACI 129
I DGL + N D Q P + ++N L P L + + + ++CI
Sbjct: 70 RFESI-----PDGLPEENKDVMQDVPTLCESTMKNCLAPFK-ELLRRINTTKDVPPVSCI 123
Query: 130 INNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQ--FPTLENPELNVQLPG 187
+++ + + D A E +P W + Y HFY + P + L+ ++
Sbjct: 124 VSDGVMSFTLDAAEELGVPDVLFWTPSACGFLAYLHFYRFIEKGLSPIKDESSLDTKINW 183
Query: 188 LPLLKP---QDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAE 244
+P +K +D+PSFI TN + K+ ++ N+F LE +V+ S+
Sbjct: 184 IPSMKNLGLKDIPSFIRATNTEDIMLNFFVHEADRAKRASAIILNTFDSLEHDVVRSIQS 243
Query: 245 TYP-VIPVGPLLPLSLLGVDENEDVG---IEMWKPQDSCLEWLNDQPPSSVIYISFGSLI 300
P V +GPL +DE D+G MW+ + CL+WL+ + P+SV+Y++FGS+
Sbjct: 244 IIPQVYTIGPLHLFVNRDIDEESDIGQIGTNMWREEMECLDWLDTKSPNSVVYVNFGSIT 303
Query: 301 VLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLP-LPQGFKEETKNRGMVVPWCPQTKV 359
V+SAK+L A L + FLWVI+ +P LP F ET NR M+ WCPQ KV
Sbjct: 304 VMSAKQLVEFAWGLAATKKDFLWVIRPDLVAGDVPMLPPDFLIETANRRMLASWCPQEKV 363
Query: 360 LVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDG 419
L HPA+ FLTH GWNS LE+++ G PM+ +P + +Q TN K D + VG+ + G
Sbjct: 364 LSHPAVGGFLTHSGWNSTLESLSGGVPMVCWPFFAEQQTNCKYCCDEWEVGMEI----GG 419
Query: 420 FVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVAD-GGSSDRNIQTFVDEIL 475
V E+E + E++ G K +++++ A E +R A EA GSS+ N Q VD++L
Sbjct: 420 DVRREEVEELVRELMDGDKGKKMRQKAEEWQRLAEEATKPIYGSSELNFQMVVDKVL 476
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,147,191
Number of Sequences: 26719
Number of extensions: 493596
Number of successful extensions: 1965
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 119
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1419
Number of HSP's gapped (non-prelim): 157
length of query: 490
length of database: 11,318,596
effective HSP length: 103
effective length of query: 387
effective length of database: 8,566,539
effective search space: 3315250593
effective search space used: 3315250593
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0327.19


