

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0323a.9
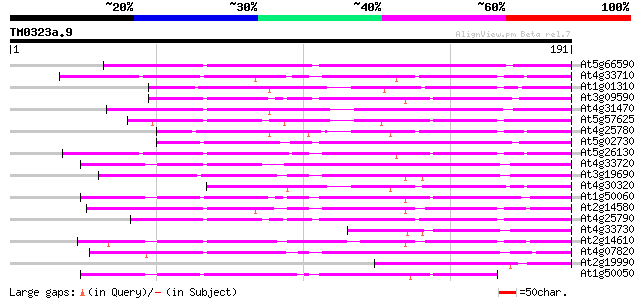
(191 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66590 Unknown protein (K1F13.27) 98 3e-21
At4g33710 pathogenesis-related protein 1 precursor, 18.9K 80 9e-16
At1g01310 pathogenesis related protein, like 77 4e-15
At3g09590 putative pathogenesis-related protein 75 2e-14
At4g31470 pathogenesis-related protein homolog 73 1e-13
At5g57625 pathogenesis-related protein - like 72 2e-13
At4g25780 putative pathogenesis-related protein 72 2e-13
At5g02730 pathogenesis related protein - like 70 7e-13
At5g26130 pathogenesis-related protein - like 68 3e-12
At4g33720 pathogenesis-related protein 1 precursor, 19.3K 68 3e-12
At3g19690 PR-1 protein, putative 64 6e-11
At4g30320 PR-1-like protein 63 8e-11
At1g50060 branched-chain amino acid aminotransferase, putative 63 1e-10
At2g14580 pathogenesis-related PR-1-like protein 62 2e-10
At4g25790 putative pathogenesis-related protein 61 4e-10
At4g33730 pathogenesis-related protein - like 60 9e-10
At2g14610 pathogenesis-related PR-1-like protein 60 9e-10
At4g07820 putative pathogenesis-related protein 55 2e-08
At2g19990 pathogenesis-related protein (PR-1) 55 2e-08
At1g50050 pathogenesis-related protein 1b precursor (pr-1b), put... 54 5e-08
>At5g66590 Unknown protein (K1F13.27)
Length = 185
Score = 97.8 bits (242), Expect = 3e-21
Identities = 55/159 (34%), Positives = 80/159 (49%), Gaps = 5/159 (3%)
Query: 33 LLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCT 92
++ S P + AK + HN R G+ L WS L AS AR++R++ C
Sbjct: 31 IVSTSPPPPPTISAAAKAFTDAHNKARAMVGVPP-LVWSQTLEAAASRLARYQRNQKKCE 89
Query: 93 PGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIV 152
+ +YG NQ + + P + WVK++ +Y + AN+ CG Y Q+V
Sbjct: 90 FASLNPGKYGA--NQLWAKGLVAVTPSLAVETWVKEKPFYNYKSDTCAANHTCGVYKQVV 147
Query: 153 WRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
WR +K LGCAQA C + L +CFY PPGNV G++P
Sbjct: 148 WRNSKELGCAQATCTKE--STVLTICFYNPPGNVIGQKP 184
>At4g33710 pathogenesis-related protein 1 precursor, 18.9K
Length = 166
Score = 79.7 bits (195), Expect = 9e-16
Identities = 63/176 (35%), Positives = 92/176 (51%), Gaps = 14/176 (7%)
Query: 18 KMFPHPFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKD 77
KMF P VL+ALAL+L + A D + YL HN R + + H++W A+
Sbjct: 2 KMFISPQTLVLLALALVLAFAVPLKAQ-DRRQDYLDVHNHARDDVSVP-HIKWHAGAARY 59
Query: 78 ASLYA-RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRF-YYEGN 135
A YA R +RD L + RYG + L + M +R WV+++ Y+ +
Sbjct: 60 AWNYAQRRKRDCRLIHSN--SRGRYG----ENLAWSSGDMSGAAAVRLWVREKSDYFHKS 113
Query: 136 NSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
N+C A CG YTQ+VW+ ++++GCA+ +C D GG + C Y PGNV G RP
Sbjct: 114 NTC-RAGKQCGHYTQVVWKNSEWVGCAKVKC--DNGGTFV-TCNYSHPGNVRGRRP 165
>At1g01310 pathogenesis related protein, like
Length = 294
Score = 77.4 bits (189), Expect = 4e-15
Identities = 49/147 (33%), Positives = 72/147 (48%), Gaps = 16/147 (10%)
Query: 48 AKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRD--KYLCTPGIFDKKRYGVCC 105
++ +L HN VR G +W +LA A +A R + + + G + + +
Sbjct: 138 SREFLIAHNLVRARVG-EPPFQWDGRLAAYARTWANQRVGDCRLVHSNGPYGENIFWAGK 196
Query: 106 NQWLEDTVKIMDPREVMRQWV-KDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQA 164
N W PR+++ W +D+FY N+C P + CG YTQIVWR + +GCA
Sbjct: 197 NNW--------SPRDIVNVWADEDKFYDVKGNTCEP-QHMCGHYTQIVWRDSTKVGCASV 247
Query: 165 ECGPDYGGVRLNVCFYYPPGNVPGERP 191
+C GGV +C Y PPGN GE P
Sbjct: 248 DC--SNGGV-YAICVYNPPGNYEGENP 271
>At3g09590 putative pathogenesis-related protein
Length = 186
Score = 75.1 bits (183), Expect = 2e-14
Identities = 52/145 (35%), Positives = 74/145 (50%), Gaps = 10/145 (6%)
Query: 48 AKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIFDKKRYGVCCNQ 107
++ +L HN R G+ L W LA+ A +A+ R+ C+ I YG N
Sbjct: 50 SREFLQAHNDARVSSGVPT-LGWDRDLARFADKWAKQRKSD--CSM-IHSGGPYGE--NI 103
Query: 108 WLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPANYPCGDYTQIVWRKTKYLGCAQAEC 166
+ K P +V+ +W ++RF Y+ N+C P CG YTQ+VWR+T +GCA+ +C
Sbjct: 104 FWHRRKKTWSPEKVVTRWFEERFNYDVKTNTCAPGKM-CGHYTQMVWRETTAVGCARVKC 162
Query: 167 GPDYGGVRLNVCFYYPPGNVPGERP 191
G L VC Y P GN GERP
Sbjct: 163 HNGRG--YLVVCEYDPRGNYEGERP 185
>At4g31470 pathogenesis-related protein homolog
Length = 185
Score = 72.8 bits (177), Expect = 1e-13
Identities = 49/160 (30%), Positives = 75/160 (46%), Gaps = 14/160 (8%)
Query: 34 LLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRD--KYLC 91
L+ S A + +L HN +R + + L+WS+ LA AS +AR RR K +
Sbjct: 37 LIFSQDKALARNTIQQQFLRPHNILRAKLRLPP-LKWSNSLALYASRWARTRRGDCKLIH 95
Query: 92 TPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQI 151
+ G + + + W PR+ + W + YY+ S AN C YTQ+
Sbjct: 96 SGGPYGENLFWGSGKGWT--------PRDAVAAWASEMKYYDRRTSHCKANGDCLHYTQL 147
Query: 152 VWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
VW+K+ +GCA + C G +C Y PPGN+ G+ P
Sbjct: 148 VWKKSSRIGCAISFCKT---GDTFIICNYDPPGNIVGQPP 184
>At5g57625 pathogenesis-related protein - like
Length = 207
Score = 71.6 bits (174), Expect = 2e-13
Identities = 55/158 (34%), Positives = 76/158 (47%), Gaps = 21/158 (13%)
Query: 41 TPATTDG--AKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCT----PG 94
TP+ G A+++L HN +R G+ L W KLA A+ +A RR Y C+ G
Sbjct: 63 TPSLPAGSIARLFLDPHNALRSGLGLPP-LIWDGKLASYATWWANQRR--YDCSLTHSTG 119
Query: 95 IFDKKRYGVCCNQWLEDTVKIMDPREVMRQW-VKDRFYYEGNNSCVPANYPCGDYTQIVW 153
+ + + + W P ++ W V+ R Y NSC + CG YTQ+VW
Sbjct: 120 PYGENLFWGSGSSWA--------PGFAVQSWIVEGRSYNHNTNSCDGSGM-CGHYTQMVW 170
Query: 154 RKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
R TK LGCA+ C + G C Y PPGN GE+P
Sbjct: 171 RDTKRLGCARVVC--ENGAGVFITCNYDPPGNYVGEKP 206
>At4g25780 putative pathogenesis-related protein
Length = 190
Score = 71.6 bits (174), Expect = 2e-13
Identities = 51/149 (34%), Positives = 74/149 (49%), Gaps = 22/149 (14%)
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRD----KYLCTPGIFDKKR---YGV 103
+LF HN VR + L W +L A +A RR ++ + G F+ +G
Sbjct: 55 FLFRHNLVRAAR-FEPPLIWDRRLQNYAQGWANQRRGDCALRHSVSNGEFNLGENIYWGY 113
Query: 104 CCNQWLEDTVKIMDPREVMRQWVKD-RFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCA 162
N W P + + W + RFY+ G+N+C A CG YTQIVW+ T+ +GCA
Sbjct: 114 GAN-W--------SPADAVVAWASEKRFYHYGSNTC-DAGQMCGHYTQIVWKSTRRVGCA 163
Query: 163 QAECGPDYGGVRLNVCFYYPPGNVPGERP 191
+ C D GG+ + C Y PPGN G++P
Sbjct: 164 RVVC--DNGGIFM-TCNYDPPGNYIGQKP 189
>At5g02730 pathogenesis related protein - like
Length = 205
Score = 70.1 bits (170), Expect = 7e-13
Identities = 47/141 (33%), Positives = 68/141 (47%), Gaps = 8/141 (5%)
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLE 110
+L HN R G S +L W LA+ AS +A+ R+ T YG N +
Sbjct: 60 FLLAHNAARVASGAS-NLRWDQGLARFASKWAKQRKSDCKMT---HSGGPYGE--NIFRY 113
Query: 111 DTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDY 170
+ PR V+ +W+ + Y+ + + CG YTQIVWR T +GCA+++C +
Sbjct: 114 QRSENWSPRRVVDKWMDESLNYDRVANTCKSGAMCGHYTQIVWRTTTAVGCARSKCDNNR 173
Query: 171 GGVRLNVCFYYPPGNVPGERP 191
G L +C Y P GN GE P
Sbjct: 174 G--FLVICEYSPSGNYEGESP 192
>At5g26130 pathogenesis-related protein - like
Length = 164
Score = 68.2 bits (165), Expect = 3e-12
Identities = 54/174 (31%), Positives = 82/174 (47%), Gaps = 12/174 (6%)
Query: 19 MFPHPFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDA 78
MF P +L + L L + A D + YL EHN R + G+ ++W + A
Sbjct: 1 MFKSPQTLILSVIVLFLAFAVPLKAQ-DQPQDYLDEHNRARTQVGVPP-MKWHAGAEQYA 58
Query: 79 SLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRF-YYEGNNS 137
YA+ R+ T YG + L + + E ++ WV ++ Y +N+
Sbjct: 59 WNYAQQRKGDCSLTHSN-SNGLYG----ENLAWSGGALSGAEAVKLWVNEKSDYIYASNT 113
Query: 138 CVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
C CG YTQ+VWR ++++GCA+ +C D GG + C YYPPGN G P
Sbjct: 114 CSDGKQ-CGHYTQVVWRTSEWVGCAKVKC--DNGGTFV-TCNYYPPGNYRGRWP 163
>At4g33720 pathogenesis-related protein 1 precursor, 19.3K
Length = 163
Score = 67.8 bits (164), Expect = 3e-12
Identities = 49/167 (29%), Positives = 74/167 (43%), Gaps = 15/167 (8%)
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F + +L+V +A + D +L HN R E G+ L W +K+A A YA
Sbjct: 11 FLAITFFLVLIVHLKAQDSPQD----FLAVHNRARAEVGVGP-LRWDEKVAAYARNYANQ 65
Query: 85 RRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYP 144
R+ G K + + + M + WV ++F Y+ +++ +
Sbjct: 66 RK-------GDCAMKHSSGSYGENIAWSSGSMTGVAAVDMWVDEQFDYDYDSNTCAWDKQ 118
Query: 145 CGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR ++ LGCA+ C G C Y PPGN GE P
Sbjct: 119 CGHYTQVVWRNSERLGCAKVRCN---NGQTFITCNYDPPGNWVGEWP 162
>At3g19690 PR-1 protein, putative
Length = 161
Score = 63.5 bits (153), Expect = 6e-11
Identities = 48/163 (29%), Positives = 74/163 (44%), Gaps = 13/163 (7%)
Query: 31 LALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYL 90
L LL ++ D + +L HN R E G+ + L W D++A A+ YA R +
Sbjct: 9 LFLLAIALFYGSLAEDLQQQFLEAHNEARNEVGL-DPLVWDDEVAAYAASYANQRINDCA 67
Query: 91 CTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCV-PANYPCGDY 148
+ +G + + + M + W+ ++ YY+ +N+C P C Y
Sbjct: 68 L---VHSNGPFG----ENIAMSSGEMSAEDAAEMWINEKQYYDYDSNTCNDPNGGTCLHY 120
Query: 149 TQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
TQ+VW+ T LGCA+ C G C Y PPGN GE+P
Sbjct: 121 TQVVWKNTVRLGCAKVVCN---SGGTFITCNYDPPGNYIGEKP 160
>At4g30320 PR-1-like protein
Length = 161
Score = 63.2 bits (152), Expect = 8e-11
Identities = 45/127 (35%), Positives = 61/127 (47%), Gaps = 15/127 (11%)
Query: 68 LEWSDKLAKDASLYARFRRDKYLCTP--GIFDKKRYGVCCNQWLEDTVKIMDPREVMRQW 125
L+W KLA+ A +A RR T G + + + N+W P + W
Sbjct: 46 LKWDAKLARYAQWWANQRRGDCALTHSNGPYGENLFWGSGNRW--------GPSQAAYGW 97
Query: 126 VKD-RFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPG 184
+ + R Y +NSC + CG YTQIVW+ T+ +GCA C GGV L C Y PPG
Sbjct: 98 LSEARSYNYRSNSC--NSEMCGHYTQIVWKNTQKIGCAHVICNGG-GGVFL-TCNYDPPG 153
Query: 185 NVPGERP 191
N G +P
Sbjct: 154 NFLGRKP 160
>At1g50060 branched-chain amino acid aminotransferase, putative
Length = 161
Score = 62.8 bits (151), Expect = 1e-10
Identities = 50/168 (29%), Positives = 76/168 (44%), Gaps = 16/168 (9%)
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F V+VA++ L+V++ A D YL HN R + G+ ++ W LA A Y+ F
Sbjct: 8 FLVIVAISFLVVATNAQNTPQD----YLNSHNTARAQVGVP-NVVWDTTLAAYALNYSNF 62
Query: 85 RRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPANY 143
R+ C + YG + + ++ WV ++ YY N+C
Sbjct: 63 RKAD--CNL-VHSNGPYG---ENLAKGSSSSFSAISAVKLWVDEKPYYSYAYNNCTGGKQ 116
Query: 144 PCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
C YTQ+VWR + +GCA+ +C + V C Y PGN GE P
Sbjct: 117 -CLHYTQVVWRDSVKIGCARVQCTNTWWFVS---CNYNSPGNWVGEYP 160
>At2g14580 pathogenesis-related PR-1-like protein
Length = 161
Score = 61.6 bits (148), Expect = 2e-10
Identities = 49/167 (29%), Positives = 79/167 (46%), Gaps = 17/167 (10%)
Query: 27 VLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA-RFR 85
+L+ LA L+ + D + Y+ HN R + G+ ++W + LA A YA + +
Sbjct: 9 ILIILAALVGALVVPLKAQDSQQDYVNAHNQARSQIGVGP-MQWDEGLAAYARNYANQLK 67
Query: 86 RDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPANYP 144
D L + + YG + L + + + WV ++ Y N+C N
Sbjct: 68 GDCRL----VHSRGPYG----ENLAKSGGDLSGVAAVNLWVNEKANYNYDTNTC---NGV 116
Query: 145 CGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + LGCA+ C + GG ++ C Y PPGN ++P
Sbjct: 117 CGHYTQVVWRNSVRLGCAKVRC--NNGGTIIS-CNYDPPGNYANQKP 160
>At4g25790 putative pathogenesis-related protein
Length = 210
Score = 60.8 bits (146), Expect = 4e-10
Identities = 51/150 (34%), Positives = 68/150 (45%), Gaps = 9/150 (6%)
Query: 42 PATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIFDKKRY 101
P T + +L HN VR G+ L W K+A A+ +A RR Y C+ Y
Sbjct: 69 PPTGSFEQQFLDPHNTVRGGLGLPP-LVWDVKIASYATWWANQRR--YDCSL-THSTGPY 124
Query: 102 GVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGC 161
G N + V V+ + Y N+C + CG YTQIVWR+T+ LGC
Sbjct: 125 GE--NLFWGSGSDFTSTFAVESWTVEAKSYNHMTNTC-EGDGMCGHYTQIVWRETRRLGC 181
Query: 162 AQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
A+ C + G C Y PPGN GE+P
Sbjct: 182 ARVVC--ENGAGVFITCNYDPPGNYVGEKP 209
>At4g33730 pathogenesis-related protein - like
Length = 172
Score = 59.7 bits (143), Expect = 9e-10
Identities = 33/78 (42%), Positives = 44/78 (56%), Gaps = 8/78 (10%)
Query: 116 MDPREVMRQWVKDRFYYEG-NNSCV-PANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGV 173
M + + WV ++ YY+ +NSC PA CG YTQ+VWR + LGC +A+C G
Sbjct: 100 MSAAQAVAMWVHEKSYYDFYSNSCHGPA---CGHYTQVVWRGSARLGCGKAKCN---NGA 153
Query: 174 RLNVCFYYPPGNVPGERP 191
+ VC Y P GN G RP
Sbjct: 154 SIVVCNYDPAGNYIGARP 171
>At2g14610 pathogenesis-related PR-1-like protein
Length = 161
Score = 59.7 bits (143), Expect = 9e-10
Identities = 54/170 (31%), Positives = 76/170 (43%), Gaps = 20/170 (11%)
Query: 24 FFAVLVALA-LLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA 82
F V VAL L++ S+A + D YL HN R G+ ++W +++A A YA
Sbjct: 9 FLIVFVALVGALVLPSKAQDSPQD----YLRVHNQARGAVGVGP-MQWDERVAAYARSYA 63
Query: 83 RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPA 141
R I YG D + + WV ++ Y N+C
Sbjct: 64 EQLRGNCRL---IHSGGPYGENLAWGSGDLSGV----SAVNMWVSEKANYNYAANTC--- 113
Query: 142 NYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
N CG YTQ+VWRK+ LGCA+ C + GG ++ C Y P GN E+P
Sbjct: 114 NGVCGHYTQVVWRKSVRLGCAKVRC--NNGGTIIS-CNYDPRGNYVNEKP 160
>At4g07820 putative pathogenesis-related protein
Length = 160
Score = 55.5 bits (132), Expect = 2e-08
Identities = 48/165 (29%), Positives = 72/165 (43%), Gaps = 14/165 (8%)
Query: 28 LVALALLLVSSQATPATT-DGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRR 86
L+ LA+ L S + P D + Y HN R G+S L WS L A YA RR
Sbjct: 8 LLVLAIALALSFSVPLKAQDQPQDYFNAHNRARVSVGVSP-LMWSQTLTAYAQAYAEKRR 66
Query: 87 DKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCG 146
D L G YG + ++ + E + ++ + Y+ + A C
Sbjct: 67 DCGLFLSG----GPYG----ETIKADIIDFSAEEFVSTFLNQKSDYDYTTNTCRAGKSCD 118
Query: 147 DYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
Y Q+++RK+ +LGCA+ +C G L +C Y P + ERP
Sbjct: 119 GYKQVLFRKSVFLGCAKVKCN---NGGFLAIC-SYDPSVILSERP 159
>At2g19990 pathogenesis-related protein (PR-1)
Length = 176
Score = 55.1 bits (131), Expect = 2e-08
Identities = 28/68 (41%), Positives = 39/68 (57%), Gaps = 4/68 (5%)
Query: 125 WVKDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPD-YGGVRLNVCFYYPP 183
W+ ++ Y+ +++ + CG YTQIVWR + LGCA C D Y V +C Y PP
Sbjct: 111 WMTEKENYDYDSNTCGGDGVCGHYTQIVWRDSVRLGCASVRCKNDEYIWV---ICSYDPP 167
Query: 184 GNVPGERP 191
GN G+RP
Sbjct: 168 GNYIGQRP 175
>At1g50050 pathogenesis-related protein 1b precursor (pr-1b),
putative
Length = 226
Score = 53.9 bits (128), Expect = 5e-08
Identities = 40/143 (27%), Positives = 66/143 (45%), Gaps = 12/143 (8%)
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F ++VA++ L++++ A A D YL HN R + G++ ++ W +A A+ YA
Sbjct: 8 FLIIVAISFLVLATNAQNAQQD----YLNTHNTARAQVGVA-NVVWDTVVAAYATNYANA 62
Query: 85 RRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPANY 143
R+ TP YG + + WV ++ YY N+C+ A
Sbjct: 63 RKVDCSLTPSTGGS--YG---ENLANGNNALFTGVAAVNLWVNEKPYYNYTANACIGAQQ 117
Query: 144 PCGDYTQIVWRKTKYLGCAQAEC 166
C YTQ+VW + +GCA+ C
Sbjct: 118 -CKHYTQVVWSNSVKIGCARVLC 139
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,898,665
Number of Sequences: 26719
Number of extensions: 202796
Number of successful extensions: 459
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 414
Number of HSP's gapped (non-prelim): 22
length of query: 191
length of database: 11,318,596
effective HSP length: 94
effective length of query: 97
effective length of database: 8,807,010
effective search space: 854279970
effective search space used: 854279970
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0323a.9


