

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0322.12
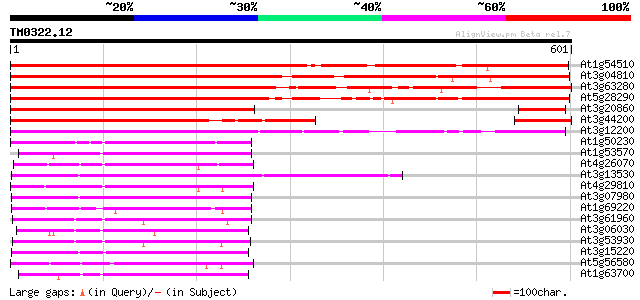
(601 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g54510 unknown protein 646 0.0
At3g04810 kinase like protein 634 0.0
At3g63280 unknown protein 626 e-180
At5g28290 serine (threonine) protein kinase - like 607 e-174
At3g20860 putative serine/threonine protein kinase 384 e-107
At3g44200 protein kinase-like protein 366 e-101
At3g12200 putative protein 324 1e-88
At1g50230 hypothetical protein 147 2e-35
At1g53570 MEK kinase MAP3Ka, putative 136 4e-32
At4g26070 MEK1 134 1e-31
At3g13530 MAP3K epsilon protein kinase 133 3e-31
At4g29810 MAP kinase kinase 2 132 6e-31
At3g07980 putative MAP3K epsilon protein kinase 132 7e-31
At1g69220 serine/threonine kinase (SIK1) 128 1e-29
At3g61960 serine/threonine-protein kinase-like protein 126 3e-29
At3g06030 NPK1-related protein kinase 3 125 7e-29
At3g53930 unknown protein 125 9e-29
At3g15220 putative MAP kinase 124 1e-28
At5g56580 protein kinase MEK1 homolog 122 6e-28
At1g63700 putative protein kinase 122 7e-28
>At1g54510 unknown protein
Length = 612
Score = 646 bits (1667), Expect = 0.0
Identities = 365/612 (59%), Positives = 431/612 (69%), Gaps = 36/612 (5%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYE LEQIGKGSFGSALLVRHKHEKK YVLKKIRLARQ+ RTRRSAHQEMELISK+R+
Sbjct: 1 MEQYEFLEQIGKGSFGSALLVRHKHEKKKYVLKKIRLARQTQRTRRSAHQEMELISKMRH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEK C+VCI++GYCE GD+A AIKK+NGV+F EEKLCKWLVQLLM L+YL
Sbjct: 61 PFIVEYKDSWVEKACYVCIVIGYCEGGDMAQAIKKSNGVHFQEEKLCKWLVQLLMGLEYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLTK QDIRLGDFGLAK+LTSDDL SS+VGTPSYMCPELLADIPY
Sbjct: 121 HSNHILHRDVKCSNIFLTKEQDIRLGDFGLAKILTSDDLTSSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCCIYEMA LKPAFKAFD+QALINKINK+IV+PLP YS FRGLVKSMLR
Sbjct: 181 GSKSDIWSLGCCIYEMAYLKPAFKAFDMQALINKINKTIVSPLPAKYSGPFRGLVKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPES-NYMRRTRFVVPDSVS 299
KNPE+RP AS+LL HPHLQPY+L + L+LN+ RR T P + P S M++ F P
Sbjct: 241 KNPEVRPSASDLLRHPHLQPYVLDVKLRLNNLRRKTLPPELPSSKRIMKKAHFSEPAVTC 300
Query: 300 TLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNTN 359
+ + SL NDR+LNP E+D+ A + S+ + ++S+ +
Sbjct: 301 PAFGERQHRSLWNDRALNPE---AEEDT------ASSIKCISRRISDLSIESSSKGTLIC 351
Query: 360 KTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTL-PVSHAPSV-KFPPPTRR 417
K S + + + K S +RR I +G + D L PVS + K P RR
Sbjct: 352 KQVSSSACKVSKYPLAKSSVTSRR-------IMETGRRSDHLHPVSGGGTTSKIIPSARR 404
Query: 418 ASHPLPTRASMAKATLYTN-VGTFPRVDS-DVSVNAPQIDKIAEFPLASCE-DLPFFPVH 474
S PL RA+ + Y VG V S + S+N PQ+DKIA FPLA E D+ F P+
Sbjct: 405 TSLPLTKRATNQEVAAYNPIVGILQNVKSPEYSINEPQVDKIAIFPLAPYEQDIFFTPMQ 464
Query: 475 VPSSTSVHCYSSSAGSAYRSITKEKCIHEDKVTVPG------GSCPKGSECS-KHVTAGV 527
+S S S+ + RSITK+KC + T G + GS S ++ TAG
Sbjct: 465 RKTS------SKSSSVSDRSITKDKCTVQTHTTWQGIQLNMVDNISDGSSSSDQNATAGA 518
Query: 528 SIHSSAELHQR-RFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPR 586
S H+++ +R RFD SSY+QRA+ALEGLLEFSARLLQ R+DEL VLLKPFG KVSPR
Sbjct: 519 SSHTTSSSSRRCRFDPSSYRQRADALEGLLEFSARLLQEGRYDELNVLLKPFGPGKVSPR 578
Query: 587 ETAIWLAKSFKE 598
ETAIW+AKS KE
Sbjct: 579 ETAIWIAKSLKE 590
>At3g04810 kinase like protein
Length = 606
Score = 634 bits (1634), Expect = 0.0
Identities = 360/619 (58%), Positives = 423/619 (68%), Gaps = 40/619 (6%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQ+ RTRRSAHQEMELISK+ N
Sbjct: 1 MENYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQTGRTRRSAHQEMELISKIHN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEKGC+VCII+GYC+ GD+A+AIKK NGV+F EEKLCKWLVQ+L+AL+YL
Sbjct: 61 PFIVEYKDSWVEKGCYVCIIIGYCKGGDMAEAIKKTNGVHFTEEKLCKWLVQILLALEYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLTK+QDIRLGDFGLAK+LTSDDLASS+VGTPSYMCPELLADIPY
Sbjct: 121 HANHILHRDVKCSNIFLTKDQDIRLGDFGLAKVLTSDDLASSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCC+YEM A+KPAFKAFD+Q LIN+IN+SIV PLP YS+AFRGLVKSMLR
Sbjct: 181 GSKSDIWSLGCCMYEMTAMKPAFKAFDMQGLINRINRSIVPPLPAQYSAAFRGLVKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP A+ELL P LQPYI KIHLK+N P + P QWPES RR F
Sbjct: 241 KNPELRPSAAELLRQPLLQPYIQKIHLKVNDPGSNVLPAQWPESESARRNSF-------- 292
Query: 301 LSDQDKCLSLSNDRSLNPS-ISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNTN 359
+ + S PS G +DS +K + + ++S T+
Sbjct: 293 --PEQRRRPAGKSHSFGPSRFRGNLEDSVSSIKKTVPAYLNRERQVDLS---------TD 341
Query: 360 KTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPP-TRRA 418
+ TV R + R + P + + S LPVS + P RRA
Sbjct: 342 ASGDGTVVRRTSEASKSSRYVPVRASASPVRPRQPRSDLGQLPVSSQLKNRKPAALIRRA 401
Query: 419 SHPLPTR-ASMAKATLYTNVGTF-PRVDS-DVSVNAPQIDKIAEFPLASCEDLPFFPV-- 473
S P + A K +LY + +F +++S DVS+NAP+IDKI EFPLAS E+ PF PV
Sbjct: 402 SMPSSRKPAKEIKDSLYISKTSFLHQINSPDVSMNAPRIDKI-EFPLASYEEEPFVPVVR 460
Query: 474 -HVPSSTSVHCYSSSAGSAYR-SITKEKCIHEDKVTVPGGSC-----------PKGSECS 520
++S YS SITK+K E G +
Sbjct: 461 GKKKKASSRGSYSPPPEPPLDCSITKDKFTLEPGQNREGAIMKAVYEEDAYLEDRSESSD 520
Query: 521 KHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGL 580
++ TAG S +S+ + ++RFD SSYQQRAEALEGLLEFSARLLQ +R+DEL VLL+PFG
Sbjct: 521 QNATAGASSRASSGVRRQRFDPSSYQQRAEALEGLLEFSARLLQDERYDELNVLLRPFGP 580
Query: 581 EKVSPRETAIWLAKSFKET 599
KVSPRETAIWL+KSFKET
Sbjct: 581 GKVSPRETAIWLSKSFKET 599
>At3g63280 unknown protein
Length = 555
Score = 626 bits (1615), Expect = e-180
Identities = 354/612 (57%), Positives = 416/612 (67%), Gaps = 68/612 (11%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+YEVLEQIGKGSFGSALLVRHK E+K YVLKKIRLARQSDR RRSAHQEMELIS VRN
Sbjct: 1 MERYEVLEQIGKGSFGSALLVRHKQERKKYVLKKIRLARQSDRARRSAHQEMELISTVRN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PF+VEYKDSWVEKGC+VCI++GYC+ GD+ D IK+A GV+FPEEKLC+WLVQLLMALDYL
Sbjct: 61 PFVVEYKDSWVEKGCYVCIVIGYCQGGDMTDTIKRACGVHFPEEKLCQWLVQLLMALDYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H NHILHRDVKCSNIFLTK QDIRLGDFGLAK+LTSDDL SS+VGTPSYMCPELLADIPY
Sbjct: 121 HSNHILHRDVKCSNIFLTKEQDIRLGDFGLAKILTSDDLTSSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCC+YEMAA KP FKA D+Q LI KI+K I+ P+P MYS +FRGL+KSMLR
Sbjct: 181 GSKSDIWSLGCCMYEMAAHKPPFKASDVQTLITKIHKLIMDPIPAMYSGSFRGLIKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP A+ELLNHPHLQPYI +++KL SPRRSTFP Q+ E + +T
Sbjct: 241 KNPELRPSANELLNHPHLQPYISMVYMKLESPRRSTFPLQFSERD-------------AT 287
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLST-SSKEVYEVSVGGACGPWNTN 359
L ++ + S SNDR LNPS+S TE S + KA + ++V EV+VG
Sbjct: 288 LKERRRS-SFSNDRRLNPSVSDTEAGSVSSSGKASPTPMFNGRKVSEVTVG--------- 337
Query: 360 KTKSPTVERTPRSRADKDSTAARRQ----TMGPSKISRSGSKHDTLPVSHAPSVKFPPPT 415
+ + A K S AAR T + R+ KH+ + VS+ +
Sbjct: 338 -VVREEIVPQRQEEAKKQSGAARTPRVAGTSAKASTQRTVFKHELMKVSNPTERR----- 391
Query: 416 RRASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEFP-----LASCEDLP 469
RR S PL T +++ ++S DVSVN P+ DKIAEFP + E
Sbjct: 392 RRVSLPLVVENPY---TYESDITALCSLNSPDVSVNTPRFDKIAEFPEDIFQNQNRETAS 448
Query: 470 FFPVHVPSSTSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGGSCPKGSECSKHVTAGVSI 529
V S +S C ++ SITK+KC V
Sbjct: 449 RREVARHSFSSPPCPPHGEDNSNGSITKDKCT-------------------------VQK 483
Query: 530 HSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETA 589
S +E+ QRRFDTSSYQQRAEALEGLLEFSA+LLQ +R+DELGVLLKPFG E+VS RETA
Sbjct: 484 RSVSEVKQRRFDTSSYQQRAEALEGLLEFSAKLLQQERYDELGVLLKPFGAERVSSRETA 543
Query: 590 IWLAKSFKETVI 601
IWL KSFKE +
Sbjct: 544 IWLTKSFKEASV 555
>At5g28290 serine (threonine) protein kinase - like
Length = 568
Score = 607 bits (1566), Expect = e-174
Identities = 354/608 (58%), Positives = 420/608 (68%), Gaps = 57/608 (9%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQ+ RTRRSAHQEMELISK+RN
Sbjct: 1 MEHYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQTGRTRRSAHQEMELISKIRN 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
PFIVEYKDSWVEKGC+VCI++GYC+ GD+A+AIKKANGV F EEKLCKWLVQLLMAL+YL
Sbjct: 61 PFIVEYKDSWVEKGCYVCIVIGYCKGGDMAEAIKKANGVEFSEEKLCKWLVQLLMALEYL 120
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H +HILHRDVKCSNIFLTK+QDIRLGDFGLAK+LTSDDLASS+VGTPSYMCPELLADIPY
Sbjct: 121 HASHILHRDVKCSNIFLTKDQDIRLGDFGLAKILTSDDLASSVVGTPSYMCPELLADIPY 180
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
GSKSDIWSLGCC+YEM ALKPAFKAFD+Q LIN+IN+SIVAPLP YS+AFR LVKSMLR
Sbjct: 181 GSKSDIWSLGCCMYEMTALKPAFKAFDMQGLINRINRSIVAPLPAQYSTAFRSLVKSMLR 240
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
KNPELRP AS+LL P LQPY+ K+ LKL+ T P S RR+ +
Sbjct: 241 KNPELRPSASDLLRQPLLQPYVQKVLLKLSFREHDTLP-----SESERRSSY-------- 287
Query: 301 LSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNTNK 360
Q + + S PS G +Q+ + K P +T
Sbjct: 288 --PQQRKRTSGKSVSFGPSRFGVDQEDSVSSVK---------------------PVHTYL 324
Query: 361 TKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPV-SHAPS----VKFPPPT 415
+ V+ S D RR + S + + SK+ +PV S+ P +K T
Sbjct: 325 HRHRPVD---LSANDTSRVVVRRPAV--SSVVSNSSKY--VPVRSNQPKSGGLLKPAVVT 377
Query: 416 RRASHPLPTR-ASMAKATLY-TNVGTFPRVDS-DVSVNAPQIDKIAEFPLASCEDLPFFP 472
RRAS P+ + A K +LY N+G +++S DVSVN+P+ID+I +FPLAS E++PF P
Sbjct: 378 RRASLPISQKPAKGTKDSLYHPNIGILHQLNSPDVSVNSPRIDRI-KFPLASYEEMPFIP 436
Query: 473 VHVPSSTSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGGSCPKGSECSKHVTAGVSIHSS 532
V S S + S + I +DK T+ K ++ TAG S +S
Sbjct: 437 VVRKKKGS----SRGSYSPPPEPPLDCSITKDKFTLEPERETKSDLSDQNATAGASSRAS 492
Query: 533 AELHQR-RFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIW 591
+ +R RFD SSY+QRAEALEGLLEFSARLL +R+DEL VLLKPFG KVSPRETAIW
Sbjct: 493 SGASRRQRFDPSSYRQRAEALEGLLEFSARLLIDERYDELNVLLKPFGPGKVSPRETAIW 552
Query: 592 LAKSFKET 599
L+KSFKE+
Sbjct: 553 LSKSFKES 560
>At3g20860 putative serine/threonine protein kinase
Length = 427
Score = 384 bits (987), Expect = e-107
Identities = 181/262 (69%), Positives = 224/262 (85%), Gaps = 1/262 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+ YEV+EQIG+G+FGSA LV HK E++ YV+KKIRLA+Q++R + +A QEM LISK+++
Sbjct: 12 MDDYEVVEQIGRGAFGSAFLVIHKSERRKYVVKKIRLAKQTERCKLAAIQEMSLISKLKS 71
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVEYKDSWVEK C VCI+ YCE GD+ IKK+ GV EEKLC+W+VQLL+A+DYL
Sbjct: 72 PYIVEYKDSWVEKDC-VCIVTSYCEGGDMTQMIKKSRGVFASEEKLCRWMVQLLLAIDYL 130
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N +LHRD+KCSNIFLTK ++RLGDFGLAK+L DDLASS+VGTP+YMCPELLADIPY
Sbjct: 131 HNNRVLHRDLKCSNIFLTKENEVRLGDFGLAKLLGKDDLASSMVGTPNYMCPELLADIPY 190
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC++E+AA +PAFKA D+ ALINKIN+S ++PLP MYSS+ + L+KSMLR
Sbjct: 191 GYKSDIWSLGCCMFEVAAHQPAFKAPDMAALINKINRSSLSPLPVMYSSSLKRLIKSMLR 250
Query: 241 KNPELRPPASELLNHPHLQPYI 262
KNPE RP A+ELL HPHLQPY+
Sbjct: 251 KNPEHRPTAAELLRHPHLQPYL 272
Score = 65.1 bits (157), Expect = 1e-10
Identities = 33/50 (66%), Positives = 39/50 (78%)
Query: 546 QQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 595
++RAEALE LLE A LL+ ++FDEL +LKPFG E VS RETAIWL KS
Sbjct: 363 EERAEALESLLELCAGLLRQEKFDELEGVLKPFGDETVSSRETAIWLTKS 412
>At3g44200 protein kinase-like protein
Length = 941
Score = 366 bits (940), Expect = e-101
Identities = 189/329 (57%), Positives = 243/329 (73%), Gaps = 19/329 (5%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+QYE++EQIG+G+FG+A+LV HK E+K YVLKKIRLARQ++R RRSAHQEM LI++V++
Sbjct: 5 MDQYELMEQIGRGAFGAAILVHHKAERKKYVLKKIRLARQTERCRRSAHQEMSLIARVQH 64
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVE+K++WVEKGC+VCI+ GYCE GD+A+ +KK+NGV FPEEKLCKW QLL+A++YL
Sbjct: 65 PYIVEFKEAWVEKGCYVCIVTGYCEGGDMAELMKKSNGVYFPEEKLCKWFTQLLLAVEYL 124
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N++LHRD+KCSNIFLTK+QD+RLGDFGLAK L +DDL SS+VGTP+YMCPELLADIPY
Sbjct: 125 HSNYVLHRDLKCSNIFLTKDQDVRLGDFGLAKTLKADDLTSSVVGTPNYMCPELLADIPY 184
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCCIYEMAA +PAFKAFD+ LI+K S +G VK R
Sbjct: 185 GFKSDIWSLGCCIYEMAAYRPAFKAFDMAGLISK-------------KSTHQGNVKEEPR 231
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVST 300
+ + ASE+L HP+LQPY+ + L++ S P + S RR+ +S S+
Sbjct: 232 VS--AKRMASEILKHPYLQPYVEQYRPTLSA--ASITPEKPLNSREGRRSMAESQNSNSS 287
Query: 301 LSDQDKCLSLSNDRSLNPSISG--TEQDS 327
+ +S N R + PS TE DS
Sbjct: 288 SEKDNFYVSDKNIRYVVPSNGNKVTETDS 316
Score = 81.6 bits (200), Expect = 1e-15
Identities = 40/61 (65%), Positives = 48/61 (78%)
Query: 541 DTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKSFKETV 600
D S++QRAEALEGLLE SA LL+ R +EL ++L+PFG KVSPRETAIWLAKS K +
Sbjct: 865 DIKSFRQRAEALEGLLELSADLLEQSRLEELAIVLQPFGKNKVSPRETAIWLAKSLKGMM 924
Query: 601 I 601
I
Sbjct: 925 I 925
>At3g12200 putative protein
Length = 571
Score = 324 bits (830), Expect = 1e-88
Identities = 209/597 (35%), Positives = 317/597 (53%), Gaps = 54/597 (9%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
++ Y V+EQ+ +G S +V H E K Y +KKI LA+ +D+ +++A QEM+L+S ++N
Sbjct: 16 LDNYHVVEQVRRGKSSSDFVVLHDIEDKKYAMKKICLAKHTDKLKQTALQEMKLLSSLKN 75
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IV Y+DSW++ CI Y E G++A+AIKKA G FPEE++ KWL QLL+A++YL
Sbjct: 76 PYIVHYEDSWIDNDNNACIFTAYYEGGNMANAIKKARGKLFPEERIFKWLAQLLLAVNYL 135
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H N ++H D+ CSNIFL K+ ++LG++GLAK++ + S + G + MCPE+L D PY
Sbjct: 136 HSNRVVHMDLTCSNIFLPKDDHVQLGNYGLAKLINPEKPVSMVSGISNSMCPEVLEDQPY 195
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
G KSDIWSLGCC+YE+ A +PAFKA D+ LINKIN+S+++PLP +YSS + ++K MLR
Sbjct: 196 GYKSDIWSLGCCMYEITAHQPAFKAPDMAGLINKINRSLMSPLPIVYSSTLKQMIKLMLR 255
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSV-- 298
K PE RP A ELL +P LQPY+L+ L+ FP + S + R +P
Sbjct: 256 KKPEYRPTACELLRNPSLQPYLLQCQ-NLSPIYLPVFPIKPVNSPKDKARRNSLPGKFGK 314
Query: 299 STLSDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNT 358
+S + +S S + +L P + TE S +Q A + + ++ + +C +T
Sbjct: 315 ERVSREKSEVSRSLE-NLYPFWTNTETGSSSSSQPASSTNGAEDKLETKRIDPSC---DT 370
Query: 359 NKTKSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPPTRRA 418
K T +++ S D D +T P
Sbjct: 371 LKISEFTSQKSDESLIDPDIAVYSTET----------------------------PAEEN 402
Query: 419 SHPLPTRASMAKATLYTNVGTFPRVDSDVSVNAPQIDKIAEFPLASCEDLPFFPVHVPSS 478
+ P T ++ + +V +V+ + P+ + AE A + P + VP S
Sbjct: 403 ALPKETENIFSEESQLRDVDVGVVSAQEVACSPPRAIEEAETQEALPK--PKEQITVPIS 460
Query: 479 TSVHCYSSSAGSAYRSITKEKCIHEDKVTVPGGSCPKGSECSKHVTAGVSIHSSAELHQR 538
+ H SS A + DK +K V S SS
Sbjct: 461 SVAH---SSTEVAAAKDHLSGSLEGDK--------------AKMVKLTASEMSSVLSKLT 503
Query: 539 RFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 595
+ ++RA+ALE LLE A L++ ++++EL LL PFG + VS R+TAIW AK+
Sbjct: 504 KLGPPQSKERADALECLLEKCAGLVKQEKYEELAGLLTPFGEDGVSARDTAIWFAKT 560
>At1g50230 hypothetical protein
Length = 269
Score = 147 bits (370), Expect = 2e-35
Identities = 91/260 (35%), Positives = 141/260 (54%), Gaps = 6/260 (2%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
+E Y V+E +G+GSFG R K+ + +K I ++D+ S QE+E++ K+++
Sbjct: 3 VEDYHVIELVGEGSFGRVYKGRRKYTGQTVAMKFIMKQGKTDKDIHSLRQEIEILRKLKH 62
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
I+E DS+ E C++ + + G+L + ++ + PEE++ QL+ ALDYL
Sbjct: 63 ENIIEMLDSF-ENAREFCVVTEFAQ-GELFEILEDDKCL--PEEQVQAIAKQLVKALDYL 118
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAK-MLTSDDLASSIVGTPSYMCPELLADIP 179
H N I+HRD+K NI + ++L DFG A+ M T+ + SI GTP YM PEL+ + P
Sbjct: 119 HSNRIIHRDMKPQNILIGAGSVVKLCDFGFARAMSTNTVVLRSIKGTPLYMAPELVKEQP 178
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y D+WSLG +YE+ +P F + ALI I K V P S+ F +K +L
Sbjct: 179 YDRTVDLWSLGVILYELYVGQPPFYTNSVYALIRHIVKDPV-KYPDEMSTYFESFLKGLL 237
Query: 240 RKNPELRPPASELLNHPHLQ 259
K P R L HP ++
Sbjct: 238 NKEPHSRLTWPALREHPFVK 257
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 136 bits (342), Expect = 4e-32
Identities = 82/255 (32%), Positives = 141/255 (55%), Gaps = 8/255 (3%)
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTR---RSAHQEMELISKVRNPFIVEY 66
+G G+FG L + + K+ +K++++ ++ + +QE+ L++++ +P IV+Y
Sbjct: 220 LGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQLNQEINLLNQLCHPNIVQY 279
Query: 67 KDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHIL 126
S + + + + + Y G + +K +F E + + Q+L L YLH + +
Sbjct: 280 YGSELSEET-LSVYLEYVSGGSIHKLLKDYG--SFTEPVIQNYTRQILAGLAYLHGRNTV 336
Query: 127 HRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPE-LLADIPYGSKSD 185
HRD+K +NI + N +I+L DFG+AK +T+ S G+P +M PE +++ Y D
Sbjct: 337 HRDIKGANILVDPNGEIKLADFGMAKHVTAFSTMLSFKGSPYWMAPEVVMSQNGYTHAVD 396
Query: 186 IWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAP-LPTMYSSAFRGLVKSMLRKNPE 244
IWSLGC I EMA KP + F+ A I KI S P +P S+ + ++ L++NP
Sbjct: 397 IWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDHLSNDAKNFIRLCLQRNPT 456
Query: 245 LRPPASELLNHPHLQ 259
+RP AS+LL HP L+
Sbjct: 457 VRPTASQLLEHPFLR 471
>At4g26070 MEK1
Length = 354
Score = 134 bits (337), Expect = 1e-31
Identities = 94/268 (35%), Positives = 146/268 (54%), Gaps = 16/268 (5%)
Query: 5 EVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFIV 64
EV++ IGKGS G+ LV+HK ++ + LK I+L + + T R+ QE+ + + P++V
Sbjct: 69 EVIKVIGKGSSGNVQLVKHKLTQQFFALKVIQLNTE-ESTCRAISQELRINLSSQCPYLV 127
Query: 65 EYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH-VN 123
S+ G V II+ + + G LAD +KK V PE L ++L L Y+H
Sbjct: 128 SCYQSFYHNG-LVSIILEFMDGGSLADLLKKVGKV--PENMLSAICKRVLRGLCYIHHER 184
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTS-DDLASSIVGTPSYMCPELLADIPYGS 182
I+HRD+K SN+ + ++++ DFG++K+LTS LA+S VGT YM PE ++ Y +
Sbjct: 185 RIIHRDLKPSNLLINHRGEVKITDFGVSKILTSTSSLANSFVGTYPYMSPERISGSLYSN 244
Query: 183 KSDIWSLGCCIYEMAALK-----PAFKA--FDIQALINKI--NKSIVAPLPTMYSSAFRG 233
KSDIWSLG + E A K P K + L++ I N AP ++S F
Sbjct: 245 KSDIWSLGLVLLECATGKFPYTPPEHKKGWSSVYELVDAIVENPPPCAP-SNLFSPEFCS 303
Query: 234 LVKSMLRKNPELRPPASELLNHPHLQPY 261
+ ++K+P R A ELL H ++ +
Sbjct: 304 FISQCVQKDPRDRKSAKELLEHKFVKMF 331
>At3g13530 MAP3K epsilon protein kinase
Length = 1368
Score = 133 bits (334), Expect = 3e-31
Identities = 103/419 (24%), Positives = 183/419 (43%), Gaps = 5/419 (1%)
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
+Y + ++IGKG++G +K++ L + QE++L+ + +
Sbjct: 19 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIVQEDLNTIMQEIDLLKNLNHKN 78
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHV 122
IV+Y S K + II+ Y E G LA+ IK FPE + ++ Q+L L YLH
Sbjct: 79 IVKYLGSSKTK-THLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHE 137
Query: 123 NHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDL-ASSIVGTPSYMCPELLADIPYG 181
++HRD+K +NI TK ++L DFG+A L D+ S+VGTP +M PE++
Sbjct: 138 QGVIHRDIKGANILTTKEGLVKLADFGVATKLNEADVNTHSVVGTPYWMAPEVIEMSGVC 197
Query: 182 SKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRK 241
+ SDIWS+GC + E+ P + + +I + P+P S ++ +K
Sbjct: 198 AASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDDNPPIPDSLSPDITDFLRQCFKK 257
Query: 242 NPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVSTL 301
+ RP A LL+HP ++ + L T + + + D+ +L
Sbjct: 258 DSRQRPDAKTLLSHPWIRNSRRALQSSLR--HSGTIKYMKEATASSEKDDEGSQDAAESL 315
Query: 302 SDQDKCLSLSNDRSLNPSISGTEQDSQCFTQKAHGLSTSSKEVYEVSVGGACGPWNTNKT 361
S ++ +S ++ +S P + + S+ L + E + P +
Sbjct: 316 SGENVGISKTDSKSKLPLVGVSSFRSEKDQSTPSDLGEEGTDNSEDDIMSDQVPTLSIHE 375
Query: 362 KSPTVERTPRSRADKDSTAARRQTMGPSKISRSGSKHDTLPVSHAPSVKFPPPTRRASH 420
KS + TP+ +D + R +T S ++ +T + H + P + SH
Sbjct: 376 KSSDAKGTPQDVSDFHGKSERGETPENLVTETSEARKNTSAIKHV-GKELSIPVDQTSH 433
>At4g29810 MAP kinase kinase 2
Length = 363
Score = 132 bits (332), Expect = 6e-31
Identities = 85/271 (31%), Positives = 144/271 (52%), Gaps = 14/271 (5%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
+ ++++ IGKGS G LV+HK + + LK I+L + R++ QE+++ +
Sbjct: 67 LSDLDMVKVIGKGSSGVVQLVQHKWTGQFFALKVIQL-NIDEAIRKAIAQELKINQSSQC 125
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P +V S+ + G + +I+ Y + G LAD +K + P+ L Q+L L YL
Sbjct: 126 PNLVTSYQSFYDNGA-ISLILEYMDGGSLADFLKSVKAI--PDSYLSAIFRQVLQGLIYL 182
Query: 121 HVN-HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADI 178
H + HI+HRD+K SN+ + ++++ DFG++ ++T+ LA++ VGT +YM PE +
Sbjct: 183 HHDRHIIHRDLKPSNLLINHRGEVKITDFGVSTVMTNTAGLANTFVGTYNYMSPERIVGN 242
Query: 179 PYGSKSDIWSLGCCIYEMAALK----PAFKAFDIQALINKINKSIVAPLPTM----YSSA 230
YG+KSDIWSLG + E A K P + ++ + + P P + +S
Sbjct: 243 KYGNKSDIWSLGLVVLECATGKFPYAPPNQEETWTSVFELMEAIVDQPPPALPSGNFSPE 302
Query: 231 FRGLVKSMLRKNPELRPPASELLNHPHLQPY 261
+ + L+K+P R A EL+ HP L Y
Sbjct: 303 LSSFISTCLQKDPNSRSSAKELMEHPFLNKY 333
>At3g07980 putative MAP3K epsilon protein kinase
Length = 1367
Score = 132 bits (331), Expect = 7e-31
Identities = 79/258 (30%), Positives = 129/258 (49%), Gaps = 2/258 (0%)
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
+Y + ++IGKG++G + +K++ L + QE++L+ + +
Sbjct: 19 KYMLGDEIGKGAYGRVYIGLDLENGDFVAIKQVSLENIGQEDLNTIMQEIDLLKNLNHKN 78
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHV 122
IV+Y S K + II+ Y E G LA+ IK FPE + ++ Q+L L YLH
Sbjct: 79 IVKYLGSLKTK-THLHIILEYVENGSLANIIKPNKFGPFPESLVTVYIAQVLEGLVYLHE 137
Query: 123 NHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDL-ASSIVGTPSYMCPELLADIPYG 181
++HRD+K +NI TK ++L DFG+A L D S+VGTP +M PE++
Sbjct: 138 QGVIHRDIKGANILTTKEGLVKLADFGVATKLNEADFNTHSVVGTPYWMAPEVIELSGVC 197
Query: 182 SKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRK 241
+ SDIWS+GC I E+ P + + +I + P+P S ++ +K
Sbjct: 198 AASDIWSVGCTIIELLTCVPPYYDLQPMPALYRIVQDDTPPIPDSLSPDITDFLRLCFKK 257
Query: 242 NPELRPPASELLNHPHLQ 259
+ RP A LL+HP ++
Sbjct: 258 DSRQRPDAKTLLSHPWIR 275
>At1g69220 serine/threonine kinase (SIK1)
Length = 836
Score = 128 bits (321), Expect = 1e-29
Identities = 82/268 (30%), Positives = 138/268 (50%), Gaps = 22/268 (8%)
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
+YE L ++GKGS+GS R ++ +K I L + + E+E++ + +P
Sbjct: 248 KYEFLNELGKGSYGSVYKARDLKTSEIVAVKVISLT-EGEEGYEEIRGEIEMLQQCNHPN 306
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLV-----QLLMAL 117
+V Y S+ + ++ I++ YC G +AD + N EE L ++ + + L L
Sbjct: 307 VVRYLGSYQGED-YLWIVMEYCGGGSVADLM------NVTEEALEEYQIAYICREALKGL 359
Query: 118 DYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLT-SDDLASSIVGTPSYMCPELLA 176
YLH + +HRD+K NI LT+ +++LGDFG+A LT + ++ +GTP +M PE++
Sbjct: 360 AYLHSIYKVHRDIKGGNILLTEQGEVKLGDFGVAAQLTRTMSKRNTFIGTPHWMAPEVIQ 419
Query: 177 DIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTM-----YSSAF 231
+ Y K D+W+LG EMA P + ++ I+ + P P + +S F
Sbjct: 420 ENRYDGKVDVWALGVSAIEMAEGLPPRSSVHPMRVLFMIS---IEPAPMLEDKEKWSLVF 476
Query: 232 RGLVKSMLRKNPELRPPASELLNHPHLQ 259
V L K P LRP A+E+L H ++
Sbjct: 477 HDFVAKCLTKEPRLRPTAAEMLKHKFVE 504
>At3g61960 serine/threonine-protein kinase-like protein
Length = 626
Score = 126 bits (317), Expect = 3e-29
Identities = 76/263 (28%), Positives = 134/263 (50%), Gaps = 10/263 (3%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y + +IG GSF L +H+ +K+I S + R + +E+ ++S + +P I
Sbjct: 10 YALGPRIGSGSFAVVWLAKHRSSGLEVAVKEIDKKLLSPKVRDNLLKEISILSTIDHPNI 69
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
+ + ++ +E G + +++ YC GDLA I + V PE ++ QL + L L
Sbjct: 70 IRFYEA-IETGDRIFLVLEYCSGGDLAGYINRHGKV--PEAVAKHFMRQLALGLQVLQEK 126
Query: 124 HILHRDVKCSNIFLTKNQD---IRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H +HRD+K N+ L+ + +++GDFG A+ LT + +A + G+P YM PE++ + Y
Sbjct: 127 HFIHRDLKPQNLLLSSKEVTPLLKIGDFGFARSLTPESMAETFCGSPLYMAPEIIRNQKY 186
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFR----GLVK 236
+K+D+WS G ++++ KP F + L + I + P + L +
Sbjct: 187 DAKADLWSAGAILFQLVTGKPPFDGNNHIQLFHNIVRDTELKFPEDTRNEIHPDCVDLCR 246
Query: 237 SMLRKNPELRPPASELLNHPHLQ 259
S+LR+NP R E NH L+
Sbjct: 247 SLLRRNPIERLTFREFFNHMFLR 269
>At3g06030 NPK1-related protein kinase 3
Length = 651
Score = 125 bits (314), Expect = 7e-29
Identities = 83/260 (31%), Positives = 140/260 (52%), Gaps = 14/260 (5%)
Query: 8 EQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQS---DRTR---RSAHQEMELISKVRNP 61
E IG G+FG + + +L +K++ +A S ++T+ R +E++L+ + +P
Sbjct: 72 ELIGCGAFGRVYMGMNLDSGELLAIKQVLIAPSSASKEKTQGHIRELEEEVQLLKNLSHP 131
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH 121
IV Y + V + + I++ + G ++ ++K +FPE + + QLL+ L+YLH
Sbjct: 132 NIVRYLGT-VRESDSLNILMEFVPGGSISSLLEKFG--SFPEPVIIMYTKQLLLGLEYLH 188
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLGDFGLAKM---LTSDDLASSIVGTPSYMCPELLADI 178
N I+HRD+K +NI + IRL DFG +K L + + A S+ GTP +M PE++
Sbjct: 189 NNGIMHRDIKGANILVDNKGCIRLADFGASKKVVELATVNGAKSMKGTPYWMAPEVILQT 248
Query: 179 PYGSKSDIWSLGCCIYEMAALKPAF-KAFDIQALINKINKSIV-APLPTMYSSAFRGLVK 236
+ +DIWS+GC + EMA KP + + + A + I ++ P+P S + +
Sbjct: 249 GHSFSADIWSVGCTVIEMATGKPPWSEQYQQFAAVLHIGRTKAHPPIPEDLSPEAKDFLM 308
Query: 237 SMLRKNPELRPPASELLNHP 256
L K P LR A+ELL HP
Sbjct: 309 KCLHKEPSLRLSATELLQHP 328
>At3g53930 unknown protein
Length = 711
Score = 125 bits (313), Expect = 9e-29
Identities = 78/261 (29%), Positives = 135/261 (50%), Gaps = 9/261 (3%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y V QIG GSF RH + +K+I +AR + + + S E+ ++ K+ +P I
Sbjct: 20 YAVGRQIGSGSFSVVWEGRHLVHGTVVAIKEIAMARLNKKLQESLMSEIIILRKINHPNI 79
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
+ + D +E + +++ YC+ GDL+ I K V PE +++QL L L N
Sbjct: 80 IRFIDM-IEAPGKINLVLEYCKGGDLSMYIHKHGSV--PEATAKHFMLQLAAGLQVLRDN 136
Query: 124 HILHRDVKCSNIFLTKNQD---IRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
+I+HRD+K N+ L+ + + +++ DFG A+ L LA ++ G+P YM PE++ Y
Sbjct: 137 NIIHRDLKPQNLLLSTDDNDAALKIADFGFARSLQPRGLAETLCGSPLYMAPEIMQLQKY 196
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPT---MYSSAFRGLVKS 237
+K+D+WS+G ++++ + F L+ I +S P S+ + L +
Sbjct: 197 DAKADLWSVGAILFQLVTGRTPFTGNSQIQLLQNIIRSTELHFPADCRDLSTDCKDLCQK 256
Query: 238 MLRKNPELRPPASELLNHPHL 258
+LR+NP R E +HP L
Sbjct: 257 LLRRNPVERLTFEEFFHHPFL 277
>At3g15220 putative MAP kinase
Length = 690
Score = 124 bits (312), Expect = 1e-28
Identities = 81/255 (31%), Positives = 131/255 (50%), Gaps = 6/255 (2%)
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
++ +E IG+GSFG K K +K I L D +E+ ++S+ R P+
Sbjct: 14 RFSQIELIGRGSFGDVYKAFDKDLNKEVAIKVIDLEESEDEIE-DIQKEISVLSQCRCPY 72
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHV 122
I EY S++ + + II+ Y G +AD ++ N ++ E + LL A++YLH
Sbjct: 73 ITEYYGSYLHQ-TKLWIIMEYMAGGSVADLLQSNNPLD--ETSIACITRDLLHAVEYLHN 129
Query: 123 NHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLT-SDDLASSIVGTPSYMCPELLADIP-Y 180
+HRD+K +NI L++N D+++ DFG++ LT + + VGTP +M PE++ + Y
Sbjct: 130 EGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQNSEGY 189
Query: 181 GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLR 240
K+DIWSLG + EMA +P ++ I + L +S + V L+
Sbjct: 190 NEKADIWSLGITVIEMAKGEPPLADLHPMRVLFIIPRETPPQLDEHFSRQVKEFVSLCLK 249
Query: 241 KNPELRPPASELLNH 255
K P RP A EL+ H
Sbjct: 250 KAPAERPSAKELIKH 264
>At5g56580 protein kinase MEK1 homolog
Length = 356
Score = 122 bits (306), Expect = 6e-28
Identities = 86/272 (31%), Positives = 144/272 (52%), Gaps = 17/272 (6%)
Query: 2 EQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELI-SKVRN 60
E E ++ IGKGS G LVRHK K + +K I++ Q + R+ QE+++ + +
Sbjct: 68 EDLETVKVIGKGSGGVVQLVRHKWVGKFFAMKVIQMNIQEE-IRKQIVQELKINQASSQC 126
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPE-EKLCKWLVQLLMALDY 119
P +V S+ G F +++ Y +RG LAD I++ + P +CK Q+L+ L Y
Sbjct: 127 PHVVVCYHSFYHNGAF-SLVLEYMDRGSLADVIRQVKTILEPYLAVVCK---QVLLGLVY 182
Query: 120 LH-VNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLAD 177
LH H++HRD+K SN+ + ++++ DFG++ L S + VGT +YM PE ++
Sbjct: 183 LHNERHVIHRDIKPSNLLVNHKGEVKISDFGVSASLASSMGQRDTFVGTYNYMSPERISG 242
Query: 178 IPYGSKSDIWSLGCCIYEMAALK-PAFKAFDIQ---ALINKINKSIVAPLPT----MYSS 229
Y SDIWSLG + E A + P ++ D Q + + + P PT +S
Sbjct: 243 STYDYSSDIWSLGMSVLECAIGRFPYLESEDQQNPPSFYELLAAIVENPPPTAPSDQFSP 302
Query: 230 AFRGLVKSMLRKNPELRPPASELLNHPHLQPY 261
F V + ++K+P R + +LL+HP ++ +
Sbjct: 303 EFCSFVSACIQKDPPARASSLDLLSHPFIKKF 334
>At1g63700 putative protein kinase
Length = 883
Score = 122 bits (305), Expect = 7e-28
Identities = 77/252 (30%), Positives = 135/252 (53%), Gaps = 10/252 (3%)
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQ---EMELISKVRNPFIVEY 66
+G GSFG L + ++ +K++ L ++R SA Q E+ ++S++R+ IV+Y
Sbjct: 406 LGMGSFGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRLRHQNIVQY 465
Query: 67 KDS-WVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHI 125
S V+ + I + Y G + +++ F E + + Q+L L YLH +
Sbjct: 466 YGSETVDDKLY--IYLEYVSGGSIYKLLQEYG--QFGENAIRNYTQQILSGLAYLHAKNT 521
Query: 126 LHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPYGSKS- 184
+HRD+K +NI + + +++ DFG+AK +T+ S G+P +M PE++ + + +
Sbjct: 522 VHRDIKGANILVDPHGRVKVADFGMAKHITAQSGPLSFKGSPYWMAPEVIKNSNGSNLAV 581
Query: 185 DIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAP-LPTMYSSAFRGLVKSMLRKNP 243
DIWSLGC + EMA KP + ++ + KI S P +P S + V+ L++NP
Sbjct: 582 DIWSLGCTVLEMATTKPPWSQYEGVPAMFKIGNSKELPDIPDHLSEEGKDFVRKCLQRNP 641
Query: 244 ELRPPASELLNH 255
RP A++LL+H
Sbjct: 642 ANRPTAAQLLDH 653
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,672,769
Number of Sequences: 26719
Number of extensions: 600844
Number of successful extensions: 3998
Number of sequences better than 10.0: 981
Number of HSP's better than 10.0 without gapping: 838
Number of HSP's successfully gapped in prelim test: 143
Number of HSP's that attempted gapping in prelim test: 1789
Number of HSP's gapped (non-prelim): 1144
length of query: 601
length of database: 11,318,596
effective HSP length: 105
effective length of query: 496
effective length of database: 8,513,101
effective search space: 4222498096
effective search space used: 4222498096
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0322.12


