

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0322.4
(153 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
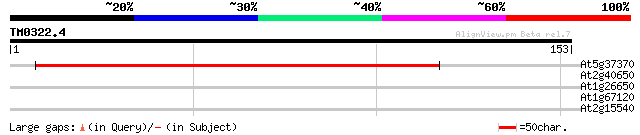
At5g37370 unknown protein 187 2e-48
At2g40650 unknown protein 38 0.002
At1g26650 unknown protein 32 0.18
At1g67120 hypothetical protein 26 7.4
At2g15540 putative non-LTR retroelement reverse transcriptase 26 9.7
>At5g37370 unknown protein
Length = 393
Score = 187 bits (475), Expect = 2e-48
Identities = 91/110 (82%), Positives = 96/110 (86%)
Query: 8 EIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRG 67
EIQ++GR ESLLEKVL MNILSSDYFKELY LKTY EVIDEIYNQV+HVEPWM GNCRG
Sbjct: 3 EIQSNGRAYESLLEKVLSMNILSSDYFKELYGLKTYHEVIDEIYNQVNHVEPWMGGNCRG 62
Query: 68 PSTFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPNTV 117
PST +CLLY FFTM LTVKQMHGLLKH DSPYIR VGFLYLRYV D T+
Sbjct: 63 PSTAYCLLYKFFTMKLTVKQMHGLLKHTDSPYIRAVGFLYLRYVADAKTL 112
>At2g40650 unknown protein
Length = 355
Score = 37.7 bits (86), Expect = 0.002
Identities = 24/93 (25%), Positives = 48/93 (50%), Gaps = 2/93 (2%)
Query: 17 ESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRGPSTFFCLLY 76
++L+EK++ I ++KE T ++D+ ++DH+ G+ R P+ F CL+
Sbjct: 18 QNLVEKIVRTKIYQHTFWKEQCFGLTAETLVDKAM-ELDHLGGTFGGS-RKPTPFLCLIL 75
Query: 77 *FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLR 109
+ + + +K+ D Y+R++G YLR
Sbjct: 76 KMLQIQPEKEIVVEFIKNDDYKYVRILGAFYLR 108
>At1g26650 unknown protein
Length = 335
Score = 31.6 bits (70), Expect = 0.18
Identities = 16/58 (27%), Positives = 29/58 (49%)
Query: 18 SLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRGPSTFFCLL 75
SLL K + + Y +E+ + +L ++ +I+ +V W+ G TFFC+L
Sbjct: 118 SLLSKAAVVYSVDCSYSREVVDISKFLVILQKIWRRVVFTYVWICILIVGCFTFFCVL 175
>At1g67120 hypothetical protein
Length = 5138
Score = 26.2 bits (56), Expect = 7.4
Identities = 25/84 (29%), Positives = 44/84 (51%), Gaps = 13/84 (15%)
Query: 8 EIQTSGRPIESL--LEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNC 65
++ T R ++SL LEK + I+ + F L +L Y EVID +H W +G
Sbjct: 2490 DVSTFTRFLDSLRVLEKKILCEIVGAPSFSVLIQL--YTEVID------NHSFFW-SGLV 2540
Query: 66 RGPSTFFCLLY*FFTMMLTVKQMH 89
+ LL+ F++++ ++K+MH
Sbjct: 2541 SSSDEY--LLFSFWSLIKSIKKMH 2562
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 25.8 bits (55), Expect = 9.7
Identities = 12/32 (37%), Positives = 19/32 (58%)
Query: 79 FTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRY 110
F ++L KQ+ L++ P+S RV+ Y RY
Sbjct: 741 FNLVLLAKQLWRLIRFPNSLLSRVLRGRYFRY 772
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.352 0.162 0.599
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,204,291
Number of Sequences: 26719
Number of extensions: 120807
Number of successful extensions: 464
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 461
Number of HSP's gapped (non-prelim): 5
length of query: 153
length of database: 11,318,596
effective HSP length: 90
effective length of query: 63
effective length of database: 8,913,886
effective search space: 561574818
effective search space used: 561574818
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0322.4


