

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0323a.7
(624 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
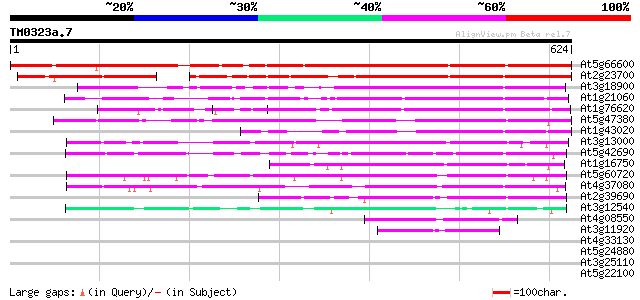
Score E
Sequences producing significant alignments: (bits) Value
At5g66600 putative protein 651 0.0
At2g23700 unknown protein 451 e-127
At3g18900 unknown protein 325 4e-89
At1g21060 unknown protein 243 2e-64
At1g76620 unknown protein 235 6e-62
At5g47380 putative protein 213 3e-55
At1g43020 unknown protein 184 2e-46
At3g13000 unknown protein 166 5e-41
At5g42690 putative protein 157 2e-38
At1g16750 unknown protein 157 2e-38
At5g60720 unknown protein 147 2e-35
At4g37080 unknown protein 138 1e-32
At2g39690 hypothetical protein 129 4e-30
At3g12540 hypothetical protein 110 2e-24
At4g08550 hypothetical protein 88 1e-17
At3g11920 hypothetical protein 82 7e-16
At4g33130 hypothetical protein 34 0.21
At5g24880 glutamic acid-rich protein 32 1.1
At3g25110 acyl-(acyl carrier protein) thioesterase 32 1.4
At5g22100 RNA 3'-terminal phosphate cyclase-like protein 31 1.8
>At5g66600 putative protein
Length = 614
Score = 651 bits (1679), Expect = 0.0
Identities = 360/632 (56%), Positives = 444/632 (69%), Gaps = 37/632 (5%)
Query: 1 MLGLSFTPRHKRSNSV--PDKNRVENDN-AESLKVSAQRMKLDEGYITECAQARKKGAPT 57
ML L HKRS S P+K RVE D + S ++QRMKLD G E K
Sbjct: 12 MLDLRIVQNHKRSKSASFPEKKRVEGDKTSNSSHEASQRMKLDMGRSNES----KHNQYH 67
Query: 58 SEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPS---LVNSSEMIMPKPATELIKE 114
S ++LKQE LE RLQDQF+VRC LEKALGY++ S L ++++ MPKPAT+LIK+
Sbjct: 68 SNTETSLKQEITHLETRLQDQFKVRCALEKALGYRTASSYVLTETNDIAMPKPATDLIKD 127
Query: 115 IAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYP-LTTPRAQVVKASLPEVL 173
+AVLEMEV++LEQ+LLSLYRKAF+QQ+SSVSP + + K P +TTPR ++ + +
Sbjct: 128 VAVLEMEVIHLEQYLLSLYRKAFEQQISSVSPNLENKKPKSPPVTTPRRRLDFSEDDDTP 187
Query: 174 TKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSA 233
+K + TV D N K+ + +D R HS S + +
Sbjct: 188 SKTDQHTVPLLDDN-------------QNQSKKTEIAAVDRDQMDPSFRRSHSQRSAFGS 234
Query: 234 FTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLS 293
R + P + K+ R+CHSQPL + N+ISLAEHLGTRISDHV P TPNKLS
Sbjct: 235 --RKASPEDSWGKASRSCHSQPLYVQN-----GDNLISLAEHLGTRISDHV-PETPNKLS 286
Query: 294 EDMVKCISAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDAR 353
E MVKC+S IYCKLA+PP L GL SP+SSLSS SAFS DQ D SPGF N+SSFD R
Sbjct: 287 EGMVKCMSEIYCKLAEPPSVLHRGLSSPNSSLSS-SAFSPSDQYDTSSPGFGNSSSFDVR 345
Query: 354 LDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKH 413
LDNSFH EG ++FSGPYS++VEV IYR+ K ++E L Q F+SLI RLEEVDPRKLKH
Sbjct: 346 LDNSFHVEGEKDFSGPYSSIVEVLCIYRDAKKASEVEDLLQNFKSLISRLEEVDPRKLKH 405
Query: 414 EEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCR 473
EEKLAFWIN+HNALVMHAFLAYGIPQNNVKRV LLLKAAYN+GG T+SA+ IQS+IL C+
Sbjct: 406 EEKLAFWINVHNALVMHAFLAYGIPQNNVKRVLLLLKAAYNIGGHTISAEAIQSSILGCK 465
Query: 474 MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQ 533
MS PGQWLR+ F+SR KF++GD R YAI++PEPL HFAL SG+HSDPAVRVYTPKR+ Q
Sbjct: 466 MSHPGQWLRLLFASR-KFKAGDERLAYAIDHPEPLLHFALTSGSHSDPAVRVYTPKRIQQ 524
Query: 534 ELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVK 593
ELE +K+EYIR L +R+ Q++L+PKLVETF KDS +CP G+ EM+ S+PES RK VK
Sbjct: 525 ELETSKEEYIRMNLSIRK--QRILLPKLVETFAKDSGLCPAGLTEMVNRSIPESSRKCVK 582
Query: 594 KCH-QLAKSRKCIEWIPHNFTFRYLISKDVLK 624
+C +K RK I+WIPH+FTFRYLI ++ K
Sbjct: 583 RCQSSTSKPRKTIDWIPHSFTFRYLILREAAK 614
>At2g23700 unknown protein
Length = 707
Score = 451 bits (1160), Expect = e-127
Identities = 239/425 (56%), Positives = 311/425 (72%), Gaps = 24/425 (5%)
Query: 201 DNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFT-RSSPPAEPLAKSLRACHSQPLSMM 259
+ L+ +C + E++ +DS V RC SSL+Q S F R SPP + S+ ACHSQPLS+
Sbjct: 306 NRLKDQC---FIEKEDIDSCVRRCQSSLNQRSTFNNRISPPED----SVFACHSQPLSIH 358
Query: 260 EYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLL 319
EY + SN SLAEH+GTRISDH+ +TPNKLSE+M+KC SAIY KLADPP ++ G
Sbjct: 359 EYIQN-GSNDASLAEHMGTRISDHIF-MTPNKLSEEMIKCASAIYSKLADPP-SINHGFS 415
Query: 320 SPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWI 379
SPSSS SS S FS DQ DMWSP FR NSSFD + EFSGPYS+M+EVS I
Sbjct: 416 SPSSSPSSTSEFSPQDQYDMWSPSFRKNSSFDDQF----------EFSGPYSSMIEVSHI 465
Query: 380 YREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQ 439
+R K D++ +++ F L+ +LE VDPRKL H+EKLAFWIN+HNALVMH FLA GIPQ
Sbjct: 466 HRNR-KRRDLDLMNRNFSLLLKQLESVDPRKLTHQEKLAFWINVHNALVMHTFLANGIPQ 524
Query: 440 NNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQT 499
NN KR LL K AY +GGR VS + IQS ILR +M RPGQWL++ + KFR+GD Q
Sbjct: 525 NNGKRFLLLSKPAYKIGGRMVSLEAIQSYILRIKMPRPGQWLKLLLIPK-KFRTGDEHQE 583
Query: 500 YAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVP 559
Y++E+ EPL +FALCSGNHSDPA+RV+TPK ++QELE AK+EYIRAT GV +KDQK+++P
Sbjct: 584 YSLEHSEPLLYFALCSGNHSDPAIRVFTPKGIYQELETAKEEYIRATFGV-KKDQKLVLP 642
Query: 560 KLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLIS 619
K++E+F+KDS + +MEMIQE LPE+++K++KK + + +EW PHNF FRYLI+
Sbjct: 643 KIIESFSKDSGLGQAALMEMIQECLPETMKKTIKKLNSGRSRKSIVEWTPHNFVFRYLIA 702
Query: 620 KDVLK 624
+++++
Sbjct: 703 RELVR 707
Score = 140 bits (354), Expect = 2e-33
Identities = 87/163 (53%), Positives = 116/163 (70%), Gaps = 12/163 (7%)
Query: 9 RHKRSNS--VPDKNRVENDNA-ESLKVSAQRMKLDEGYITECA----QARKKGAPTSEIH 61
RHKRS S VP+K ++E++N+ +S ++QR+KLD + C + +K +P +
Sbjct: 12 RHKRSKSCTVPEKKKLEDENSIDSSLDASQRLKLD---LPRCGDKSFEMKKDLSPDVKFK 68
Query: 62 STLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEME 121
S+LKQE +LE+RLQ+QF+VR LEKALGYK+PS + PKP TELIKEIAVLE+E
Sbjct: 69 SSLKQEIQELEKRLQNQFDVRGALEKALGYKTPSRDIKGDST-PKPPTELIKEIAVLELE 127
Query: 122 VVYLEQHLLSLYRKAFDQQLSSVSPPT-KEESLKYPLTTPRAQ 163
V +LEQ+LLSLYRKAFDQQ SSVSPPT K++S P +T R +
Sbjct: 128 VSHLEQYLLSLYRKAFDQQTSSVSPPTSKQQSSCSPKSTLRGK 170
>At3g18900 unknown protein
Length = 496
Score = 325 bits (834), Expect = 4e-89
Identities = 215/546 (39%), Positives = 308/546 (56%), Gaps = 91/546 (16%)
Query: 76 QDQFEVRCTLEKALGYKSPSLVNSSE-MIMPKPATELIKEIAVLEMEVVYLEQHLLSLYR 134
+D+ VR ++ LG K + +E +PK A +L++EIA LE++V+YLE +LL LYR
Sbjct: 39 EDEVLVRREMKTVLGRKLFLEDSKNEDSSIPKEAKKLVEEIAGLELQVMYLETYLLLLYR 98
Query: 135 KAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQSNDHELEAMRKE 194
+ F+ +++S K E + ++ LE +
Sbjct: 99 RFFNNKITS-------------------------------KLESEEKERSEDLLECTKL- 126
Query: 195 HDRNELDNLRKE-CNGGWPEEKHLDSGVYRCHSSLSQYSAFT-RSSPPAEPLAKSLRACH 252
+D+ +K C+ P++ DSG++R HSSLS S ++ R SP A +
Sbjct: 127 -----IDSPKKGVCS---PQKLVEDSGIFRSHSSLSHCSGYSFRMSPHAMDSSYH----R 174
Query: 253 SQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPM 312
S P SM+E + + E +GT +S++V +PN LSE+MVKCIS + +L DP
Sbjct: 175 SLPFSMLEQS--------DIDELIGTYVSENVHK-SPNSLSEEMVKCISELCRQLVDP-- 223
Query: 313 TLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYST 372
G L SSP FR E ++ S PY
Sbjct: 224 ----GSLDNDLESSSP---------------FRGK-------------EPLKIISRPYDK 251
Query: 373 MVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAF 432
++ V I R+ K +E + FRSL+ +LE V+PRKL HEEKLAFWINIHN+LVMH+
Sbjct: 252 LLMVKSISRDSEKLNAVEPALKHFRSLVNKLEGVNPRKLNHEEKLAFWINIHNSLVMHSI 311
Query: 433 LAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFR 492
L YG P+N++KRV LLKAAYNVGGR+++ DTIQ++IL CR+SRPG R F+SR+K R
Sbjct: 312 LVYGNPKNSMKRVSGLLKAAYNVGGRSLNLDTIQTSILGCRVSRPGLVFRFLFASRSKGR 371
Query: 493 SGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRK 552
+GD + YAI + E L HFALCSG+ SDP+VR+YTPK V ELE ++EY+R+ LG+ K
Sbjct: 372 AGDLGRDYAITHRESLLHFALCSGSLSDPSVRIYTPKNVMMELECGREEYVRSNLGI-SK 430
Query: 553 DQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNF 612
D K+L+PKLVE + KD+++C GV++MI + LP R ++KC R I+WI H+F
Sbjct: 431 DNKILLPKLVEIYAKDTELCNVGVLDMIGKCLPCEARDRIQKCRNKKHGRFSIDWIAHDF 490
Query: 613 TFRYLI 618
F L+
Sbjct: 491 RFGLLL 496
>At1g21060 unknown protein
Length = 505
Score = 243 bits (621), Expect = 2e-64
Identities = 190/561 (33%), Positives = 283/561 (49%), Gaps = 104/561 (18%)
Query: 62 STLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEME 121
ST + +L +E+RL + C + PK + +L KEIA LE E
Sbjct: 37 STYTKPSLSIEKRLLTYQDSNCN------------------VTPKSSEDLRKEIASLEFE 78
Query: 122 VVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTV 181
++ EQ+LLSLYR AFD+Q+SS SP T+ + + L K E V
Sbjct: 79 ILRTEQYLLSLYRTAFDEQVSSFSPHTETSLVSN---------------QFLPKSEQSDV 123
Query: 182 QSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPA 241
S V+ H S S + PP
Sbjct: 124 TS-------------------------------------VFSYHYQASPASECSSLCPPR 146
Query: 242 EPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLG-TRISDHVLPVTPNKLSEDMVKCI 300
A SL+A LS E +R SS+ +L + LG T I D++ P++LSED+++CI
Sbjct: 147 SFQA-SLKA-----LSAREKSRYVSSSHTTLGDLLGSTLIVDNI--ANPSRLSEDILRCI 198
Query: 301 SAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHG 360
++YC L+ SPSS PS+ S + ++FD+ NS H
Sbjct: 199 CSVYCTLSSKARINSCLQASPSS----PSSVS-------------SKATFDSL--NSRHE 239
Query: 361 EGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFW 420
E +E + P ++E ++ + + + Q FRSL+ +LE+VDP ++K EEKLAFW
Sbjct: 240 ER-KEANVPGVVVIESLELHLDDGSFNHAAVMLQNFRSLVQKLEKVDPSRMKREEKLAFW 298
Query: 421 INIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQW 480
INIHNAL MHA+LAYG +N R +LKAAY+VGG +V+ IQS+IL R
Sbjct: 299 INIHNALTMHAYLAYGT--HNRARNTSVLKAAYDVGGYSVNPYIIQSSILGIRPHFSQPL 356
Query: 481 LRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKD 540
L+ FS K ++ + + YA+E PE L+HFAL SG +DP VRVYT VF++L +K+
Sbjct: 357 LQTLFSPSRKSKTCNVKHIYALEYPEALAHFALSSGFSTDPPVRVYTADCVFRDLRKSKE 416
Query: 541 EYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAK 600
E+IR + + + K+L+PK+V + KD + P +ME + LP+S +++ +K L K
Sbjct: 417 EFIRNNVRI-HNETKILLPKIVHYYAKDMSLEPSALMETTVKCLPDSTKRTAQKL--LKK 473
Query: 601 SRKCIEWIPHNFTFRYLISKD 621
+ IE+ P N +FRY+I ++
Sbjct: 474 KSRNIEYSPENSSFRYVIIQE 494
>At1g76620 unknown protein
Length = 527
Score = 235 bits (599), Expect = 6e-62
Identities = 159/403 (39%), Positives = 227/403 (55%), Gaps = 21/403 (5%)
Query: 226 SSLSQYSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGT-RISDHV 284
+SL+ Y A+ + SL+A LS E R S SL E LG+ I DH
Sbjct: 135 TSLTHYQAYQKPISYPRSFNTSLKA-----LSSREGTRVVSGTH-SLGELLGSSHIVDHN 188
Query: 285 LPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLL--SPSSSLSSPSAFSIGDQGDMWSP 342
+ PNKLSED+++CIS++YC L+ + SP SS +S S + D WS
Sbjct: 189 NFINPNKLSEDIMRCISSVYCTLSRGSTSTTSTCFPASPVSSNASTIFSSKFNYEDKWS- 247
Query: 343 GFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICR 402
N +S D L+ H + + ++E ++ + +G + Q FRSL+
Sbjct: 248 --LNGASEDHFLN---HCQDQDNVLPCGVVVIEALRVHLDDGSFGYAALMLQNFRSLVQN 302
Query: 403 LEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSA 462
LE+VDP ++K EEKLAFWINIHNALVMHA+LAYG +N R +LKAAY++GG ++
Sbjct: 303 LEKVDPSRMKREEKLAFWINIHNALVMHAYLAYG--THNRARNTSVLKAAYDIGGYRINP 360
Query: 463 DTIQSTIL--RCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSD 520
IQS+IL R + P L+ FS K ++ R YA+E PE L+HFA+ SG +D
Sbjct: 361 YIIQSSILGIRPHYTSPSPLLQTLFSPSRKSKTCSVRHIYALEYPEALAHFAISSGAFTD 420
Query: 521 PAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMI 580
P VRVYT R+F++L AK EYIR+ + V K K+L+PK+ + + KD M +ME
Sbjct: 421 PTVRVYTADRIFRDLRQAKQEYIRSNVRV-YKGTKILLPKIFQHYVKDMSMDVSKLMEAT 479
Query: 581 QESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDVL 623
+ LPE RK +KC + KS K EW+P N +FRY+I+ +++
Sbjct: 480 SQCLPEDARKIAEKCLKEKKS-KNFEWLPENLSFRYVIAGELV 521
Score = 60.8 bits (146), Expect = 2e-09
Identities = 68/216 (31%), Positives = 100/216 (45%), Gaps = 44/216 (20%)
Query: 98 NSSEMIMPKPATELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQL---------------- 141
NSS ++PK + EL KEIA +E+E++++E++LLSLYRK+F+QQL
Sbjct: 70 NSSLCLIPKSSEELKKEIASIEIEILHMERYLLSLYRKSFEQQLPNSFSNLSVTTTLPRS 129
Query: 142 --SSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQSNDHEL-EAMRKEH--D 196
+S + T ++ + P++ PR+ SL + L+ E V S H L E + H D
Sbjct: 130 VTTSPTSLTHYQAYQKPISYPRS--FNTSL-KALSSREGTRVVSGTHSLGELLGSSHIVD 186
Query: 197 RNELDNLRKECNGGWPEEKHLDSGVYRCHSS----LSQYSAFTRSSP-PAEPLAKSLRAC 251
N N K L + RC SS LS+ S T S+ PA P++ +
Sbjct: 187 HNNFINPNK-----------LSEDIMRCISSVYCTLSRGSTSTTSTCFPASPVSSNASTI 235
Query: 252 HSQPLSMMEYARGCSSNIISLAEHLG-TRISDHVLP 286
S S Y S N S L + D+VLP
Sbjct: 236 FS---SKFNYEDKWSLNGASEDHFLNHCQDQDNVLP 268
>At5g47380 putative protein
Length = 598
Score = 213 bits (542), Expect = 3e-55
Identities = 165/585 (28%), Positives = 278/585 (47%), Gaps = 98/585 (16%)
Query: 49 QARKKGAPTSEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPA 108
Q K +S ++L+++ QL RLQ + +R LE+A+G S SL + A
Sbjct: 77 QMLTKNNVSSNDRASLERDVEQLHLRLQQEKSMRMVLERAMGRASSSL-SPGHRHFAGQA 135
Query: 109 TELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKAS 168
ELI EI +LE EV E H+LSLYR F+Q +S P+++ S
Sbjct: 136 NELITEIELLEAEVTNREHHVLSLYRSIFEQTVSRA--PSEQSS---------------- 177
Query: 169 LPEVLTKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSL 228
++ S H ++ ++ D N + N N +P + H+ +
Sbjct: 178 -----------SISSPAHHIKQPPRKQDPNVISNAFCSSNN-FPLKPW--------HAMV 217
Query: 229 SQYSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARG-CSSNIISLAEHLGTRISDHVLPV 287
+ + ++S + R C S A+ + +++ + DH+
Sbjct: 218 TLKDSSRKTSKKDQSSQFQFRNCIPSTTSCSSQAKSHFLKDSVTVKSPSQRTLKDHLYQC 277
Query: 288 TPNKLSEDMVKCISAIYCKLADPPMTLPPG--LLSPSSSLSSPSAFSIGDQGDMWSPGFR 345
PNKLSEDMVKC+S++Y L M+ P +LS SS+ + +I ++ WS
Sbjct: 278 -PNKLSEDMVKCMSSVYFWLCCSAMSADPEKRILSRSSTSNVIIPKNIMNEDRAWS---- 332
Query: 346 NNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEE 405
+MVEVSWI + ++ + +R L+ +LE
Sbjct: 333 ------------------------CRSMVEVSWISSDKKRFSQVTYAINNYRLLVEQLER 368
Query: 406 VDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTI 465
V +++ KLAFWINI+NAL+MH +AYN+GG ++A+TI
Sbjct: 369 VTINQMEGNAKLAFWINIYNALLMH--------------------SAYNIGGHIINANTI 408
Query: 466 QSTILRCRMSRPGQWLRMFFSS--RTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAV 523
+ +I + R G+WL S+ R K + ++++ PEPL FALC G SDP +
Sbjct: 409 EYSIFCFQTPRNGRWLETIISTALRKKPAEDKVKSMFSLDKPEPLVCFALCIGALSDPVL 468
Query: 524 RVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQES 583
+ YT V +EL+ +K E++ A + V+ + +KVL+PK++E FTK++ + +M + ++
Sbjct: 469 KAYTASNVKEELDASKREFLGANVVVKMQ-KKVLLPKIIERFTKEASLSFDDLMRWLIDN 527
Query: 584 LPESLRKSVKKCHQ----LAKSRKCIEWIPHNFTFRYLISKDVLK 624
E L +S++KC Q K+ + +EW+P++ FRY+ SKD+++
Sbjct: 528 ADEKLGESIQKCVQGKPNNKKASQVVEWLPYSSKFRYVFSKDLME 572
>At1g43020 unknown protein
Length = 406
Score = 184 bits (466), Expect = 2e-46
Identities = 121/368 (32%), Positives = 183/368 (48%), Gaps = 72/368 (19%)
Query: 257 SMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPP 316
S + +G S SLA+ LG ++PNKLSE++++ I I+ KL+D
Sbjct: 106 SKTDKIKGSDSGHPSLADLLGLNT------LSPNKLSEEILRSICVIHYKLSD------- 152
Query: 317 GLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEV 376
+ + + NS + E +E V +
Sbjct: 153 -------------------------------NGHNRLVKNSKNEEYGQELG------VGI 175
Query: 377 SWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYG 436
+Y + +E + Q FRSL+ +LE+VDP +L EEKLAFWINIHNALVMHA
Sbjct: 176 HKLYLDDYNLKSVESMLQNFRSLVQKLEKVDPARLGREEKLAFWINIHNALVMHA----- 230
Query: 437 IPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDG 496
A+N+GG V+A IQS+IL R LR FS ++ G
Sbjct: 231 ---------------AFNIGGEWVNAYDIQSSILGIRPCHSPSRLRTLFSPAKSSKTSSG 275
Query: 497 RQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKV 556
R TYA++ EPL HFAL +G +DP VRVYT + +FQEL A+D YI+ ++G K+ K+
Sbjct: 276 RHTYALDYAEPLLHFALSTGASTDPMVRVYTSEGIFQELRQARDSYIQTSVGF-EKETKI 334
Query: 557 LVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRY 616
L+PK++ + KD+ + G + + E L ES R ++++ K +CI W+ FRY
Sbjct: 335 LLPKIIYNYAKDTSLDMGELFSTVSECLMESQRTAMRRIVN-KKQERCIRWVHDESKFRY 393
Query: 617 LISKDVLK 624
+I ++++
Sbjct: 394 VIHSELVR 401
Score = 38.5 bits (88), Expect = 0.011
Identities = 22/42 (52%), Positives = 29/42 (68%), Gaps = 3/42 (7%)
Query: 110 ELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQL---SSVSPPT 148
ELI EIA L+ E++ LE++LLSLYR +F L SS+ P T
Sbjct: 41 ELIGEIAELDTEILQLERYLLSLYRSSFGDHLPDNSSLPPCT 82
>At3g13000 unknown protein
Length = 582
Score = 166 bits (419), Expect = 5e-41
Identities = 167/589 (28%), Positives = 267/589 (44%), Gaps = 94/589 (15%)
Query: 64 LKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEMEVV 123
L+++ +L+ +LQ + ++ LE + L SS + P PA EL+ I LE V
Sbjct: 52 LEEDVKRLQLQLQQEIDLHTFLESVMEKDPWELSYSSSV--PHPAQELLSNIVTLETAVT 109
Query: 124 YLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQS 183
LEQ ++ + + QLS + +Y LT + + +S L + + QS
Sbjct: 110 KLEQEMM-----SLNFQLSQER--NERRLAEYQLTHSASPLNSSSSLRYLNQSDSELHQS 162
Query: 184 NDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPAEP 243
+ S Q + SS + P
Sbjct: 163 AEDS--------------------------------------PSQDQIVHYQESSSESSP 184
Query: 244 LAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAI 303
++ +E +N A L + L PN LSE+MV+C+ I
Sbjct: 185 AESTVEQTLDPSNDFLEKRLMRKTN----ARKLPRGMPPKYLWDQPNLLSEEMVRCMKNI 240
Query: 304 YCKLADPPMT---------LPPGLLSPSSSLSSPSAFSIGDQGDMWSP-------GFRNN 347
+ LADP T L P +SP LSS +++ + M S +NN
Sbjct: 241 FMSLADPTATSKASSNESHLSP--VSPRGHLSSSASWWPSTERSMISSWVQSPQIDIQNN 298
Query: 348 SSFDARLD--NSFHGEGVEEFS--GPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRL 403
++ A D + + G ++ G YS EVSW+ + +KFR+L+ +L
Sbjct: 299 ANVLATGDVFDPYRVRGKLSWAEIGNYSLASEVSWMSVGKKQLEYASGALKKFRTLVEQL 358
Query: 404 EEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSAD 463
V+P L EKLAFWIN++NAL+MHA+LAYG+P++++K L+ KAAY VGG + +A
Sbjct: 359 ARVNPIHLSCNEKLAFWINLYNALIMHAYLAYGVPKSDLKLFSLMQKAAYTVGGHSYTAA 418
Query: 464 TIQSTILRCR--MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDP 521
T++ IL+ + M RP L + K + + ++ +I+ EPL FAL G +S P
Sbjct: 419 TMEYVILKMKPPMHRPQIALLLAIH---KMKVSEEQRRASIDTHEPLLGFALSCGMYSSP 475
Query: 522 AVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTK----DSDMCPGGVM 577
AVR+Y+ K V +E+ A+ ++I+A++G+ K K+L+PK++ + K DS++ GV
Sbjct: 476 AVRIYSAKGVKEEMLEAQRDFIQASVGLSSKG-KLLLPKMLHCYAKSLVEDSNL---GV- 530
Query: 578 EMIQESLPESLRKSVKKC-----HQLAKSRKCIEWIPHNFTFRYLISKD 621
I LP V++C L SR C +P + FRYL D
Sbjct: 531 -WISRYLPPHQAAFVEQCISQRRQSLLASRNC-GILPFDSRFRYLFLPD 577
>At5g42690 putative protein
Length = 512
Score = 157 bits (396), Expect = 2e-38
Identities = 148/561 (26%), Positives = 258/561 (45%), Gaps = 80/561 (14%)
Query: 63 TLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEMEV 122
TL+++ +L ++L+ + + +E+A +L + P EL+ E+AVLE E+
Sbjct: 27 TLQEDVEKLRKKLRLEENIHRAMERAFSRPLGALPRLPPFLPPS-VLELLAEVAVLEEEL 85
Query: 123 VYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQ 182
V LE+H++ ++ + + +V + E+LK P+ K+ +E +
Sbjct: 86 VRLEEHIVHCRQELYQE---AVFTSSSIENLKCSPAFPKHWQTKSKSASTSARESESPLS 142
Query: 183 SNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPAE 242
+ RK + N+L S+ S + +++
Sbjct: 143 RAPCSVSVCRKGKE-NKL-------------------------SATSIKTPMKKTTIAHT 176
Query: 243 PLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISA 302
L KSL A ++ + C + H G PNK+SED+VKC+S
Sbjct: 177 QLNKSLEA------QKLKDSHRCRKTNAERSSHGGGD--------EPNKISEDLVKCLSN 222
Query: 303 IYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEG 362
I+ +++ ++ +++ S +AF D + + +SF
Sbjct: 223 IFMRMS----SIKRSMVTKSQENDKDTAFR-----DPYG------------ICSSFRRRD 261
Query: 363 VEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWIN 422
+ + +S + E S + L ++ + L+ RL V+ +KL +EKLAFWIN
Sbjct: 262 IGRYKN-FSDVEEAS--LNQNRTSSSSLFLIRQLKRLLGRLSLVNMQKLNQQEKLAFWIN 318
Query: 423 IHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLR 482
I+N+ +M+ FL +GIP++ V L+ KA NVGG ++A TI+ ILR + +++
Sbjct: 319 IYNSCMMNGFLEHGIPESP-DMVTLMQKATINVGGHFLNAITIEHFILR--LPHHSKYIS 375
Query: 483 MFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEY 542
S + + R + +E EPL FAL G+ S PAVRVYT +V +ELEVAK EY
Sbjct: 376 PKGSKKNEMAV---RSKFGLELSEPLVTFALSCGSWSSPAVRVYTASKVEEELEVAKREY 432
Query: 543 IRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSR 602
+ A++G+ K+ +PKL++ ++ D +++ I LP L K C + S+
Sbjct: 433 LEASVGI--SVVKIGIPKLMDWYSHDFAKDIESLLDWIFLQLPTELGKDALNCVEQGMSQ 490
Query: 603 K----CIEWIPHNFTFRYLIS 619
+ IP++FTFRYL S
Sbjct: 491 SPSSTLVHIIPYDFTFRYLFS 511
>At1g16750 unknown protein
Length = 529
Score = 157 bits (396), Expect = 2e-38
Identities = 121/359 (33%), Positives = 184/359 (50%), Gaps = 41/359 (11%)
Query: 290 NKLSEDMVKCISAIYCKLADPPMTLPPGLLSPS-SSLSSPSAFSIGDQGDMWSPGFRNNS 348
N+LS++M++C+ I+ L + + S SS +P + S WSP +
Sbjct: 178 NELSKEMIRCMRNIFVSLGETSAGSKSSQETASVSSRENPPSSSTS----WWSPSEHSRI 233
Query: 349 SFDA---RLDNSFHGEGVEEFS-----------------GPYSTMVEVSWIYREGLKWGD 388
S A R+D + + + S G Y + EV+ + E + G
Sbjct: 234 SRWAQSPRIDIQKNSDVLATESDVFDLYTVQGKLSWADIGSYRSATEVASMSVEEKRLGY 293
Query: 389 IEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLL 448
+FR+L+ RL V+P +L H EKLAFWINI+NA++MHA+LAYG+P+ ++K L+
Sbjct: 294 ASDELWRFRNLVERLARVNPAELSHNEKLAFWINIYNAMIMHAYLAYGVPKTDLKLFSLM 353
Query: 449 LKAAYNVGGRTVSADTIQSTILRCR--MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPE 506
KAAY VGG + +A TI+ L+ + RP L + S K + D ++ I PE
Sbjct: 354 QKAAYTVGGHSYNAATIEYMTLKMSPPLHRPQIALLL---SILKLKVSDEQRQAGISTPE 410
Query: 507 PLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFT 566
PL FAL G HS PAVR+Y+ + V +ELE A+ +YI+A++GV + K++VP+++ F
Sbjct: 411 PLVSFALSCGMHSSPAVRIYSAENVGEELEEAQKDYIQASVGVSPRG-KLIVPQMLHCFA 469
Query: 567 KDS-DMCPGGVMEMIQESLPESLRKSVKKC-HQ------LAKSRKCIEWIPHNFTFRYL 617
K S D C V I LP V++C H+ L S +P + FRYL
Sbjct: 470 KKSVDDCK--VALWISRHLPPRQAAFVEQCIHRRQWWGFLGSSSSKCGIVPFDSRFRYL 526
Score = 38.5 bits (88), Expect = 0.011
Identities = 38/125 (30%), Positives = 60/125 (47%), Gaps = 22/125 (17%)
Query: 64 LKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEMEVV 123
L+ + +L+ +LQ + +R L KA +S + S +P+ EL+ IA +E V
Sbjct: 47 LEHDVKRLKNQLQKETAMRALLLKASD-QSHKIELSHASSLPRSVQELLTNIAAMEATVS 105
Query: 124 YLEQHLLSLY---------RKAFDQQLS-SVSPPT---------KEESL--KYPLTTPRA 162
LEQ ++SL+ RK + L+ S+SPP K ESL K PR+
Sbjct: 106 KLEQEIMSLHFLLIQERNERKLAEYNLTHSLSPPNALDLVRLSEKNESLRPKDHKAQPRS 165
Query: 163 QVVKA 167
+V K+
Sbjct: 166 KVAKS 170
>At5g60720 unknown protein
Length = 645
Score = 147 bits (370), Expect = 2e-35
Identities = 162/632 (25%), Positives = 279/632 (43%), Gaps = 96/632 (15%)
Query: 64 LKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEM--IMPKPATELIKEIAVLEME 121
+++E L L + + R LE+ ++ PS +S + +P ELI E++++E E
Sbjct: 33 IEKEVWMLREMLDQEEKTREILEQVQKHQLPSSSSSLTLPASLPPKMKELITELSIVEGE 92
Query: 122 VVYLE---QHLLSLYRKAFDQQLSSVSPPT-----------KEESL---------KYPLT 158
+ LE HL ++ D+ L + + K++++ KYP
Sbjct: 93 ISRLEIQISHLQINLKQEQDETLKQATTSSSRRAWQTSETYKDDNINPYQAPTLPKYPKL 152
Query: 159 TPRAQVVKASLPEVLTKEEYPTVQSN--------------------DHELEAMRKEHDRN 198
P + +V + + T + D+ +E+ RK +++
Sbjct: 153 PPPSPMVNKGMMKSENNNTKSTTSHHQENATFGTKTLHFINKAIKGDYAIESFRKSNEK- 211
Query: 199 ELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPAEPLAKSLRACHSQPLSM 258
+ + KE + E + + + + SP EP S + ++
Sbjct: 212 -VGVVEKENHRSVQHENKVQENM-----KMKKIRTMKSPSPLREPRYTSPNKPNKDRVAA 265
Query: 259 MEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLP--- 315
++ + SL+ + ++ PNKL+E+++KC++ IY +L +
Sbjct: 266 LDASLDIPPK--SLSSTILMEDGQNIQKWHPNKLAENIMKCLNFIYVRLLRTTRVMELEK 323
Query: 316 PGLLSPSSSLS-SPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFS----GPY 370
G +S S++ S S +F + + S S ++R + + VE GPY
Sbjct: 324 TGPISRSTNFSLSSRSFRVDNATSSLSKSMNLVSYKESRQQDPYGIFDVESSLARDIGPY 383
Query: 371 STMV--EVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALV 428
+V S + + + L QK R L+ LE VD + L H++KLAFWIN+ NA V
Sbjct: 384 KNLVIFTSSSMDSKCISSSSSVSLIQKLRVLMNNLETVDLKVLSHQQKLAFWINMFNACV 443
Query: 429 MHAFLAYGIPQNNVKRVFLLL-KAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSS 487
MH +L +G+P+ + L+ KA NVGG+ +SA TI+ ILR S S+
Sbjct: 444 MHGYLQHGVPKTAERLQSLVYNKATMNVGGKNISAHTIEHCILRKSTS----------ST 493
Query: 488 RTKFRSGDG--RQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRA 545
T+ R + R+ Y +E +P FAL G S PAVR+YT + V ELE +K EY++A
Sbjct: 494 MTQDRHEEMIIRKLYGVEATDPNITFALSCGTRSSPAVRIYTGEGVTTELEKSKLEYLQA 553
Query: 546 TLGVRRKDQKVLVPKLVETFTKDSDM--CPGGVMEMIQ---------ESLPE--SLRKSV 592
+L V +++ +P+L+ D + GG EM Q LP SLRKS+
Sbjct: 554 SLVVTAA-KRIGLPELLLKHATDFVVLTADGGTGEMEQLGSLVKWVCNQLPTSGSLRKSM 612
Query: 593 KKC-----HQLAKSRKCIEWIPHNFTFRYLIS 619
C + + S +E IP++F F+YL++
Sbjct: 613 VDCFKNPNSKASSSSSAVEKIPYDFEFQYLLA 644
>At4g37080 unknown protein
Length = 597
Score = 138 bits (347), Expect = 1e-32
Identities = 150/601 (24%), Positives = 259/601 (42%), Gaps = 97/601 (16%)
Query: 64 LKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEMEVV 123
L Q+ +L+R+L+ + V LE+A +L + P+ EL+ E+AVLE EVV
Sbjct: 46 LLQDVDKLKRKLRQEENVHRALERAFTRPLGALPRLPSYL-PRHTLELLAEVAVLEEEVV 104
Query: 124 YLEQHLLS----LYRKAF------------DQQLSSVSP--PTKEESLKY-------PLT 158
LE+ +++ LY++A + L+ SP TK + K +
Sbjct: 105 RLEEQVVNFRQGLYQEAVYISSKRNLESPNNNSLNENSPVRSTKHQRSKSMSQHEFNSMI 164
Query: 159 TPRAQVVKASLPEVLTKEEYPTVQS-NDHELEAMRKEHDRNELDNLRKECNGGWPEEKHL 217
TP + ++ + +++ + + Q+ ND + + + NL N K +
Sbjct: 165 TPTKKHQQSLSRSISSRKLFSSDQTVNDRSGQRVVSGKQASPKSNLSSVTN-----TKPV 219
Query: 218 DSGVYRCHSSLSQYSAFTRSSPPAEPLAKSLRACHSQ-PLSMMEYA--RGCSSNIISLAE 274
D +S + P + L + L + + PL E A + S + L +
Sbjct: 220 DVRGKENQTSSNASKDKKNKESPEKKLGRFLTSVKKKKPLIKPEAAADKHSESTKLQLDD 279
Query: 275 HL-----------GTRISDHVLPV--TPNKLSEDMVKCISAIYCKLADPPMTLPPGLLSP 321
L G+ D L N++SED++KC+ I +
Sbjct: 280 RLADQDKAQESVSGSSSEDKTLQSGNVANRVSEDLLKCLVTIILR--------------- 324
Query: 322 SSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSF-HGEGVEEFSGPYSTMVEVSWIY 380
I D+ + N S + R ++ H V+ S + S++
Sbjct: 325 -----------ISSSKDIVLDPYNNCSEWRTRELGAYKHFSSVDTSSVDLGRRINASFLI 373
Query: 381 REGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQN 440
+ + L+ +L V+ L H++KLAFWIN +N+ VM+AFL +GIP
Sbjct: 374 H-------------RLKFLLNKLSVVNLDGLSHQQKLAFWINTYNSCVMNAFLEHGIPAT 420
Query: 441 NVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQTY 500
V L+ KA VGG +++A TI+ ILR L+ T+
Sbjct: 421 PEMVVALMQKATIIVGGHSLNAITIEHFILRLPYH-----LKFTCPKTATHEEMRAHSTF 475
Query: 501 AIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPK 560
+E EPL FAL G+ S PAVRVYT V +ELE AK +Y++A++G+ K+ K+++PK
Sbjct: 476 GLEWSEPLVTFALACGSWSSPAVRVYTAANVEEELEAAKRDYLQASVGI-SKNNKLMLPK 534
Query: 561 LVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEW---IPHNFTFRYL 617
+++ + D +++ + LP+ LR+ KC + +E +P++F+FR L
Sbjct: 535 VLDWYLLDFAKDLESLLDWVCLQLPDKLREEATKCMERKNKESLMELVQVVPYDFSFRLL 594
Query: 618 I 618
+
Sbjct: 595 L 595
>At2g39690 hypothetical protein
Length = 503
Score = 129 bits (325), Expect = 4e-30
Identities = 111/357 (31%), Positives = 172/357 (48%), Gaps = 37/357 (10%)
Query: 277 GTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQ 336
G R+ + PN++SE ++ C+ IY +L + + G +S S PS+ S
Sbjct: 169 GLRLVEAKTKDDPNEVSEQLINCLIGIYLEL-NHVSSKTKGDVSLSRR---PSSCSRKSN 224
Query: 337 GDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLH--- 393
+ N + D+S GV GPY + +S I+ H
Sbjct: 225 TYSYYQNAMNLDPYHVLPDSS---GGVTRDIGPYKNFIHISR--------SSIDVTHFTH 273
Query: 394 ------QKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFL 447
+ L+ +L EVD L +++KLAFWINI+NA +MHAFL YG+P ++ + + L
Sbjct: 274 YCSPAVPRLSVLMEKLSEVDLSFLTYKQKLAFWINIYNACIMHAFLEYGLPSSHNRLLTL 333
Query: 448 LLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEP 507
+ KA+ NVGG ++A I+ +LR P + R TY + EP
Sbjct: 334 MNKASLNVGGIVLNALAIEHFVLR-HPCEPED------KDSLDEKETLLRHTYGLGYSEP 386
Query: 508 LSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTK 567
FALC G+ S PA+RVYT V +L A+ EY+ A++GV K +K++VP+L++ K
Sbjct: 387 NVTFALCRGSWSSPALRVYTADEVVNDLGRARVEYLEASVGVSSK-KKIVVPQLLQWHMK 445
Query: 568 DSDMCPGGVMEMIQESLPES--LRKSVKKC-HQLAK--SRKCIEWIPHNFTFRYLIS 619
D ++E I LP S L+ + +C + AK K +E + FRYL+S
Sbjct: 446 DFADDIESLLEWIYSQLPRSGNLKGMIMECLKRKAKVPLAKIVEIQTYGHEFRYLLS 502
>At3g12540 hypothetical protein
Length = 561
Score = 110 bits (276), Expect = 2e-24
Identities = 128/582 (21%), Positives = 238/582 (39%), Gaps = 108/582 (18%)
Query: 63 TLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEMEV 122
+L++E ++++ L+D+ + L S S +++P ELI+E+A +E E+
Sbjct: 62 SLQREVMKMQGELEDEQALNKALRDMHRGPVMSQPRLSLLLLPPQVQELIEELATVEAEI 121
Query: 123 VYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQ 182
+ LE+ + L + + K+E+ + + + + P+ L + +
Sbjct: 122 LCLEKRIQDLKLDVYSE---------KKENKELEDSIDEGEEERMMNPKRLLQRQNHLPC 172
Query: 183 SNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPAE 242
D++L MR E + + + G K + R H+S+ F
Sbjct: 173 DADNDLIKMRSEDLKQRS---KSQSYGDHHVVKDIQMNSPRTHASIGSTMEF-------- 221
Query: 243 PLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISA 302
S + ++ G S T+ ++V TPN +SED+VKC+
Sbjct: 222 ----------SSRIHSSTFSDGMSR----------TQEKNNVQETTPNGVSEDLVKCLMG 261
Query: 303 IYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDN-----S 357
IY +L SL+ + + F+ S +D N +
Sbjct: 262 IYLELNRSSREREGSKTVSKLSLTH-----------LKNASFKRKSVYDHNASNLDPYGA 310
Query: 358 FHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKL 417
G + + G Y + ++ + + D R L +L +VD L H++K+
Sbjct: 311 VMGTSLRDI-GEYKNFIHITRTSIDVSRLSDCSTSLVNLRVLKEKLSKVDLSFLNHKKKM 369
Query: 418 AFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRP 477
AFWIN +NA VM+ FL +G+P + K + +L A +VGG +SA I+ +IL+
Sbjct: 370 AFWINTYNACVMNGFLEHGLPSSKEKLLTILKMATIDVGGTQLSALDIEGSILQSPC--- 426
Query: 478 GQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVF----- 532
+ R++ + EN + L + P VR + + VF
Sbjct: 427 -----------------EPRESRS-ENTDTLR----IQMRRAKPNVRALSWRLVFTCTED 464
Query: 533 --QELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRK 590
EL A+ EY+ A++GV + +K+++P+ + +D G ++E I LP +
Sbjct: 465 VVNELIKARTEYLEASIGVSGR-KKIVIPRFLHKRLRDFAEDEGSLIEWICSQLPPA--- 520
Query: 591 SVKKCHQLAKS-------------RKCIEWIPHNFTFRYLIS 619
++C QL ++ +K IE H + FRYL++
Sbjct: 521 --QRCFQLKETAMEGLNKKSESQLKKLIEVRSHEYEFRYLLA 560
>At4g08550 hypothetical protein
Length = 587
Score = 88.2 bits (217), Expect = 1e-17
Identities = 54/171 (31%), Positives = 84/171 (48%), Gaps = 9/171 (5%)
Query: 395 KFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYN 454
++ +I L V+ ++ EEKLAF+IN++N + +H+ L +G P R + + Y
Sbjct: 424 RYLRIIQELHRVELEDMQREEKLAFFINLYNMMAIHSILVWGHPAGTFDRTKMFMDFKYV 483
Query: 455 VGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALC 514
+GG T S IQ+ ILR R F+ F D R A+ EPL+HF L
Sbjct: 484 IGGYTYSLSAIQNGILRGNQ-------RPMFNPMKPFGVKDKRSKVALPYAEPLTHFTLV 536
Query: 515 SGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETF 565
G S P +R +TP + +EL A +++R G R D V ++ + F
Sbjct: 537 CGTRSGPPLRCFTPGEIDKELMEAARDFLRC--GGLRVDLNAKVAEISKIF 585
>At3g11920 hypothetical protein
Length = 630
Score = 82.4 bits (202), Expect = 7e-16
Identities = 49/135 (36%), Positives = 73/135 (53%), Gaps = 6/135 (4%)
Query: 410 KLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTI 469
+L EEKLAF++N++NA+V+HA ++ G P+ + R Y VGG + S +I++ I
Sbjct: 430 ELSTEEKLAFFLNLYNAMVIHALISIGRPEGLIARRSFFTDFQYVVGGYSYSLSSIRNDI 489
Query: 470 LRCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPK 529
LR R +P + R F +G R + PL HF LC G S P VR +TP+
Sbjct: 490 LR-RGRKPS-----YPFIRPPFNNGKTRHELGLLKLNPLVHFGLCDGTKSSPVVRFFTPQ 543
Query: 530 RVFQELEVAKDEYIR 544
V EL+ A E+ +
Sbjct: 544 GVEAELKRAAREFFQ 558
>At4g33130 hypothetical protein
Length = 125
Score = 34.3 bits (77), Expect = 0.21
Identities = 20/63 (31%), Positives = 37/63 (57%), Gaps = 3/63 (4%)
Query: 70 QLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEMEVVYLEQHL 129
+L+++LQ++ +R L A+ + S ++S + P EL+ +A+LE+ V LEQ
Sbjct: 6 RLQQQLQEEINLRLALTSAVEHSSSPFMDSPCEL---PDKELLDSLAILEITVSKLEQES 62
Query: 130 LSL 132
+SL
Sbjct: 63 VSL 65
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 32.0 bits (71), Expect = 1.1
Identities = 25/109 (22%), Positives = 47/109 (42%), Gaps = 6/109 (5%)
Query: 142 SSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQSNDHELEAMRKEHDRNELD 201
SS S +K+E + ++ KAS E+ + DH+ E ++ E D ++
Sbjct: 163 SSTSSKSKKEG------SENVRIKKASDKEIALDSASMSSAQEDHQEEILKVESDHLQVS 216
Query: 202 NLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPAEPLAKSLRA 250
+ E EEK + V + + S+ + + + +P A P+ K A
Sbjct: 217 DHDIEEPKYEKEEKEVQEKVVQANESVEEKAESSGPTPVASPVGKDCNA 265
>At3g25110 acyl-(acyl carrier protein) thioesterase
Length = 362
Score = 31.6 bits (70), Expect = 1.4
Identities = 26/80 (32%), Positives = 36/80 (44%), Gaps = 3/80 (3%)
Query: 44 ITECAQARKKG-APTSEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEM 102
I C+Q +K G P + S + +Q L DQ + E L YK +V S E+
Sbjct: 36 IVSCSQTKKTGLVPLRAVVSADQGSVVQGLATLADQLRLGSLTEDGLSYKEKFVVRSYEV 95
Query: 103 IMPKPATELIKEIAVLEMEV 122
K AT ++ IA L EV
Sbjct: 96 GSNKTAT--VETIANLLQEV 113
>At5g22100 RNA 3'-terminal phosphate cyclase-like protein
Length = 375
Score = 31.2 bits (69), Expect = 1.8
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query: 364 EEFSGPYSTMVEV-SWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLK 412
+E S T VEV SW+ +E K G ++ HQ L+C L E D K++
Sbjct: 275 KERSPAEDTGVEVASWLLQEIEKGGVVDSTHQGLLFLLCALSEQDVSKVR 324
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,579,057
Number of Sequences: 26719
Number of extensions: 641925
Number of successful extensions: 2068
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1966
Number of HSP's gapped (non-prelim): 54
length of query: 624
length of database: 11,318,596
effective HSP length: 105
effective length of query: 519
effective length of database: 8,513,101
effective search space: 4418299419
effective search space used: 4418299419
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0323a.7


