

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.5
(1411 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
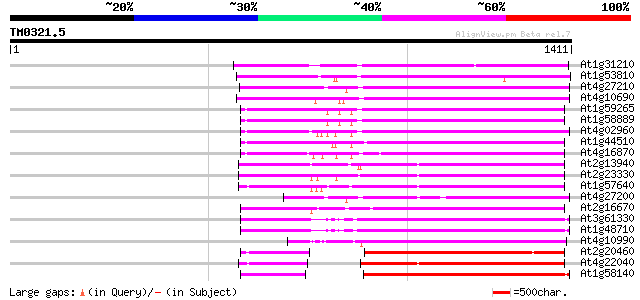
Score E
Sequences producing significant alignments: (bits) Value
At1g31210 putative reverse transcriptase 711 0.0
At1g53810 701 0.0
At4g27210 putative protein 698 0.0
At4g10690 retrotransposon like protein 678 0.0
At1g59265 polyprotein, putative 649 0.0
At1g58889 polyprotein, putative 648 0.0
At4g02960 putative polyprotein of LTR transposon 647 0.0
At1g44510 polyprotein, putative 636 0.0
At4g16870 retrotransposon like protein 634 0.0
At2g13940 putative retroelement pol polyprotein 573 e-163
At2g23330 putative retroelement pol polyprotein 563 e-160
At1g57640 548 e-156
At4g27200 putative protein 541 e-153
At2g16670 putative retroelement pol polyprotein 528 e-150
At3g61330 copia-type polyprotein 497 e-140
At1g48710 hypothetical protein 496 e-140
At4g10990 putative retrotransposon polyprotein 439 e-123
At2g20460 putative retroelement pol polyprotein 429 e-120
At4g22040 LTR retrotransposon like protein 422 e-118
At1g58140 hypothetical protein 422 e-118
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 711 bits (1835), Expect = 0.0
Identities = 379/850 (44%), Positives = 510/850 (59%), Gaps = 41/850 (4%)
Query: 562 FL*CSCSLQSVCGKSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGR 621
FL S Q + ++ IKVFQSD G EF +NK+ T + G+ HR SCP+T QNG
Sbjct: 556 FLSVFISFQKLVENQLNTKIKVFQSDGGGEFVSNKLKTHLSEHGIHHRISCPYTPQQNGL 615
Query: 622 VEQKHRHVLELGLAMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPT 681
E+KHRH++ELGL+ML+HSH P ++WV++F TA YIINR+PS VL N P++ LF P
Sbjct: 616 AERKHRHLVELGLSMLFHSHTPQKFWVESFFTANYIINRLPSSVLKNLSPYEALFGEKPD 675
Query: 682 YANFHPFGCRVFPCLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQF 741
Y++ FG +PCLRP NKF PRS C+F GY+S +KG++CF P Y+S + F
Sbjct: 676 YSSLRVFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYISRNVIF 735
Query: 742 DEFCFPFAGSHSSPHDLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDV 801
+E PF + S + P+ S P+ +
Sbjct: 736 NESELPFKEKYQS-------------------------LVPQYSTPLLQAWQHNKISEIS 770
Query: 802 LPALPVPPADAP---PSPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAA--- 855
+PA PV P + A T+ T P T S PV ++
Sbjct: 771 VPAAPVQLFSKPIDLNTYAGSQVTEQLTDPEPTSNNEGSDEEVNPVAEEIAANQEQVINS 830
Query: 856 -RPRTRSQNGIFRPNPRYALVHAQPTGILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSA 914
TRS+ GI +PN RYAL+ T+ +PK SAMKHP W A+ +E++
Sbjct: 831 HAMTTRSKAGIQKPNTRYALI--------TSRMNTAEPKTLASAMKHPGWNEAVHEEINR 882
Query: 915 LHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFS 974
+H TW+LVP N++ KWVF+TK H DG++++LKARLVA+GF Q G DY TFS
Sbjct: 883 VHMLHTWSLVPPTDDMNILSSKWVFKTKLHPDGSIDKLKARLVAKGFDQEEGVDYLETFS 942
Query: 975 PVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRL 1034
PVV+ T+RL+L W + QLDV NAFLHG L E V+M QP GF+DP+ PTHVCRL
Sbjct: 943 PVVRTATIRLVLDVSTSKGWPIKQLDVSNAFLHGELQEPVFMYQPSGFIDPQKPTHVCRL 1002
Query: 1035 NKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGS 1094
KA+YGLKQAPRAWF S+FLL +GF CS++DPSLF ++ LYLL+YVDDI+LTGS
Sbjct: 1003 TKAIYGLKQAPRAWFDTFSNFLLDYGFVCSKSDPSLFVCHQDGKILYLLLYVDDILLTGS 1062
Query: 1095 DPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEAS 1154
D SLL + L F++K LG YFLG++I +GLFL Q YA D+L +A M + +
Sbjct: 1063 DQSLLEDLLQALKNRFSMKDLGPPRYFLGIQIEDYANGLFLHQTAYATDILQQAGMSDCN 1122
Query: 1155 HVSTPLAAGSHLVSSGEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEH 1214
+ TPL ++S E + +PT++RSL G LQYLTITRPD+ +AVN + Q + +PT
Sbjct: 1123 PMPTPLPQQLDNLNS-ELFAEPTYFRSLAGKLQYLTITRPDIQFAVNFICQRMHSPTTSD 1181
Query: 1215 FQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLS 1274
F +KRILRY+ GT GL +R S+ + YSD+D A C +TRRST G+ I LG NL+S
Sbjct: 1182 FGLLKRILRYIKGTIGMGLPIKRNSTLTLSAYSDSDHAGCKNTRRSTTGFCILLGSNLIS 1241
Query: 1275 WSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQ 1334
WSAK+QPTV+ SS E+EYRA+ A E+ W+ LL +L + + DN SA++++
Sbjct: 1242 WSAKRQPTVSNSSTEAEYRALTYAAREITWISFLLRDLGIPQYLPTQVYCDNLSAVYLSA 1301
Query: 1335 NPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
NP H +KH D D H++ E VA G + +H+ + QLAD+FTK+LPR F RSKL V
Sbjct: 1302 NPALHNRSKHFDTDYHYIREQVALGLIETQHISATFQLADVFTKSLPRRAFVDLRSKLGV 1361
Query: 1395 GLNPTLSLKG 1404
+PT SL+G
Sbjct: 1362 SGSPTPSLRG 1371
>At1g53810
Length = 1522
Score = 701 bits (1810), Expect = 0.0
Identities = 383/892 (42%), Positives = 519/892 (57%), Gaps = 66/892 (7%)
Query: 570 QSVCGKSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHV 629
Q + + IK+FQ D G EF +++ G+ SCP+T QNG E+KHRH+
Sbjct: 568 QKLVENQLGHKIKIFQCDGGGEFISSQFLKHLQDHGIQQNMSCPYTPQQNGMAERKHRHI 627
Query: 630 LELGLAMLYHSHVPTRYWVDAFSTAVYIINRVPSKVL-SNQIPFQLLFHVAPTYANFHPF 688
+ELGL+M++ S +P +YW+++F TA ++IN +P+ L +N+ P+Q L+ AP Y+ F
Sbjct: 628 VELGLSMIFQSKLPLKYWLESFFTANFVINLLPTSSLDNNESPYQKLYGKAPEYSALRVF 687
Query: 689 GCRVFPCLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPF 748
GC +P LR Y + KF PRS C+F GY+ +KG++C P Y+S H FDE PF
Sbjct: 688 GCACYPTLRDYASTKFDPRSLKCVFLGYNEKYKGYRCLYPPTGRIYISRHVVFDENTHPF 747
Query: 749 AG--SHSSPHD---LVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLP 803
SH P D L+ + F +P P +S P+ S+ P D P
Sbjct: 748 ESIYSHLHPQDKTPLLEAWFKSFHHVTPTQPD-------QSRYPVSSIPQPETTDLSAAP 800
Query: 804 ALPVPPADAPP---------------SPAPP---------------SPTQTATPPVGTQA 833
A P S +P SPT ++ P ++
Sbjct: 801 ASVAAETAGPNASDDTSQDNETISVVSGSPERTTGLDSASIGDSYHSPTADSSHPSPARS 860
Query: 834 PPASPPTTPPVV---AQVPLAPIAARPR--TRSQNGIFRPNPRYALVHAQPTGILTAFHT 888
PAS P P+ AQ AP+ TR + GI +PN RY L LT +
Sbjct: 861 SPASSPQGSPIQMAPAQQVQAPVTNEHAMVTRGKEGISKPNKRYVL--------LTHKVS 912
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGT 948
+ +PK A+KHP W AM++E+ + TWTLVP NV+G WVFRTK H+DG+
Sbjct: 913 IPEPKTVTEALKHPGWNNAMQEEMGNCKETETWTLVPYSPNMNVLGSMWVFRTKLHADGS 972
Query: 949 VERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHG 1008
+++LKARLVA+GF Q G DY T+SPVV+ TVRLIL + W+L Q+DVKNAFLHG
Sbjct: 973 LDKLKARLVAKGFKQEEGIDYLETYSPVVRTPTVRLILHVATVLKWELKQMDVKNAFLHG 1032
Query: 1009 HLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADP 1068
LTETVYM QP GFVD P HVC L+K+LYGLKQ+PRAWF R S+FLL GF CS DP
Sbjct: 1033 DLTETVYMRQPAGFVDKSKPDHVCLLHKSLYGLKQSPRAWFDRFSNFLLEFGFICSLFDP 1092
Query: 1069 SLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITY 1128
SLF + + + LL+YVDD+++TG++ LT +A LN EF +K +G++ YFLG++I
Sbjct: 1093 SLFVYSSNNDVILLLLYVDDMVITGNNSQSLTHLLAALNKEFRMKDMGQVHYFLGIQIQT 1152
Query: 1129 TPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSS-GEAYFDPTHYRSLVGALQ 1187
GLF+ Q KYA DLL A M S + TPL VS+ E + DPT++RSL G LQ
Sbjct: 1153 YDGGLFMSQQKYAEDLLITASMANCSPMPTPLPLQLDRVSNQDEVFSDPTYFRSLAGKLQ 1212
Query: 1188 YLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAV---- 1243
YLT+TRPD+ +AVN V Q + P++ F +KRILRY+ GT G+ + SS V
Sbjct: 1213 YLTLTRPDIQFAVNFVCQKMHQPSVSDFNLLKRILRYIKGTVSMGIQYNSNSSSVVSAYE 1272
Query: 1244 -----LGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANT 1298
YSD+D+A C +TRRS GY F+G N++SWS+KKQPTV+RSS E+EYR+++ T
Sbjct: 1273 SDYDLSAYSDSDYANCKETRRSVGGYCTFMGQNIISWSSKKQPTVSRSSTEAEYRSLSET 1332
Query: 1299 ASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVAS 1358
ASE+ W+ ++L E+ V L TP L DN SA+++ NP H KH D+D H++ E VA
Sbjct: 1333 ASEIKWMSSILREIGVSLPDTPELFCDNLSAVYLTANPAFHARTKHFDVDHHYIRERVAL 1392
Query: 1359 GRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRVGLNPTLSLKGVLEMNS 1410
L V+H+P LQLADIFTK+LP F R KL V PT SL+G + N+
Sbjct: 1393 KTLVVKHIPGHLQLADIFTKSLPFEAFTRLRFKLGVDFPPTPSLRGCISTNN 1444
>At4g27210 putative protein
Length = 1318
Score = 698 bits (1801), Expect = 0.0
Identities = 372/862 (43%), Positives = 504/862 (58%), Gaps = 50/862 (5%)
Query: 577 ISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAM 636
+S+ I VFQ D G EF ++K S G+ + SCPHT QNG E+KHRH++ELGL+M
Sbjct: 386 LSQKISVFQCDGGGEFVSHKFLQHLQSHGIQQQLSCPHTPQQNGLAERKHRHLVELGLSM 445
Query: 637 LYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQI-PFQLLFHVAPTYANFHPFGCRVFPC 695
L+ SHVP ++WV+AF TA ++IN +P+ L I P++ L+ P Y + FG FP
Sbjct: 446 LFQSHVPHKFWVEAFFTANFLINLLPTSALKESISPYEKLYDKKPDYTSLRSFGSACFPT 505
Query: 696 LRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSP 755
LR Y NKF+P S C+F GY+ +KG++C P Y+S H FDE +PF+ ++
Sbjct: 506 LRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPFSHTYKHL 565
Query: 756 HDLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPS 815
H + S SP P +P S P+ + S P LP P+
Sbjct: 566 HPQPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFT--------SADFPPLPQRKTPLLPT 617
Query: 816 PAPPSPTQTATPPVGTQAPPASPPTTPPV----------------------------VAQ 847
P S A+ Q+P T V Q
Sbjct: 618 LVPISSVSHASNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSDSEETIQQASVNVHQ 677
Query: 848 VPLAPIAARPRTRSQNGIFRPNPRYALVHAQPTGILTAFHTVT--KPKGFKSAMKHP*WL 905
+ TR++ GI +PNPRY + H V+ +PK +A+KHP W
Sbjct: 678 THASTNVHPMVTRAKVGISKPNPRYVFLS----------HKVSYPEPKTVTAALKHPGWT 727
Query: 906 AAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIP 965
AM +E+ + TW+LVP S +V+G KWVFRTK H+DGT+ +LKAR+VA+GF Q
Sbjct: 728 GAMTEEIGNCSETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGFLQEE 787
Query: 966 GFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDP 1025
G DY T+SPVV+ TVRL+L +W + Q+DVKNAFLHG L ETVYM QP GFVDP
Sbjct: 788 GIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAGFVDP 847
Query: 1026 RFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVY 1085
P HVC L+K++YGLKQ+PRAWF + S+FLL GF CS++DPSLF + + + LL+Y
Sbjct: 848 SKPDHVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCSKSDPSLFIYAHNNNLILLLLY 907
Query: 1086 VDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLL 1145
VDD+++TG+ LT +A LN EF + +G+L YFLG+++ +GLF+ Q KYA DLL
Sbjct: 908 VDDMVITGNSSQTLTSLLAALNKEFRMTDMGQLHYFLGIQVQRQQNGLFMSQQKYAEDLL 967
Query: 1146 SRAMMLEASHVSTPLAAGSHLVSSGEAYF-DPTHYRSLVGALQYLTITRPDLSYAVNTVS 1204
A M + + TPL V E F DPT++RS+ G LQYLT+TRPD+ +AVN V
Sbjct: 968 IAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVC 1027
Query: 1205 QFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGY 1264
Q + PT+ F +KRILRY+ GT G+++ R S + YSD+DW C TRRS G
Sbjct: 1028 QKMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYSDSDWGNCKQTRRSVGGL 1087
Query: 1265 AIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLS 1324
F+G NL+SWS+KK PTV+RSS E+EY+++++ ASE++WL LL ELR+ L TP L
Sbjct: 1088 CTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPDTPELFC 1147
Query: 1325 DNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPL 1384
DN SA+++ NP H KH D+D HFV E VA L V+H+P S Q+ADIFTK+LP
Sbjct: 1148 DNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTKSLPYEA 1207
Query: 1385 FEIFRSKLRVGLNPTLSLKGVL 1406
F R KL V L+PT SL+G +
Sbjct: 1208 FIHLRGKLGVTLSPTPSLRGTI 1229
>At4g10690 retrotransposon like protein
Length = 1515
Score = 678 bits (1749), Expect = 0.0
Identities = 367/879 (41%), Positives = 514/879 (57%), Gaps = 50/879 (5%)
Query: 570 QSVCGKSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHV 629
Q + I +FQ D G EF + K AS G+ SCPHT QNG E++HR++
Sbjct: 565 QQLVENQYQHKIAMFQCDGGGEFVSYKFVAHLASCGIKQLISCPHTPQQNGIAERRHRYL 624
Query: 630 LELGLAMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLS-NQIPFQLLFHVAPTYANFHPF 688
ELGL++++HS VP + WV+AF T+ ++ N +PS LS N+ P+++L P Y F
Sbjct: 625 TELGLSLMFHSKVPHKLWVEAFFTSNFLSNLLPSSTLSDNKSPYEMLHGTPPVYTALRVF 684
Query: 689 GCRVFPCLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPF 748
G +P LRPY NKF P+S C+F GY++ +KG++C +P Y+ H FDE FP+
Sbjct: 685 GSACYPYLRPYAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLFDERKFPY 744
Query: 749 AGSHSSPHDLVVSTFYEP---------ASGSPPSPHPVVLVAPE----SSLPIGSL---- 791
+ +S + S + S PS + ++ P SS+P G
Sbjct: 745 SDIYSQFQTISGSPLFTAWQKGFSSTALSRETPSTNVEDIIFPSATVSSSVPTGCAPNIA 804
Query: 792 SCPSCDDSDVLPA--LPVPPADAPPSPAPPSPTQTATPP----------VGTQAPPASP- 838
+ D DV A + VPP+ + P P ++ + + + P S
Sbjct: 805 ETATAPDVDVAAAHDMVVPPSPITSTSLPTQPEESTSDQNHYSTDSETAISSAMTPQSIN 864
Query: 839 ---------PTTPPVVAQVPLAPIAARPR-TRSQNGIFRPNPRYALVHAQPTGILTAFHT 888
P V++ AP + P TR+++GI +PNP+YAL +
Sbjct: 865 VSLFEDSDFPPLQSVISSTTAAPETSHPMITRAKSGITKPNPKYALFSVKSN-------- 916
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGT 948
+PK K A+K W AM +E+ +H+ TW LVP ++G KWVF+TK +SDG+
Sbjct: 917 YPEPKSVKEALKDEGWTNAMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVFKTKLNSDGS 976
Query: 949 VERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHG 1008
++RLKARLVA+G+ Q G DY T+SPVV++ TVR IL +N W L QLDVKNAFLH
Sbjct: 977 LDRLKARLVARGYEQEEGVDYVETYSPVVRSATVRSILHVATINKWSLKQLDVKNAFLHD 1036
Query: 1009 HLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADP 1068
L ETV+M QPPGF DP P +VC+L KA+Y LKQAPRAWF + SS+LL++GF CS +DP
Sbjct: 1037 ELKETVFMTQPPGFEDPSRPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKYGFICSFSDP 1096
Query: 1069 SLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITY 1128
SLF + KG ++LL+YVDD+ILTG++ LL Q + L+ EF +K +G L YFLG++ Y
Sbjct: 1097 SLFVYLKGRDVMFLLLYVDDMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQAHY 1156
Query: 1129 TPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAYFDPTHYRSLVGALQY 1188
DGLFL Q KY DLL A M + S + TPL L + + + +PT++R L G LQY
Sbjct: 1157 HNDGLFLSQEKYTSDLLVNAGMSDCSSMPTPLQL-DLLQGNNKPFPEPTYFRRLAGKLQY 1215
Query: 1189 LTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSD 1248
LT+TRPD+ +AVN V Q + APTM F +KRIL Y+ GT G+ + + YSD
Sbjct: 1216 LTLTRPDIQFAVNFVCQKMHAPTMSDFHLLKRILHYLKGTMTMGINLSSNTDSVLRCYSD 1275
Query: 1249 ADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNL 1308
+DWA C DTRRST G+ FLG N++SWSAK+ PTV++SS E+EYR ++ ASE+ W+ L
Sbjct: 1276 SDWAGCKDTRRSTGGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRTLSFAASEVSWIGFL 1335
Query: 1309 LHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPT 1368
L E+ + P + DN SA++++ NP H +KH +D ++V E VA G L V+H+P
Sbjct: 1336 LQEIGLPQQQIPEMYCDNLSAVYLSANPALHSRSKHFQVDYYYVRERVALGALTVKHIPA 1395
Query: 1369 SLQLADIFTKALPRPLFEIFRSKLRVGLNPTLSLKGVLE 1407
S QLADIFTK+LP+ F R KL V L P SL+G ++
Sbjct: 1396 SQQLADIFTKSLPQAPFCDLRFKLGVVLPPDTSLRGCIK 1434
>At1g59265 polyprotein, putative
Length = 1466
Score = 649 bits (1675), Expect = 0.0
Identities = 367/885 (41%), Positives = 497/885 (55%), Gaps = 81/885 (9%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
I F SDNG EF + F+ G+ H S PHT NG E+KHRH++E GL +L H+
Sbjct: 586 IGTFYSDNGGEFV--ALWEYFSQHGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLSHA 643
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
+P YW AF+ AVY+INR+P+ +L + PFQ LF +P Y FGC +P LRPY
Sbjct: 644 SIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNYDKLRVFGCACYPWLRPYN 703
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPHDLVV 760
+K +S C+F GYS + C + S Y+S H +FDE CFPF+ ++ +
Sbjct: 704 QHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDENCFPFSNYLATLSPVQE 763
Query: 761 STFYEPASGSPPSPHPV---VLVAPESSLPIGSLSCPSCDD-----------------SD 800
SP + P VL AP S P + + PS S
Sbjct: 764 QRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHAATPPSSPSAPFRNSQVSSSNLDSSFSS 823
Query: 801 VLPALPVPPADAPPSPAPPS-PTQTAT--------------------------------- 826
P+ P P A P P + PTQT T
Sbjct: 824 SFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSS 883
Query: 827 --PPVGTQAPPASPPTTPPVVAQVPLAPIAARPR-------------TRSQNGIFRPNPR 871
P T A +S TPP + P P+A TR++ GI +PNP+
Sbjct: 884 SSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPLNTHSMGTRAKAGIIKPNPK 943
Query: 872 YALVHAQPTGILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRP-STT 930
Y+L + ++P+ A+K W AM E++A N TW LVP P S
Sbjct: 944 YSLA--------VSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDLVPPPPSHV 995
Query: 931 NVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVV 990
+VG +W+F K++SDG++ R KARLVA+G+ Q PG DY+ TFSPV+K+T++R++L V
Sbjct: 996 TIVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAV 1055
Query: 991 LNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQ 1050
W + QLDV NAFL G LT+ VYM QPPGF+D P +VC+L KALYGLKQAPRAW+
Sbjct: 1056 DRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLKQAPRAWYV 1115
Query: 1051 RLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEF 1110
L ++LL GF S +D SLF +G + +Y+LVYVDDI++TG+DP+LL + L+ F
Sbjct: 1116 ELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNTLDNLSQRF 1175
Query: 1111 AIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHL-VSS 1169
++K +L YFLG+E P GL L Q +Y DLL+R M+ A V+TP+A L + S
Sbjct: 1176 SVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAPSPKLSLYS 1235
Query: 1170 GEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQ 1229
G DPT YR +VG+LQYL TRPD+SYAVN +SQF+ PT EH QA+KRILRY+ GT
Sbjct: 1236 GTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRILRYLAGTP 1295
Query: 1230 HFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCE 1289
+ G+ ++ ++ ++ YSDADWA D ST GY ++LG + +SWS+KKQ V RSS E
Sbjct: 1296 NHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTE 1355
Query: 1290 SEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDC 1349
+EYR++ANT+SE+ W+ +LL EL +RL+ P++ DN A ++ NPV H KHI +D
Sbjct: 1356 AEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDY 1415
Query: 1350 HFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
HF+ V SG L V HV T QLAD TK L R F+ F SK+ V
Sbjct: 1416 HFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
>At1g58889 polyprotein, putative
Length = 1466
Score = 648 bits (1671), Expect = 0.0
Identities = 366/885 (41%), Positives = 496/885 (55%), Gaps = 81/885 (9%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
I F SDNG EF + F+ G+ H S PHT NG E+KHRH++E GL +L H+
Sbjct: 586 IGTFYSDNGGEFV--ALWEYFSQHGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLSHA 643
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
+P YW AF+ AVY+INR+P+ +L + PFQ LF +P Y FGC +P LRPY
Sbjct: 644 SIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNYDKLRVFGCACYPWLRPYN 703
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPHDLVV 760
+K +S C+F GYS + C + S Y+S H +FDE CFPF+ ++ +
Sbjct: 704 QHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDENCFPFSNYLATLSPVQE 763
Query: 761 STFYEPASGSPPSPHPV---VLVAPESSLPIGSLSCPSCDD-----------------SD 800
SP + P VL AP S P + + PS S
Sbjct: 764 QRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHAATPPSSPSAPFRNSQVSSSNLDSSFSS 823
Query: 801 VLPALPVPPADAPPSPAPPS-PTQTAT--------------------------------- 826
P+ P P A P P + PTQT T
Sbjct: 824 SFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSS 883
Query: 827 --PPVGTQAPPASPPTTPPVVAQVPLAPIAARPR-------------TRSQNGIFRPNPR 871
P T A +S TPP + P P+A TR++ GI +PNP+
Sbjct: 884 SSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPLNTHSMGTRAKAGIIKPNPK 943
Query: 872 YALVHAQPTGILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRP-STT 930
Y+L + ++P+ A+K W AM E++A N TW LVP P S
Sbjct: 944 YSLA--------VSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDLVPPPPSHV 995
Query: 931 NVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVV 990
+VG +W+F K++SDG++ R KAR VA+G+ Q PG DY+ TFSPV+K+T++R++L V
Sbjct: 996 TIVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAV 1055
Query: 991 LNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQ 1050
W + QLDV NAFL G LT+ VYM QPPGF+D P +VC+L KALYGLKQAPRAW+
Sbjct: 1056 DRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLKQAPRAWYV 1115
Query: 1051 RLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEF 1110
L ++LL GF S +D SLF +G + +Y+LVYVDDI++TG+DP+LL + L+ F
Sbjct: 1116 ELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNTLDNLSQRF 1175
Query: 1111 AIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHL-VSS 1169
++K +L YFLG+E P GL L Q +Y DLL+R M+ A V+TP+A L + S
Sbjct: 1176 SVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAPSPKLSLYS 1235
Query: 1170 GEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQ 1229
G DPT YR +VG+LQYL TRPD+SYAVN +SQF+ PT EH QA+KRILRY+ GT
Sbjct: 1236 GTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRILRYLAGTP 1295
Query: 1230 HFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCE 1289
+ G+ ++ ++ ++ YSDADWA D ST GY ++LG + +SWS+KKQ V RSS E
Sbjct: 1296 NHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTE 1355
Query: 1290 SEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDC 1349
+EYR++ANT+SE+ W+ +LL EL +RL+ P++ DN A ++ NPV H KHI +D
Sbjct: 1356 AEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDY 1415
Query: 1350 HFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
HF+ V SG L V HV T QLAD TK L R F+ F SK+ V
Sbjct: 1416 HFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 647 bits (1669), Expect = 0.0
Identities = 377/908 (41%), Positives = 505/908 (55%), Gaps = 96/908 (10%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
I SDNG EF + + G+ H S PHT NG E+KHRH++E+GL +L H+
Sbjct: 565 IGTLYSDNGGEFV--VLRDYLSQHGISHFTSPPHTPEHNGLSERKHRHIVEMGLTLLSHA 622
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
VP YW AFS AVY+INR+P+ +L Q PFQ LF P Y FGC +P LRPY
Sbjct: 623 SVPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACYPWLRPYN 682
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPHDLVV 760
+K +S C F GYS + C + Y S H QFDE CFPF+ ++ V
Sbjct: 683 RHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNFG-----V 737
Query: 761 STFYEPASGSPPS--------PHPVVLVAP--------------------------ESSL 786
ST E S S P+ P+VL AP S+L
Sbjct: 738 STSQEQRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSPLCTTQVSSSNL 797
Query: 787 PIGSLSCPSCDD------------------------SDVL----PALPVPPADAPPSPAP 818
P S+S PS + S +L P P P + SP P
Sbjct: 798 PSSSISSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPNPNSPSPNSPNQNSPLP 857
Query: 819 ---------PSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARP-------RTRSQ 862
P+P+ + + P + S P PPV+ P+ + A+ TR++
Sbjct: 858 QSPISSPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQVNAQAPVNTHSMATRAK 917
Query: 863 NGIFRPNPRYALVHAQPTGILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWT 922
+GI +PN +Y+ T+ ++P+ AMK W AM E++A N TW
Sbjct: 918 DGIRKPNQKYSYA--------TSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWD 969
Query: 923 LVPRPS-TTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATT 981
LVP P + +VG +W+F KF+SDG++ R KARLVA+G+ Q PG DY+ TFSPV+K+T+
Sbjct: 970 LVPPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTS 1029
Query: 982 VRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGL 1041
+R++L V W + QLDV NAFL G LT+ VYM QPPGFVD P +VCRL KA+YGL
Sbjct: 1030 IRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGL 1089
Query: 1042 KQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQ 1101
KQAPRAW+ L ++LL GF S +D SLF +G + +Y+LVYVDDI++TG+D LL
Sbjct: 1090 KQAPRAWYVELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKH 1149
Query: 1102 FIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLA 1161
+ L+ F++K L YFLG+E P GL L Q +Y DLL+R ML A V+TP+A
Sbjct: 1150 TLDALSQRFSVKEHEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMA 1209
Query: 1162 AGSHL-VSSGEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKR 1220
L + SG DPT YR +VG+LQYL TRPDLSYAVN +SQ++ PT +H+ A+KR
Sbjct: 1210 TSPKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKR 1269
Query: 1221 ILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQ 1280
+LRY+ GT G+ ++ ++ ++ YSDADWA TD ST GY ++LG + +SWS+KKQ
Sbjct: 1270 VLRYLAGTPDHGIFLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQ 1329
Query: 1281 PTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHK 1340
V RSS E+EYR++ANT+SEL W+ +LL EL ++LS P++ DN A ++ NPV H
Sbjct: 1330 KGVVRSSTEAEYRSVANTSSELQWICSLLTELGIQLSHPPVIYCDNVGATYLCANPVFHS 1389
Query: 1341 HAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRVGLNPTL 1400
KHI LD HF+ V SG L V HV T QLAD TK L R F+ F K+ V P
Sbjct: 1390 RMKHIALDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRVAFQNFSRKIGVIKVPP- 1448
Query: 1401 SLKGVLEM 1408
S GVL +
Sbjct: 1449 SCGGVLRI 1456
>At1g44510 polyprotein, putative
Length = 1459
Score = 636 bits (1640), Expect = 0.0
Identities = 384/871 (44%), Positives = 489/871 (56%), Gaps = 66/871 (7%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
I+ SDNG EF + S+G+ H S PHT NG E+KHRH++E GL +L +
Sbjct: 592 IRTLYSDNGGEFI--ALRDFLVSNGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLTQA 649
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
VP YW AF+TAVY+INR+P+ VL Q PFQ LF +P Y FGC FP LRPY
Sbjct: 650 SVPREYWTYAFATAVYLINRMPTPVLCLQSPFQKLFGSSPNYQRLRVFGCLCFPWLRPYT 709
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPHDLVV 760
NK RS C+F GYS + C + + Y S H FDE +PFA S +
Sbjct: 710 RNKLEERSKRCVFLGYSLTQTAYLCLDVDNNRLYTSRHVMFDESTYPFAASIREQSQSSL 769
Query: 761 STFYEPASGSPPS----PHPVV-LVAPESSLPIGSLSCPSCDDSDVLPALPVPPA----- 810
T E +S S P+ P V+ L +P +S P +DS V P P
Sbjct: 770 VTPPESSSSSSPANSGFPCSVLRLQSPPASSPETPSPPQQQNDSPVSPRQTGSPTPSHHS 829
Query: 811 ---DAPPSPAP------PSPTQTATPPVGTQAPPASP---PTTPPVVAQVPLAPIAARP- 857
D+ SP+P P+ P Q+ P SP P P P + I RP
Sbjct: 830 QVRDSTLSPSPSVSNSEPTAPHENGPEPEAQSNPNSPFIGPLPNPNPETNPSSSIEQRPV 889
Query: 858 --------------------------------RTRSQNGIFRPNPRYALVHAQPTGILTA 885
+TRS+N I +P + +L A L+
Sbjct: 890 DKSTTTALPPNQTTIAATSNSRSQPPKNNHQMKTRSKNNITKPKTKTSLTVALTQPHLSE 949
Query: 886 FHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHS 945
+TVT+ A+K W AM DE A +N TW LVP T ++VG +WVF+ K+
Sbjct: 950 PNTVTQ------ALKDKKWRFAMSDEFDAQQRNHTWDLVPPNPTQHLVGCRWVFKLKYLP 1003
Query: 946 DGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAF 1005
+G +++ KARLVA+GF Q G DY+ TFSPV+KATT+R++L V +W L QLDV NAF
Sbjct: 1004 NGLIDKYKARLVAKGFNQQYGVDYAETFSPVIKATTIRVVLDVAVKKNWPLKQLDVNNAF 1063
Query: 1006 LHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSR 1065
L G LTE VYM QPPGFVD P+HVCRL KA+YGLKQAPRAW+ L LL GF S
Sbjct: 1064 LQGTLTEEVYMAQPPGFVDKDRPSHVCRLRKAIYGLKQAPRAWYMELKQHLLNIGFVNSL 1123
Query: 1066 ADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLE 1125
AD SLF + G T LYLLVYVDDII+TGSD ++ ++ L F+IK L YFLG+E
Sbjct: 1124 ADTSLFIYSHGTTLLYLLVYVDDIIVTGSDHKSVSAVLSSLAERFSIKDPTDLHYFLGIE 1183
Query: 1126 ITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHL-VSSGEAYFDPTHYRSLVG 1184
T T GL L Q KY DLL++ ML+A V+TPL L + G D + YRS+VG
Sbjct: 1184 ATRTNTGLHLMQRKYMTDLLAKHNMLDAKPVATPLPTSPKLTLHGGTKLNDASEYRSVVG 1243
Query: 1185 ALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVL 1244
+LQYL TRPD+++AVN +SQF+ PT +H+QA KR+LRY+ GT G+ F +SSP L
Sbjct: 1244 SLQYLAFTRPDIAFAVNRLSQFMHQPTSDHWQAAKRVLRYLAGTTTHGI-FLNSSSPIHL 1302
Query: 1245 -GYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELV 1303
+SDADWA + ST Y I+LG N +SWS+KKQ V+RSS ESEYRA+AN ASE+
Sbjct: 1303 HAFSDADWAGDSADYVSTNAYVIYLGRNPISWSSKKQRGVSRSSTESEYRAVANAASEIR 1362
Query: 1304 WLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAV 1363
WL +LL EL +RL P + DN A ++ NPV H KHI LD HFV ++ S L V
Sbjct: 1363 WLCSLLTELHIRLPHGPTIFCDNIGATYICANPVFHSRMKHIALDYHFVRGMIQSRALRV 1422
Query: 1364 RHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
HV T+ QLAD TK+L RP F RSK+ V
Sbjct: 1423 SHVSTNDQLADALTKSLSRPHFLSARSKIGV 1453
>At4g16870 retrotransposon like protein
Length = 1474
Score = 634 bits (1635), Expect = 0.0
Identities = 373/892 (41%), Positives = 481/892 (53%), Gaps = 88/892 (9%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
I+ SDNG EF + S+G+ H S PHT NG E+KHRH++E GL +L +
Sbjct: 587 IRTLYSDNGGEFI--ALREFLVSNGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLTQA 644
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
VP YW AF+ AVY+INR+P+ VLS + PFQ LF P Y FGC FP LRPY
Sbjct: 645 SVPREYWPYAFAAAVYLINRMPTPVLSMESPFQKLFGSKPNYERLRVFGCLCFPWLRPYT 704
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPHDLVV 760
+NK RS C+F GYS + CF+ Y S H FDE FPF+ + +S + L
Sbjct: 705 HNKLEERSRRCVFLGYSLTQTAYLCFDVEHKRLYTSRHVVFDEASFPFS-NLTSQNSLPT 763
Query: 761 STF-------------------------------YEPASGSPPSPH---PVVLVAPES-- 784
TF +P +P SPH P AP S
Sbjct: 764 VTFEQSSSPLVTPILSSSSVLPSCLSSPCTVLHQQQPPVTTPNSPHSSQPTTSPAPLSPH 823
Query: 785 -------SLPIGSLSCPSCDDSDVLPALPVPPADAPPSP---APP--------------- 819
+P S P S L + P P + P P +PP
Sbjct: 824 RSTTMDFQVPQVRSSSPLLSSSSSLNSEPTAPNENGPEPEAQSPPIGPLSNPTHEAFIGP 883
Query: 820 ------SPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARP----------RTRSQN 863
+PT P P P TT + ++ + +P +TR++N
Sbjct: 884 LPNPNRNPTNEIEPTPAPHPKPVKPTTTTTTPNRTTVSDASHQPTAPQQNQHNMKTRAKN 943
Query: 864 GIFRPNPRYALVHAQPTGILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTL 923
I +PN +++L P + ++P A+K W AM DE A +N TW L
Sbjct: 944 NIKKPNTKFSLTATLPN------RSPSEPTNVTQALKDKKWRFAMSDEFDAQQRNHTWDL 997
Query: 924 VPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVR 983
VP S +VG KWVF+ K+ +G +++ KARLVA+GF Q G DY+ TFSPV+K+TT+R
Sbjct: 998 VPHESQL-LVGCKWVFKLKYLPNGAIDKYKARLVAKGFNQQYGVDYAETFSPVIKSTTIR 1056
Query: 984 LILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQ 1043
L+L V DW++ QLDV NAFL G LTE VYM QPPGF+D PTHVCRL KA+YGLKQ
Sbjct: 1057 LVLDVAVKKDWEIKQLDVNNAFLQGTLTEEVYMAQPPGFIDKDRPTHVCRLRKAIYGLKQ 1116
Query: 1044 APRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFI 1103
APRAW+ L L GF S +D SLF + G T +Y+LVYVDDII+TGSD S + +
Sbjct: 1117 APRAWYMELKQHLFNIGFVNSLSDASLFIYCHGTTFVYVLVYVDDIIVTGSDKSSIDAVL 1176
Query: 1104 ARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAG 1163
L F+IK L YFLG+E T T GL L Q KY DLL++ M +A V TPL
Sbjct: 1177 TSLAERFSIKDPTDLHYFLGIEATRTKQGLHLMQRKYIKDLLAKHNMADAKPVLTPLPTS 1236
Query: 1164 SHL-VSSGEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRIL 1222
L + G D + YRS+VG+LQYL TRPD++YAVN +SQ + PT +H+QA KR+L
Sbjct: 1237 PKLTLHGGTKLNDASEYRSVVGSLQYLAFTRPDIAYAVNRLSQLMPQPTEDHWQAAKRVL 1296
Query: 1223 RYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPT 1282
RY+ GT G+ TS + +SDADWA +D ST Y I+LG N +SWS+KKQ
Sbjct: 1297 RYLAGTSTHGIFLDTTSPLNLHAFSDADWAGDSDDYVSTNAYVIYLGKNPISWSSKKQRG 1356
Query: 1283 VARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHA 1342
VARSS ESEYRA+AN ASE+ WL +LL +L +RL P + DN A ++ NPV H
Sbjct: 1357 VARSSTESEYRAVANAASEVKWLCSLLSKLHIRLPIRPSIFCDNIGATYLCANPVFHSRM 1416
Query: 1343 KHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
KHI +D HFV ++ SG L V HV T QLAD TK L R F+ R K+ V
Sbjct: 1417 KHIAIDYHFVRNMIQSGALRVSHVSTRDQLADALTKPLSRAHFQSARFKIGV 1468
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 573 bits (1476), Expect = e-163
Identities = 338/866 (39%), Positives = 474/866 (54%), Gaps = 58/866 (6%)
Query: 575 KSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGL 634
K +++K+ +SDNGTEF + + F +G++H+ SC T QNGRVE+KHRH+L +
Sbjct: 641 KQFGKTVKMVRSDNGTEFMC--LSSYFRENGIIHQTSCVGTPQQNGRVERKHRHILNVAR 698
Query: 635 AMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFP 694
A+L+ + +P ++W ++ TA Y+INR PS +LS + P+++L P Y+ FG +
Sbjct: 699 ALLFQASLPIKFWGESILTAAYLINRTPSSILSGRTPYEVLHGSKPVYSQLRVFGSACYV 758
Query: 695 CLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSS 754
+KF RS CIF GY KG+K ++ + VS F E FP+AG +SS
Sbjct: 759 HRVTRDKDKFGQRSRSCIFVGYPFGKKGWKVYDIERNEFLVSRDVIFREEVFPYAGVNSS 818
Query: 755 PHDLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPP 814
+ ST S P + V S+ C +V+ V ++ P
Sbjct: 819 T---LASTSLPTVSEDDDWAIPPLEV--RGSIDSVETERVVCTTDEVVLDTSVSDSEIPN 873
Query: 815 SPAPPSPTQTATPPVGTQAPPASPPTTP---PVVAQVPLAPIAARPRTRSQNGIFRPNPR 871
P T ++P + + + PTTP PV + +P++P P+ R P P+
Sbjct: 874 QEFVPDDTPPSSPLSVSPSGSPNTPTTPIVVPVASPIPVSP----PKQRKSKRATHPPPK 929
Query: 872 ---YAL---------VHAQPT------------------------------GILTAFHTV 889
Y L +HA P L A
Sbjct: 930 LNDYVLYNAMYTPSSIHALPADPSQSSTVPGKSLFPLTDYVSDAAFSSSHRAYLAAITDN 989
Query: 890 TKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTV 949
+PK FK A++ W AM E+ AL N TW +V P +G +WVF+TK++SDGTV
Sbjct: 990 VEPKHFKEAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFKTKYNSDGTV 1049
Query: 950 ERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGH 1009
ER KARLV QG Q+ G DY TF+PVV+ TTVR +L +V N W+++Q+DV NAFLHG
Sbjct: 1050 ERYKARLVVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMDVHNAFLHGD 1109
Query: 1010 LTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPS 1069
L E VYM+ PPGF P VCRL K+LYGLKQAPR WF++LS LLR GF S D S
Sbjct: 1110 LEEEVYMKLPPGFRHSH-PDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGFVQSYEDYS 1168
Query: 1070 LFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYT 1129
LF + + + L +L+YVDD+++ G+D +L +F L+ F++K LGKL YFLG+E++
Sbjct: 1169 LFSYTRNNIELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIEVSRG 1228
Query: 1130 PDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSS-GEAYFDPTHYRSLVGALQY 1188
P+G+FL Q KYA D+++ + L + TPL HL S G DP YR LVG L Y
Sbjct: 1229 PEGIFLSQRKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVGRLLY 1288
Query: 1189 LTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSD 1248
L TRP+LSY+V+ ++QF+Q P HF A R++RY+ G+ G+ + Y D
Sbjct: 1289 LLHTRPELSYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILLNADPDLTLEVYCD 1348
Query: 1249 ADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNL 1308
+DW C TRRS Y + LG + +SW KKQ TV+ SS E+EYRAM+ E+ WL L
Sbjct: 1349 SDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSYALKEIKWLRKL 1408
Query: 1309 LHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPT 1368
L EL + S L D+++A+ +A NPV H+ KHI+ DCH V + V G + +HV T
Sbjct: 1409 LKELGIEQSTPARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDAVRDGIITTQHVRT 1468
Query: 1369 SLQLADIFTKALPRPLFEIFRSKLRV 1394
+ QLAD+FTKAL R F SKL V
Sbjct: 1469 TEQLADVFTKALGRNQFLYLMSKLGV 1494
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 563 bits (1450), Expect = e-160
Identities = 335/883 (37%), Positives = 467/883 (51%), Gaps = 70/883 (7%)
Query: 575 KSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGL 634
+ + +K F++DNGTEF + F + G+LH+ SC T QNGRVE+KHRH+L +
Sbjct: 614 RQFGKQVKAFRTDNGTEFMC--LTPYFQTHGILHQTSCVDTPQQNGRVERKHRHILNVAR 671
Query: 635 AMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFP 694
A L+ ++P ++W ++ TA ++INR PS VL + P++LLF P+Y FGC +
Sbjct: 672 ACLFQGNLPVKFWGESILTATHLINRTPSAVLKGKTPYELLFGERPSYDMLRSFGCLCYA 731
Query: 695 CLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSS 754
+RP +KF+ RS C+F GY K ++ ++ + S +F E +P+A + S
Sbjct: 732 HIRPRNKDKFTSRSRKCVFIGYPHGKKAWRVYDLETGKIFASRDVRFHEDIYPYATATQS 791
Query: 755 PHDL-------------------VVSTFYEPASGSPPS---------------------- 773
L V ST + +S S P+
Sbjct: 792 NVPLPPPTPPMVNDDWFLPISTQVDSTNVDSSSSSSPAQSGSIDQPPRSIDQSPSTSTNP 851
Query: 774 -PHPVVLVAPESSLPIGSLSCPSCD--DSDVLPALPVPPADAPPSPAPPS---------- 820
P + + P SS P S+ + D SD L + P SP P
Sbjct: 852 VPEEIGSIVPSSS-PSRSIDRSTSDLSASDTTELLSTGESSTPSSPGLPELLGKGCREKK 910
Query: 821 --------PTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPRY 872
T T + SP P L+ + +T F N Y
Sbjct: 911 KSVLLKDFVTNTTSKKKTASHNIHSPSQVLPSGLPTSLSADSVSGKTLYPLSDFLTNSGY 970
Query: 873 ALVHAQPTGILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNV 932
+ H + A +PK FK A+ W AM E+ AL N TW + P
Sbjct: 971 SANHI---AFMAAILDSNEPKHFKDAILIKEWCEAMSKEIDALEANHTWDITDLPHGKKA 1027
Query: 933 VGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLN 992
+ KWV++ K++SDGT+ER KARLV G Q G D+ TF+PV K TTVR IL+
Sbjct: 1028 ISSKWVYKLKYNSDGTLERHKARLVVMGNHQKEGVDFKETFAPVAKLTTVRTILAVAAAK 1087
Query: 993 DWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRL 1052
DW++HQ+DV NAFLHG L E VYM PPGF P+ VCRL K+LYGLKQAPR WF +L
Sbjct: 1088 DWEVHQMDVHNAFLHGDLEEEVYMRLPPGFKCSD-PSKVCRLRKSLYGLKQAPRCWFSKL 1146
Query: 1053 SSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAI 1112
S+ L GF+ S D SLF G T +++LVYVDD+I+ G++ + +F ++L+ F +
Sbjct: 1147 STALRNIGFTQSYEDYSLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDRFKSQLHKCFHM 1206
Query: 1113 KYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVS-SGE 1171
K LGKL YFLGLE++ PDG L Q KYA D++ +L + P+A L S +G
Sbjct: 1207 KDLGKLKYFLGLEVSRGPDGFCLSQRKYALDIVKETGLLGCKPSAVPIALNHKLASITGP 1266
Query: 1172 AYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHF 1231
+ +P YR LVG YLTITRPDLSYAV+ +SQF+QAP + H++A R++RY+ G+
Sbjct: 1267 VFTNPEQYRRLVGRFIYLTITRPDLSYAVHILSQFMQAPLVAHWEAALRLVRYLKGSPAQ 1326
Query: 1232 GLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESE 1291
G+ R SS + Y D+D+ C TRRS Y ++LGD+ +SW KKQ TV+ SS E+E
Sbjct: 1327 GIFLRSDSSLIINAYCDSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTVSYSSAEAE 1386
Query: 1292 YRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHF 1351
YRAMA T EL WL LL +L V S+ L D+++A+ +A NPV H+ KHI+ DCH
Sbjct: 1387 YRAMAYTLKELKWLKALLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTKHIESDCHK 1446
Query: 1352 VCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
V + V + H+ T Q+AD+ TK+LPRP FE S L V
Sbjct: 1447 VRDAVLDKLITTEHIYTEDQVADLLTKSLPRPTFERLLSTLGV 1489
>At1g57640
Length = 1444
Score = 548 bits (1412), Expect = e-156
Identities = 323/846 (38%), Positives = 463/846 (54%), Gaps = 38/846 (4%)
Query: 575 KSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGL 634
+ IK+ +SDNGTEF + + F G+ H SC T QNGRVE+KHRH+L +
Sbjct: 604 RQFDTEIKIVRSDNGTEFLCMREY--FLHKGIAHETSCVGTPHQNGRVERKHRHILNIAR 661
Query: 635 AMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFP 694
A+ + S++P ++W + +A Y+INR PS +L + P+++L+ AP Y++ FG +
Sbjct: 662 ALRFQSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYA 721
Query: 695 CLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSS 754
+ + +KF+ RS C+F GY KG++ F+ +VS F E FP++ +
Sbjct: 722 HNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSCN 781
Query: 755 PHD------LVVSTFYEPASG---------SPPSPHPVVLVAP-------ESSLPIGSLS 792
D V F E A G + P V P ESS P +S
Sbjct: 782 EEDERVLVDCVGPPFIEEAIGPRTIIGRNIGEATVGPNVATGPIIPEINQESSSPSEFVS 841
Query: 793 CPSCDDSDVLPALPVPPADAPPSPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAP 852
S D L + V AD P S P+P Q TQ P V+ ++P
Sbjct: 842 LSSLDP--FLASSTVQTADLPLSSTTPAPIQLRRSSRQTQKPMKLKNFVTNTVSVESISP 899
Query: 853 IAARPRTRSQNGIFRPNPRYALVH---AQPTGILTAFHTVTKPKGFKSAMKHP*WLAAME 909
A S + ++ P +Y H + L A +P + AM W AM
Sbjct: 900 EA------SSSSLY-PIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMS 952
Query: 910 DELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDY 969
E+ +L N T+++V P +G KWV++ K+ SDG +ER KARLV G Q G DY
Sbjct: 953 AEIESLRVNQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDY 1012
Query: 970 SLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPT 1029
TF+PV K +TVRL L DW +HQ+DV NAFLHG L E VYM+ P GF P+
Sbjct: 1013 DETFAPVAKMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDD-PS 1071
Query: 1030 HVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDI 1089
VCRL+K+LYGLKQAPR WF +LSS L ++GF+ S +D SLF + +++LVYVDD+
Sbjct: 1072 KVCRLHKSLYGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGIFVHVLVYVDDL 1131
Query: 1090 ILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAM 1149
I++GS P + QF + L + F +K LG L YFLG+E++ G +L Q KY D++S
Sbjct: 1132 IISGSCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMG 1191
Query: 1150 MLEASHVSTPLAAGSHL-VSSGEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQ 1208
+L A + PL L +S+ D + YR LVG L YL +TRP+LSY+V+T++QF+Q
Sbjct: 1192 LLGARPSAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPELSYSVHTLAQFMQ 1251
Query: 1209 APTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFL 1268
P +H+ A R++RY+ G+ TS+ + G+ D+D+A C TRRS GY + L
Sbjct: 1252 NPRQDHWNAAIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQL 1311
Query: 1269 GDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQS 1328
GD +SW KKQPTV+RSS E+EYRAMA EL+WL +L++L V + SD++S
Sbjct: 1312 GDTPISWKTKKQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKS 1371
Query: 1329 ALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIF 1388
A+ ++ NPV H+ KH+++DCHF+ + + G +A VP+ QLADI TKAL F
Sbjct: 1372 AIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYF 1431
Query: 1389 RSKLRV 1394
KL +
Sbjct: 1432 LRKLGI 1437
>At4g27200 putative protein
Length = 819
Score = 541 bits (1393), Expect = e-153
Identities = 308/750 (41%), Positives = 413/750 (55%), Gaps = 66/750 (8%)
Query: 689 GCRVFPCLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPF 748
G FP LR Y NKF+P S C+F GY+ +KG++C P Y+S H FDE +PF
Sbjct: 15 GSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPF 74
Query: 749 AGSHSSPHDLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVP 808
+ ++ H + S SP P +P S P+ + S P LP
Sbjct: 75 SHTYKHLHPQPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFT--------SADFPPLPQR 126
Query: 809 PADAPPSPAPPSPTQTATPPVGTQAPPASPPTTPPV------------------------ 844
P+ P S A+ Q+P T
Sbjct: 127 KTPLLPTLVPISSVSHASNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSDSEETIQQ 186
Query: 845 ----VAQVPLAPIAARPRTRSQNGIFRPNPRYALVHAQPTGILTAFHTVT--KPKGFKSA 898
V Q P + TR++ GI +PNPRY + H V+ +PK +A
Sbjct: 187 ASVNVHQTPASTNVHPMVTRAKVGISKPNPRYVFLS----------HKVSYPEPKTVTAA 236
Query: 899 MKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVA 958
+KHP W AM +E+ + TW+LVP S +V+G KWVFRTK H+DGT+ +LKAR+VA
Sbjct: 237 LKHPGWTGAMTEEIGNCSETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVA 296
Query: 959 QGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQ 1018
+GF Q G DY T+SPVV+ TVRL+L +W + Q+DVKNAFLHG L ETVYM Q
Sbjct: 297 KGFLQEEGIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQ 356
Query: 1019 PPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHT 1078
P D HVC L+K++YGLKQ+PRAWF + S+FLL GF C ++DPSLF + +
Sbjct: 357 PAANRD-----HVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCRKSDPSLFIYAHNNN 411
Query: 1079 TLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGY-FLGLEITYTPDGLFLGQ 1137
+ LL+ LT +A LN EF + +G+ FLG+++ +GLF+ Q
Sbjct: 412 LILLLL-----------SQTLTSLLAALNKEFRMTDMGQHSLTFLGIQVQRQQNGLFMSQ 460
Query: 1138 AKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAYF-DPTHYRSLVGALQYLTITRPDL 1196
KYA DLL A M + + TPL V E F DPT++RS+ G LQYLT+TRPD+
Sbjct: 461 QKYAEDLLIAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDI 520
Query: 1197 SYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTD 1256
+AVN V Q + PT+ F +KRILRY+ GT G+++ R S + YSD+DW C
Sbjct: 521 QFAVNFVCQKMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYSDSDWGNCKQ 580
Query: 1257 TRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRL 1316
TRRS G F+G NL+SWS+KK PTV+RSS E+EY+++++ ASE++WL LL ELR+ L
Sbjct: 581 TRRSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPL 640
Query: 1317 SATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIF 1376
TP L DN SA+++ NP H KH D+D HFV E VA L V+H+P S Q+ADIF
Sbjct: 641 PDTPELFCDNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIF 700
Query: 1377 TKALPRPLFEIFRSKLRVGLNPTLSLKGVL 1406
TK+LP F R KL V L PT SL+G +
Sbjct: 701 TKSLPYEAFIHLRGKLGVTLPPTPSLRGTI 730
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 528 bits (1361), Expect = e-150
Identities = 312/832 (37%), Positives = 450/832 (53%), Gaps = 33/832 (3%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
IK +SDNGTEF + F GV+H SC T +N RVE+KHRH+L + A+ + +
Sbjct: 510 IKTVRSDNGTEFLC--LTKFFQEQGVIHERSCVATPERNDRVERKHRHLLNVARALRFQA 567
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
++P ++W + TA Y+INR PS VL++ P++ L P + + FG + R
Sbjct: 568 NLPIQFWGECVLTAAYLINRTPSSVLNDSTPYERLHKKQPRFDHLRVFGSLCYAHNRNRG 627
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPH---- 756
+KF+ RS C+F GY KG++ F+ + +VS F E FPF SH
Sbjct: 628 GDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVFSELEFPFRISHEQNVIEEE 687
Query: 757 ---------DLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPV 807
D ++ + P+P P+ + +P S S S S +P
Sbjct: 688 EEALWAPIVDGLIEEEVHLGQNAGPTP-PICVSSPISPSATSSRSEHSTSSPLDTEVVPT 746
Query: 808 PPADAPPSPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFR 867
P + +P SPT P+ P + PP V P R TR++
Sbjct: 747 PATSTTSASSPSSPTNLQFLPLSRAKPTTAQAVAPPAV------PPPRRQSTRNKAPPVT 800
Query: 868 PNP---RYALVHAQPTGILTAFHTVTKPKGFKS-AMKHP*WLAAMEDELSALHKNCTWTL 923
+ P+ + + + + K + + H ++A + A +N TWT+
Sbjct: 801 LKDFVVNTTVCQESPSKLNSILYQLQKRDDTRRFSASHTTYVA-----IDAQEENHTWTI 855
Query: 924 VPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVR 983
P +G +WV++ K +SDG+VER KARLVA G Q G DY TF+PV K TVR
Sbjct: 856 EDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKEGEDYGETFAPVAKMATVR 915
Query: 984 LILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQ 1043
L L V +W++HQ+DV NAFLHG L E VYM+ PPGF + P VCRL KALYGLKQ
Sbjct: 916 LFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGF-EASHPNKVCRLRKALYGLKQ 974
Query: 1044 APRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFI 1103
APR WF++L++ L R+GF S AD SLF KG + +L+YVDD+I+TG+ QF
Sbjct: 975 APRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYVDDLIITGNSQRATQQFK 1034
Query: 1104 ARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAG 1163
L + F +K LG L YFLG+E+ + G+++ Q KYA D++S +L + PL
Sbjct: 1035 EYLASCFHMKDLGPLKYFLGIEVARSTTGIYICQRKYALDIISETGLLGVKPANFPLEQN 1094
Query: 1164 SHL-VSSGEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRIL 1222
L +S+ DP YR LVG L YL +TR DL+++V+ +++F+Q P +H+ A R++
Sbjct: 1095 HKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSVHILARFMQEPREDHWAAALRVV 1154
Query: 1223 RYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPT 1282
RY+ G+ RR+ + G+ D+DWA +RRS GY + GD+ +SW KKQ T
Sbjct: 1155 RYLKADPGQGVFLRRSGDFQITGWCDSDWAGDPMSRRSVTGYFVQFGDSPISWKTKKQDT 1214
Query: 1283 VARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHA 1342
V++SS E+EYRAM+ ASEL+WL LL L V ++ D++SA+++A NPV H+
Sbjct: 1215 VSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCDSKSAIYIATNPVFHERT 1274
Query: 1343 KHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
KHI++D HFV + G + RHV T+ QLADIFTK L R F FR KL +
Sbjct: 1275 KHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFTKPLGRDCFSAFRIKLGI 1326
>At3g61330 copia-type polyprotein
Length = 1352
Score = 497 bits (1280), Expect = e-140
Identities = 288/828 (34%), Positives = 435/828 (51%), Gaps = 67/828 (8%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
IK +SD G EFT+ + +G+ + + P + QNG VE+K+R +LE+ +ML
Sbjct: 590 IKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVVERKNRTILEMARSMLKSK 649
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
+P W +A + AVY++NR P+K +S + P + P ++ FG +
Sbjct: 650 RLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEK 709
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPHDLVV 760
+K +S IF GY ++ KG+K +NP T +S + FDE S+ ++
Sbjct: 710 RSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFF- 768
Query: 761 STFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPS 820
P ++ + P P + PPS P
Sbjct: 769 ---------------------------------PHFEEDE-----PEPTREEPPSEEP-- 788
Query: 821 PTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPRYALVHAQPT 880
TPP T P +Q+ + PR RS + Y + Q
Sbjct: 789 ----TTPP------------TSPTSSQIEESSSERTPRFRSIQEL------YEVTENQEN 826
Query: 881 GILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFR 940
L +P F+ A++ W AM++E+ ++ KN TW L P+ +G+KWV++
Sbjct: 827 LTLFCLFAECEPMDFQKAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYK 886
Query: 941 TKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLD 1000
K +S G VER KARLVA+G++Q G DY F+PV + TVRLI+S N W++HQ+D
Sbjct: 887 AKKNSKGEVERYKARLVAKGYSQRVGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMD 946
Query: 1001 VKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHG 1060
VK+AFL+G L E VY+EQP G++ V RL K LYGLKQAPRAW R+ +
Sbjct: 947 VKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKD 1006
Query: 1061 FSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGY 1120
F + +L+ + L +YVDD+I TG++PS+ +F + EF + +G + Y
Sbjct: 1007 FIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFEMTDIGLMSY 1066
Query: 1121 FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAY-FDPTHY 1179
+LG+E+ +G+F+ Q YA ++L + + +++ V TP+ G L E DPT +
Sbjct: 1067 YLGIEVKQEDNGIFITQEGYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEEGEGVDPTTF 1126
Query: 1180 RSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTS 1239
+SLVG+L+YLT TRPD+ YAV VS++++ PT HF+A KRILRY+ GT +FGL + TS
Sbjct: 1127 KSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTS 1186
Query: 1240 SPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTA 1299
++GYSD+DW D R+ST G+ ++GD +W +KKQP V S+CE+EY A +
Sbjct: 1187 DYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCV 1246
Query: 1300 SELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASG 1359
+WL NLL EL + + DN+SA+ +A+NPV H +KHID H++ E V+
Sbjct: 1247 CHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKK 1306
Query: 1360 RLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRVGLNPTLSLKGVLE 1407
+ + +V T Q+AD FTK L R F RS L V + SL+G +E
Sbjct: 1307 DVQLEYVKTHDQVADFFTKPLKRENFIKMRSLLGVAKS---SLRGGVE 1351
>At1g48710 hypothetical protein
Length = 1352
Score = 496 bits (1278), Expect = e-140
Identities = 289/828 (34%), Positives = 434/828 (51%), Gaps = 67/828 (8%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
IK +SD G EFT+ + +G+ + + P + QNG E+K+R +LE+ +ML
Sbjct: 590 IKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSK 649
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
+P W +A + AVY++NR P+K +S + P + ++ FG +
Sbjct: 650 RLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKSGVSHLRVFGSIAHAHVPDEK 709
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPHDLVV 760
+K +S IF GY ++ KG+K +NP T +S + FDE S+ ++
Sbjct: 710 RSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFF- 768
Query: 761 STFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPS 820
P ++ + P P + PPS P
Sbjct: 769 ---------------------------------PHFEEDE-----PEPTREEPPSEEP-- 788
Query: 821 PTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPRYALVHAQPT 880
TPP T P +Q+ + PR RS + Y + Q
Sbjct: 789 ----TTPP------------TSPTSSQIEESSSERTPRFRSIQEL------YEVTENQEN 826
Query: 881 GILTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFR 940
L +P F+ A++ W AM++E+ ++ KN TW L P+ +G+KWV++
Sbjct: 827 LTLFCLFAECEPMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKTIGVKWVYK 886
Query: 941 TKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLD 1000
K +S G VER KARLVA+G+ Q G DY F+PV + TVRLI+S N W++HQ+D
Sbjct: 887 AKKNSKGEVERYKARLVAKGYIQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMD 946
Query: 1001 VKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHG 1060
VK+AFL+G L E VY+EQP G++ V RL KALYGLKQAPRAW R+ +
Sbjct: 947 VKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKD 1006
Query: 1061 FSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGY 1120
F + +L+ + L +YVDD+I TG++PS+ +F + EF + +G + Y
Sbjct: 1007 FIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSY 1066
Query: 1121 FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAY-FDPTHY 1179
+LG+E+ +G+F+ Q YA ++L + M +++ V TP+ G L E DPT +
Sbjct: 1067 YLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTF 1126
Query: 1180 RSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTS 1239
+SLVG+L+YLT TRPD+ YAV VS++++ PT HF+A KRILRY+ GT +FGL + TS
Sbjct: 1127 KSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTS 1186
Query: 1240 SPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTA 1299
++GYSD+DW D R+ST G+ ++GD +W +KKQP V S+CE+EY A +
Sbjct: 1187 DYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVVLSTCEAEYVAATSCV 1246
Query: 1300 SELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASG 1359
+WL NLL EL + + DN+SA+ +A+NPV H +KHID H++ E V+
Sbjct: 1247 CHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKK 1306
Query: 1360 RLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRVGLNPTLSLKGVLE 1407
+ + +V T Q+ADIFTK L R F RS L V + SL+G +E
Sbjct: 1307 DVQLEYVKTHDQVADIFTKPLKREDFIKMRSLLGVAKS---SLRGGVE 1351
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 439 bits (1128), Expect = e-123
Identities = 281/736 (38%), Positives = 389/736 (52%), Gaps = 47/736 (6%)
Query: 700 MNNKFSPRSTP---CIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSPH 756
M ++FS TP + Y S +KG+K + ++ + F E FPF S
Sbjct: 316 MIHQFSCAYTPQQNSVVERYPSGYKGYKVLDLESHSISITRNVVFHETKFPFKTSKFLKE 375
Query: 757 DLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADA-PPS 815
V F + P P P+ V S+P+ DD++ + A + PP
Sbjct: 376 S--VDMF---PNSILPLPAPLHFV---ESMPLDDDL--RADDNNASTSNSASSASSIPPL 425
Query: 816 PAPPSPTQTATPPVGTQAPPASPPT----TPPVVAQ-----VP----LAPIAARPRTRSQ 862
P+ + T + T + P + P P +++ VP L+P +
Sbjct: 426 PSTVNTQNTDALDIDTNSVPIARPKRNAKAPAYLSEYHCNSVPFLSSLSPTTSTSIETPS 485
Query: 863 NGIFRPNPRYALVHAQPTGI------------LTAFHTVTKPKGFKSAMKHP*WLAAMED 910
+ I P + + T I + A++ T+PK F AMK W A +
Sbjct: 486 SSI--PPKKITTPYPMSTAISYDKLTPLFHSYICAYNVETEPKAFTQAMKSEKWTRAANE 543
Query: 911 ELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYS 970
EL AL +N TW + NVVG KWVF K++ DG++ER KARLVAQGFTQ G DY
Sbjct: 544 ELHALEQNKTWIVESLTEGKNVVGCKWVFTIKYNPDGSIERYKARLVAQGFTQQEGIDYM 603
Query: 971 LTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPR---F 1027
TFSPV K +V+L+L W L Q+DV NAFLHG L E +YM P G+ P
Sbjct: 604 ETFSPVAKFGSVKLLLGLAAATGWSLTQMDVSNAFLHGELDEEIYMSLPQGYTPPTGISL 663
Query: 1028 PTH-VCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYV 1086
P+ VCRL K+LYGLKQA R W++RLSS L F S AD ++F + + +LVYV
Sbjct: 664 PSKPVCRLLKSLYGLKQASRQWYKRLSSVFLGANFIQSPADNTMFVKVSCTSIIVVLVYV 723
Query: 1087 DDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDLLS 1146
DD+++ +D S + L +EF IK LG +FLGLEI + +G+ + Q KYA +LL
Sbjct: 724 DDLMIASNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVCQRKYAQNLLE 783
Query: 1147 RAMMLEASHVSTPLAAGSHLVSS-GEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVSQ 1205
+ S P+ HL G + T YR LVG L YL ITRPD+++AV+T+SQ
Sbjct: 784 DVGLSGCKPSSIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDITFAVHTLSQ 843
Query: 1206 FLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYA 1265
FL APT H QA ++LRY+ G GL + +S + G+SDADW C D+RRS G+
Sbjct: 844 FLSAPTDIHMQAAHKVLRYLKGNPGQGLMYSASSELCLNGFSDADWGTCKDSRRSVTGFC 903
Query: 1266 IFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSD 1325
I+LG +L++W +KKQ V+RSS ESEYR++A E++WL LL +L V ++ L D
Sbjct: 904 IYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCD 963
Query: 1326 NQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLF 1385
N+SAL +A NPV H+ KHI++DCH V + + +G+L HVPT QLADI TK L
Sbjct: 964 NKSALHLATNPVFHERTKHIEIDCHTVRDQIKAGKLKTLHVPTGNQLADILTKPLHPVQS 1023
Query: 1386 EIFRSKLR-VGLNPTL 1400
IF R G +P L
Sbjct: 1024 PIFSLLFRFTGTSPVL 1039
Score = 32.3 bits (72), Expect = 2.0
Identities = 17/44 (38%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQ 624
IK +SDN E K G++H+FSC +T QN VE+
Sbjct: 294 IKAIRSDNVKELAFTK---FVKEQGMIHQFSCAYTPQQNSVVER 334
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 429 bits (1102), Expect = e-120
Identities = 227/508 (44%), Positives = 309/508 (60%), Gaps = 6/508 (1%)
Query: 892 PKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVER 951
P+ + A W A++ E+ A+ + TW + P VG KWVF KFH+DG++ER
Sbjct: 947 PQSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWVFTVKFHADGSLER 1006
Query: 952 LKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLT 1011
KAR+VA+G+TQ G DY+ TFSPV K TV+L+L W L+QLD+ NAFL+G L
Sbjct: 1007 FKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQLDISNAFLNGDLE 1066
Query: 1012 ETVYMEQPPGFVDPRF----PTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRAD 1067
ET+YM+ P G+ D + P VCRL K++YGLKQA R WF + S+ LL GF D
Sbjct: 1067 ETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSNSLLALGFEKQHGD 1126
Query: 1068 PSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEIT 1127
+LF G + LLVYVDDI++ + L A F ++ LG L YFLGLE+
Sbjct: 1127 HTLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYFLGLEVA 1186
Query: 1128 YTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSS-GEAYFDPTHYRSLVGAL 1186
T +G+ L Q KYA +LL+ A ML+ S P+ L + G D YR LVG L
Sbjct: 1187 RTSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYRRLVGKL 1246
Query: 1187 QYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGY 1246
YLTITRPD+++AVN + QF AP H AV ++L+Y+ GT GL + + GY
Sbjct: 1247 MYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGLFYSAEDDLTLKGY 1306
Query: 1247 SDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLL 1306
+DADW C D+RRST G+ +F+G +L+SW +KKQPTV+RSS E+EYRA+A + E+ WL
Sbjct: 1307 TDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALASCEMAWLS 1366
Query: 1307 NLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHV 1366
LL LRV S P+L SD+ +A+++A NPV H+ KHI++DCH V E + +G+L + HV
Sbjct: 1367 TLLLALRVH-SGVPILYSDSTAAVYIATNPVFHERTKHIEIDCHTVREKLDNGQLKLLHV 1425
Query: 1367 PTSLQLADIFTKALPRPLFEIFRSKLRV 1394
T Q+ADI TK L F SK+ +
Sbjct: 1426 KTKDQVADILTKPLFPYQFAHLLSKMSI 1453
Score = 128 bits (322), Expect = 2e-29
Identities = 62/174 (35%), Positives = 96/174 (54%), Gaps = 3/174 (1%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
+K +SDN E + + + G++ SCP T QN VE+KH+H+L + A+++ S
Sbjct: 688 VKSVRSDNAKELAFTE---FYKAKGIVSFHSCPETPEQNSVVERKHQHILNVARALMFQS 744
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
++ YW D TAV++INR PS +LSN+ PF++L P Y+ FGC +
Sbjct: 745 NMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLPDYSQLKTFGCLCYSSTSSKQ 804
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSS 754
+KF PRS C+F GY KG+K + ++ ++S + +F E FP A S S
Sbjct: 805 RHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVEFHEELFPLASSQQS 858
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 422 bits (1086), Expect = e-118
Identities = 225/513 (43%), Positives = 315/513 (60%), Gaps = 2/513 (0%)
Query: 883 LTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTK 942
L A +P + AM W AM E+ +L N T+++V P +G KWV++ K
Sbjct: 591 LAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFSIVNLPPGKRALGNKWVYKIK 650
Query: 943 FHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVK 1002
+ SDG +ER KARLV G Q G DY TF+PV K +TVRL L DW +HQ+DV
Sbjct: 651 YRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTVRLFLGVAAARDWHVHQMDVH 710
Query: 1003 NAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFS 1062
NAFLHG L E VYM+ P GF P+ VCRL+K+LYGLKQAPR WF +LSS L ++GF+
Sbjct: 711 NAFLHGDLKEEVYMKLPQGFQCDD-PSKVCRLHKSLYGLKQAPRCWFSKLSSALKQYGFT 769
Query: 1063 CSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFL 1122
S +D SLF + +++LVYVDD+I++GS P + QF + L + F +K LG L YFL
Sbjct: 770 QSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDAVAQFKSYLESCFHMKDLGLLKYFL 829
Query: 1123 GLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHL-VSSGEAYFDPTHYRS 1181
G+E++ G +L Q KY D++S +L A + PL L +S+ D + YR
Sbjct: 830 GIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFPLEQNHKLSLSTSPLLSDSSRYRR 889
Query: 1182 LVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSP 1241
LVG L YL +TRP+LSY+V+T++QF+Q P +H+ A R++RY+ G+ TS+
Sbjct: 890 LVGRLIYLAVTRPELSYSVHTLAQFMQNPRQDHWNAAIRVVRYLKSNPGQGILLSSTSTL 949
Query: 1242 AVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASE 1301
+ G+ D+D+A C TRRS GY + LGD +SW KKQPT++RSS E+EYRAMA E
Sbjct: 950 QINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKTKKQPTISRSSAEAEYRAMAFLTQE 1009
Query: 1302 LVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRL 1361
L+WL +L++L V + SD++SA+ ++ NPV H+ KH+++DCHF+ + + G +
Sbjct: 1010 LMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGII 1069
Query: 1362 AVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
A VP+ QLADI TKAL F KL +
Sbjct: 1070 ATSFVPSHKQLADILTKALGEKEVRYFLRKLGI 1102
Score = 129 bits (323), Expect = 2e-29
Identities = 62/175 (35%), Positives = 99/175 (56%), Gaps = 2/175 (1%)
Query: 575 KSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGL 634
+ IK +SDNGTEF + + F G+ H SC T QNGRVE+KHRH+L +
Sbjct: 404 RQFDTEIKTVRSDNGTEFLCMREY--FLHKGIAHETSCVGTPHQNGRVERKHRHILNIAR 461
Query: 635 AMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFP 694
A+ + S++P ++W + +A Y+INR PS +L + P+++L+ AP Y++ FG +
Sbjct: 462 ALRFQSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYA 521
Query: 695 CLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFA 749
+ + +KF+ RS C+F GY KG++ F+ +VS F E FP++
Sbjct: 522 HNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYS 576
>At1g58140 hypothetical protein
Length = 1320
Score = 422 bits (1084), Expect = e-118
Identities = 218/518 (42%), Positives = 319/518 (61%), Gaps = 4/518 (0%)
Query: 891 KPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVE 950
+P F+ A++ W AM++E+ ++ KN TW L P+ +G+KWV++ K +S G VE
Sbjct: 805 EPMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVE 864
Query: 951 RLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHL 1010
R KARLVA+G++Q G DY F+PV + TVRLI+S N W++HQ+DVK+AFL+G L
Sbjct: 865 RYKARLVAKGYSQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDL 924
Query: 1011 TETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSL 1070
E VY+EQP G++ V RL KALYGLKQAPRAW R+ + F + +L
Sbjct: 925 EEEVYIEQPQGYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHAL 984
Query: 1071 FFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTP 1130
+ + L +YVDD+I TG++PS+ +F + EF + +G + Y+LG+E+
Sbjct: 985 YIKIQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQED 1044
Query: 1131 DGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAY-FDPTHYRSLVGALQYL 1189
+G+F+ Q YA ++L + M +++ V TP+ G L E DPT ++SLVG+L+YL
Sbjct: 1045 NGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYL 1104
Query: 1190 TITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDA 1249
T TRPD+ YAV VS++++ PT HF+A KRILRY+ GT +FGL + TS ++GYSD+
Sbjct: 1105 TCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDS 1164
Query: 1250 DWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLL 1309
DW D R+ST G+ ++GD +W +KKQP V S+CE+EY A + +WL NLL
Sbjct: 1165 DWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLL 1224
Query: 1310 HELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTS 1369
EL + + DN+SA+ +A+NPV H +KHID H++ E V+ + + +V T
Sbjct: 1225 KELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTH 1284
Query: 1370 LQLADIFTKALPRPLFEIFRSKLRVGLNPTLSLKGVLE 1407
Q+ADIFTK L R F RS L V + SL+G +E
Sbjct: 1285 DQVADIFTKPLKREDFIKMRSLLGVAKS---SLRGGVE 1319
Score = 99.0 bits (245), Expect = 2e-20
Identities = 51/163 (31%), Positives = 84/163 (51%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
IK +SD G EFT+ + +G+ + + P + QNG E+K+R +LE+ +ML
Sbjct: 590 IKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSK 649
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
+P W +A + AVY++NR P+K +S + P + P ++ FG +
Sbjct: 650 RLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEK 709
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDE 743
+K +S IF GY ++ KG+K +NP T +S + FDE
Sbjct: 710 RSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDE 752
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.347 0.151 0.556
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,470,547
Number of Sequences: 26719
Number of extensions: 1309462
Number of successful extensions: 17633
Number of sequences better than 10.0: 413
Number of HSP's better than 10.0 without gapping: 243
Number of HSP's successfully gapped in prelim test: 183
Number of HSP's that attempted gapping in prelim test: 7951
Number of HSP's gapped (non-prelim): 2382
length of query: 1411
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1299
effective length of database: 8,326,068
effective search space: 10815562332
effective search space used: 10815562332
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0321.5


