

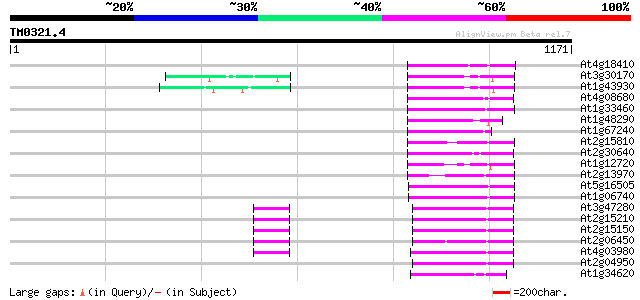
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.4
(1171 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g18410 MuDR transposable element - like protein 123 7e-28
At3g30170 unknown protein 121 3e-27
At1g43930 hypothetical protein 120 5e-27
At4g08680 putative MuDR-A-like transposon protein 119 1e-26
At1g33460 mutator transposase MUDRA, putative 116 7e-26
At1g48290 hypothetical protein 106 7e-23
At1g67240 hypothetical protein 104 3e-22
At2g15810 Mutator-like transposase 101 3e-21
At2g30640 Mutator-like transposase 94 5e-19
At1g12720 hypothetical protein 90 7e-18
At2g13970 Mutator-like transposase 88 3e-17
At5g16505 mutator-like transposase-like protein (MQK4.25) 88 3e-17
At1g06740 mudrA-like protein 85 2e-16
At3g47280 putative protein 81 4e-15
At2g15210 putaive transposase of transposon FARE2.7 (cds1) 81 4e-15
At2g15150 putative transposase of FARE2.2 (CDS1) 81 4e-15
At2g06450 putative transposase of FARE2.11 80 5e-15
At4g03980 putative transposon protein 79 2e-14
At2g04950 putative transposase of transposon FARE2.9 (cds1) 79 2e-14
At1g34620 Mutator-like protein 79 2e-14
>At4g18410 MuDR transposable element - like protein
Length = 633
Score = 123 bits (308), Expect = 7e-28
Identities = 69/230 (30%), Positives = 121/230 (52%), Gaps = 11/230 (4%)
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSE--LKVALWKAARASTVQEWKQEMEL 887
+GL+ +Q +P ++HR+CV+HIYGN +K Y GS+ +K LW A + E+KQ +E
Sbjct: 303 KGLIKAVQLELPKIEHRMCVQHIYGNLKKTY-GSKTMIKPLLWNLAWSYNETEYKQHLEK 361
Query: 888 MKAFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEG 947
++ + + Y +M+ + W R+ + C+ NN E+FN ++ + R+K +++LE
Sbjct: 362 IRCYDTKVYESVMKTNPRSWVRAFQKIGSFCEDVDNNSVESFNGSLNKAREKQFVAMLET 421
Query: 948 LKFYINNRIVKQRQLMLRWKTSIICPLIQQRL--EFSKEAADKCVAVWNGSDNFRHFQVK 1005
++ RI K R + T + P + + L E + K NG ++V+
Sbjct: 422 IRRMAMVRIAK-RSVESHTHTGVCTPYVMKFLAGEHKVASTAKVAPGTNGM-----YEVR 475
Query: 1006 RDSDSYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSYR 1055
D++ VDL TC C +WQ+ GIPC H+ + + PE++V +R
Sbjct: 476 HGGDTHRVDLAAYTCTCIKWQICGIPCEHAYGVILHKKLQPEDFVCQWFR 525
>At3g30170 unknown protein
Length = 995
Score = 121 bits (303), Expect = 3e-27
Identities = 69/232 (29%), Positives = 119/232 (50%), Gaps = 20/232 (8%)
Query: 831 GLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKA 890
GLV + ++P +HR C KHI N+++ EL+ WK AR+ TV+E+ M ++
Sbjct: 480 GLVKAIHTILPHAEHRQCSKHIMDNWKRDSHDLELQRLFWKIARSYTVEEFNNHMMELQQ 539
Query: 891 FSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKF 950
++ +AY + W+R+ + T C+ +NN+ E+FN + + R KPI+ +LE ++
Sbjct: 540 YNRQAYESLQLTSPVTWSRAFFRIGTCCNDNLNNLSESFNRTIRQARRKPILDMLEDIR- 598
Query: 951 YINNRIVKQRQLMLR-WKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKR--- 1006
RQ M+R K +I +Q R F+K A ++ + G + +
Sbjct: 599 ---------RQCMVRSAKRFLIAEKLQTR--FTKRAHEEIEKMIVGLRQCERYMARENLH 647
Query: 1007 ----DSDSYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
++ SY VD+E++TC CR+ Q+ GIPC H+ + + E+YV Y
Sbjct: 648 EIYVNNVSYFVDMEQKTCDCRKCQMVGIPCIHATCVIIGKKEKVEDYVSDYY 699
Score = 43.5 bits (101), Expect = 7e-04
Identities = 62/290 (21%), Positives = 115/290 (39%), Gaps = 40/290 (13%)
Query: 326 QVEKEAEKEIGSVV----LECERQVEKEAEKENQVVGEQAGKEKVVVEEAEKENQVVGEQ 381
+V E E +G V +E +E A E+ V + K+ V + E E ++ +
Sbjct: 27 EVAVELESVVGDGVEVPPVEAPVGIELVARGEDCVPSRHRKRRKIRVGD-EDEAEIDPDP 85
Query: 382 AGKEKVVV---EQAEKENQGVGGEEDDESYHPNSDDS------------DDSSVVSLDDS 426
++ V E + VGG++D N D++ +D + DD
Sbjct: 86 DYEDDCAVHGDEDCDAYTDAVGGDDDATGGEGNDDEAVGGGDDATGCGGNDDEALGYDDD 145
Query: 427 DYEE--NWDWTTVLPRESLLDIPHDAPPPDLYELSLPVDTNDPGATYSDFEDEDGDSDDL 484
+ N D + ++ L++ + P + D +D D D+ + D +
Sbjct: 146 ATSDGGNDDDDGGVEEDANLNLAEEFPE----FAGVEEDCSD-----DDPPDDVWEEDKI 196
Query: 485 ESP--SEGEGEGRARRYPRFRPAEDGDGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKI 542
P S+ E E R R R A + + V L + +++ D F AV ++L+ + D+K+
Sbjct: 197 PDPFSSDDEDESRTREVHGHRDAANEE-VLLELKKTYNTPDDFKLAVLRYSLKTRYDIKL 255
Query: 543 VKNDKKRVVVNCVK------GCPFNLRISKTNSRHYFLLVTYEAAHNCCR 586
+++ + V C CP+ + S + H + TY H C R
Sbjct: 256 FRSEARIVAAKCSYVDKDGVQCPWKVYCSYEKTMHKMQIRTYVNNHICVR 305
>At1g43930 hypothetical protein
Length = 946
Score = 120 bits (301), Expect = 5e-27
Identities = 67/233 (28%), Positives = 119/233 (50%), Gaps = 21/233 (9%)
Query: 831 GLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKA 890
GLV + +IP +HR C +HI N+++ EL+ WK R+ T E+ M +K+
Sbjct: 508 GLVKAIHSVIPQAEHRQCARHIMENWKRNSHDMELQRLFWKIVRSYTEGEFGAHMRALKS 567
Query: 891 FSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKF 950
++ A+ +++ W+R+ + + C+ +NN+ E+FN + E R KP++ +LE ++
Sbjct: 568 YNASAFELLLKTLPVTWSRAFFKIGSCCNDNLNNLSESFNRTIREARRKPLLDMLEDIR- 626
Query: 951 YINNRIVKQRQLMLR-WKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDS- 1008
RQ M+R K +I ++ R F+K A + + GS + + + +
Sbjct: 627 ---------RQCMVRNEKRYVIADRLRTR--FTKRAHAEIEKMIAGSQVCQRWMARHNKH 675
Query: 1009 -------DSYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
DS+ VD+ TC C +WQ+TGIPC H+ + + R+ E+YV Y
Sbjct: 676 EIKVGPVDSFTVDMNNNTCGCMKWQMTGIPCIHAASVIIGKRQKVEDYVSDWY 728
Score = 44.3 bits (103), Expect = 4e-04
Identities = 60/295 (20%), Positives = 114/295 (38%), Gaps = 29/295 (9%)
Query: 314 KDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAEKENQVVGEQAGKEKVVVEEAEK 373
+D+ +V + + +++ K V++CE E ++ + V G++ E +E
Sbjct: 26 EDVPQLVDVVQPKSNRKSRKRKKDDVVDCEEDREALSDDDCAVDGDEDCDEVCYADERVG 85
Query: 374 ENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPNSDDSDDSSVVSLD-----DSDY 428
N+ +GE+ V VE + GV E DD D +D +V + +++
Sbjct: 86 NNKQIGEEGECHIVCVEAEVNDGGGVEVEVDDGG--GLERDVNDGGIVEAEVNNGGNAEA 143
Query: 429 EENWDWTTVLPRESLLDIPHDAPPP-DLYELSLPVDTNDPGATYSDFEDE-DGDSDD--- 483
E N + + + D+ E + GA + E+E DGDS D
Sbjct: 144 EVNDGGGVEAEVNNGESVEEEGNGGLDMEEEDCEDFEAEFGAEAREEENESDGDSGDDIW 203
Query: 484 ---------LESPSEGEGEGRARRYPRFRPAEDGDGVNFVLGQVFDSKDIFIHAVKEFAL 534
+ S EG+ + P D + L + F++ + F AV ++L
Sbjct: 204 DDERIPDPLVHSDDEGDNDDEDG-----APQTDDPEESLRLAKTFNNPEDFKIAVLRYSL 258
Query: 535 QHKKDLKIVKNDKKRVVVNCVK---GCPFNLRISKTNSRHYFLLVTYEAAHNCCR 586
+ + D+K+ ++ RV C C + S + + Y+ H+C R
Sbjct: 259 KTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSYDKKKQKMQVKVYKDGHSCVR 313
Score = 32.7 bits (73), Expect = 1.2
Identities = 48/185 (25%), Positives = 73/185 (38%), Gaps = 47/185 (25%)
Query: 271 SDDDVAIVDRLDVICGKLLQGEKDVEKDKVIQEE----VVSVEKEVEKDIGSVVLECERQ 326
SDDD A+ D C ++ ++ V +K I EE +V VE EV D G V +E +
Sbjct: 62 SDDDCAVDGDED--CDEVCYADERVGNNKQIGEEGECHIVCVEAEVN-DGGGVEVEVDDG 118
Query: 327 VEKEAEKEIGSVVL-------ECERQVEK----EAEKENQVVGEQAGKEKVVVEEAEKEN 375
E + G +V E +V EAE N E+ G + +EE + E+
Sbjct: 119 GGLERDVNDGGIVEAEVNNGGNAEAEVNDGGGVEAEVNNGESVEEEGNGGLDMEEEDCED 178
Query: 376 QVVGEQAGKEKVVVEQAEKENQGVG----------------------GEEDDESYHPNSD 413
+ + E E+EN+ G G+ DDE P +D
Sbjct: 179 F-------EAEFGAEAREEENESDGDSGDDIWDDERIPDPLVHSDDEGDNDDEDGAPQTD 231
Query: 414 DSDDS 418
D ++S
Sbjct: 232 DPEES 236
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 119 bits (298), Expect = 1e-26
Identities = 67/222 (30%), Positives = 119/222 (53%), Gaps = 5/222 (2%)
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSE-LKVALWKAARASTVQEWKQEMELM 888
+GL+ +++ +P+ +HR+CVKHI N +K + + LK +WK A + +E+ + + +
Sbjct: 376 KGLLSAVKQELPNAEHRMCVKHIVENLKKNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNL 435
Query: 889 KAFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGL 948
+ + E YND++ W+R Y + C+ NN E+FN + + R K +I +LE +
Sbjct: 436 RCYDEALYNDVLNEEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETI 495
Query: 949 KFYINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDS 1008
+ RIVK+ + LR + ++ L K AD+ V+ + F+V D
Sbjct: 496 RRQGMTRIVKRNKKSLRHEGRFTKYALKM-LALEKTDADRS-KVYRCTHGV--FEVYIDE 551
Query: 1009 DSYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYV 1050
+ + VD+ + C+C +WQ++GIPC HS M A + ENY+
Sbjct: 552 NGHRVDIPKTQCSCGKWQISGIPCEHSYGAMIEAGLDAENYI 593
Score = 34.3 bits (77), Expect = 0.43
Identities = 30/115 (26%), Positives = 49/115 (42%), Gaps = 4/115 (3%)
Query: 472 SDFEDEDGDSDDLESPSEGEGEGRARRYPRFRPAEDGDGVNFVLGQVFDSKDIFIHAVKE 531
++FEDE D+ S E E R R G FV+G F + F A+ E
Sbjct: 70 ANFEDEIPIGRDIIPESSDEDEDPI--LVRDRRIRRAMGDKFVIGSTFFTGIEFKEAILE 127
Query: 532 FALQHKKDLKIVKNDKKRVVVNCVKG--CPFNLRISKTNSRHYFLLVTYEAAHNC 584
L+H ++ + +K+++ C G C + + S R F++ T H+C
Sbjct: 128 VTLKHGHNVVQDRWEKEKISFKCGMGGKCKWRVYCSYDPDRQLFVVKTSCTWHSC 182
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 116 bits (291), Expect = 7e-26
Identities = 66/228 (28%), Positives = 120/228 (51%), Gaps = 7/228 (3%)
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSEL-KVALWKAARASTVQEWKQEMELM 888
+G++ ++ +P+ +HR CVKHI N +K++ +L K +W A + + ++K + M
Sbjct: 442 KGIISAVKNALPNAEHRPCVKHIVENLKKRHGSLDLLKKFVWNLAWSYSDTQYKANLNEM 501
Query: 889 KAFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGL 948
+A+ Y D+M+ W RS + + C+ NN E+FN +++ R K ++ ++E +
Sbjct: 502 RAYIMSLYEDVMKEEPNTWCRSWFRIGSYCEDVDNNATESFNATIVKARAKALVPMMETI 561
Query: 949 KFYINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDS 1008
+ RI K++ + +WK I + + L+ E A KC F+V D
Sbjct: 562 RRQAMARISKRKAKIGKWKKK-ISEYVSEILKEEWELAVKCEVT---KGTHEKFEVWFDG 617
Query: 1009 DSYVVDL--EERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
+S V L EE C+C +WQ+TGIPC H+ A + ++ E++V P +
Sbjct: 618 NSNAVSLKTEEWDCSCCKWQVTGIPCEHAYAAINDVGKDVEDFVIPMF 665
>At1g48290 hypothetical protein
Length = 444
Score = 106 bits (265), Expect = 7e-23
Identities = 57/205 (27%), Positives = 100/205 (47%), Gaps = 18/205 (8%)
Query: 831 GLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKA 890
GLV + IP +HR C KHI N+++ EL+ WK AR+ T+ E+ +E +K
Sbjct: 164 GLVKAIHNRIPAAEHRQCAKHIMDNWKRNSHDMELQRLFWKIARSYTIGEYTANLEELKT 223
Query: 891 FSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKF 950
++ A +M +W+R+ + + C+ +NN+ E+FN + + R KP++ +LE ++
Sbjct: 224 YNPGAAASLMNTKPMEWSRAFFRIGSCCNDNLNNLSESFNRTIRQARRKPLLDMLEDIRS 283
Query: 951 YINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSD-------NFRHFQ 1003
R K+ + RWK+ F+K A ++ + GS +
Sbjct: 284 QCMVRNEKRYIIAGRWKS-----------RFTKRAHEEIEKMIAGSQFCERSMARHNKHE 332
Query: 1004 VKRDSDSYVVDLEERTCACRRWQLT 1028
+ Y VD+ + TC CR+WQ+T
Sbjct: 333 ISHFGRKYSVDMNDNTCGCRKWQMT 357
>At1g67240 hypothetical protein
Length = 851
Score = 104 bits (259), Expect = 3e-22
Identities = 53/177 (29%), Positives = 101/177 (56%), Gaps = 2/177 (1%)
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMK 889
+GL+ + +L+P +HR C +HIY N+RK Y + W A +ST +++ M+ ++
Sbjct: 507 KGLINVVADLLPQAEHRHCARHIYANWRKVYSDYSHESYFWAIAYSSTNGDYRWNMDALR 566
Query: 890 AFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLK 949
+ +A++D+++ + W R+ +ST +RC+ NN+CE+FN + + R+ P+I++LE ++
Sbjct: 567 LYDPQAHDDLLKTDPRTWCRAFFSTHSRCEDVSNNLCESFNRTIRDARNLPVINMLEEVR 626
Query: 950 FYINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKR 1006
RI K ++ +T P I + LE ++++A C +V S+N KR
Sbjct: 627 RTSMKRIAKLCGKTMKCETR-FPPKIMEILEGNRKSAKFC-SVLKSSENIYEVPDKR 681
>At2g15810 Mutator-like transposase
Length = 545
Score = 101 bits (251), Expect = 3e-21
Identities = 60/224 (26%), Positives = 109/224 (47%), Gaps = 22/224 (9%)
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMK 889
+GL+ + +L+P+ +HR C +HIY N++K Y + W A +ST + M +K
Sbjct: 126 KGLLNAVADLLPEAEHRHCARHIYSNWKKVYGDYCHESYFWAIAYSSTDGTYTYNMRALK 185
Query: 890 AFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLK 949
++ AY D+++ K + ++ + + R KP+IS+LE ++
Sbjct: 186 SYDPLAYEDLLKTEPKTEFQQGHN------------------GLRDARKKPVISMLEDVR 227
Query: 950 FYINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSD 1009
RI K+R + K+ P I L+ ++++A C + N ++V +
Sbjct: 228 RQAMKRISKRRDQTAKCKSKFP-PHIMGVLKANRKSAKLCTVLKNSE---HVYEVMEGNG 283
Query: 1010 SYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPS 1053
+ ++ RTCAC +W LTGIPC H+I + R+PE+ + S
Sbjct: 284 GFTANILHRTCACNQWNLTGIPCPHAIYVINEHNRDPEDTISSS 327
>At2g30640 Mutator-like transposase
Length = 754
Score = 94.0 bits (232), Expect = 5e-19
Identities = 59/222 (26%), Positives = 108/222 (48%), Gaps = 5/222 (2%)
Query: 830 QGLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMK 889
Q +V + P H LCV + + R ++ S L +W+AA+ T E++ +M +
Sbjct: 447 QSIVDGVDTNFPTAFHGLCVHCLTESVRTQFNNSILVNLVWEAAKCLTDFEFEGKMGEIA 506
Query: 890 AFSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLK 949
S EA + + + QWA + TR N+ E+ N V + PII +LE ++
Sbjct: 507 QISPEAASWIRNIQHSQWATYCFE-GTRFGHLTANVSESLNSWVQDASGLPIIQMLESIR 565
Query: 950 FYINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSD 1009
+ ++R+ ++W + ++ P ++ + EA ++C N F+V
Sbjct: 566 RQLMTLFNERRETSMQW-SGMLVPSAERHV---LEAIEECRLYPVHKANEAQFEVMTSEG 621
Query: 1010 SYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
++VD+ RTC CR W+L G+PCSH++A + R+N + +
Sbjct: 622 KWIVDIRCRTCYCRGWELYGLPCSHAVAALLACRQNVYRFTE 663
Score = 38.1 bits (87), Expect = 0.030
Identities = 19/74 (25%), Positives = 36/74 (47%), Gaps = 1/74 (1%)
Query: 512 NFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNC-VKGCPFNLRISKTNSR 570
N ++G F A+++ A+ + +++ VK+DK R C +GCP+ + +K
Sbjct: 179 NLIVGMEFSDVLACRRALRDAAIALRFEMQTVKSDKTRFTAKCNSEGCPWRIHCAKLPGL 238
Query: 571 HYFLLVTYEAAHNC 584
F + T +H C
Sbjct: 239 PTFTIRTIHGSHTC 252
>At1g12720 hypothetical protein
Length = 585
Score = 90.1 bits (222), Expect = 7e-18
Identities = 59/232 (25%), Positives = 105/232 (44%), Gaps = 44/232 (18%)
Query: 831 GLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKA 890
GLV + ++P +HR C KHI N+++ EL+ WK +R+ T++E+ M +K+
Sbjct: 190 GLVKAIHNVLPQAEHRQCSKHIMDNWKRDSHDMELQRLFWKISRSYTIEEFNTHMANLKS 249
Query: 891 FSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKF 950
++ +AY + W + + R KP++ +LE ++
Sbjct: 250 YNPQAYASLQLTSPMTW------------------------TIRQARRKPLLDMLEDIR- 284
Query: 951 YINNRIVKQRQLMLRW-KTSIICPLIQQRLEFSKEA-ADKCVAVWNGSDNFRHF------ 1002
RQ M+R K II ++ R A +K +A G + RH
Sbjct: 285 ---------RQCMVRTAKRFIIAERLKSRFTPRAHAEIEKMIAGSAGCE--RHLARNNLH 333
Query: 1003 QVKRDSDSYVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
++ + Y VD++++TC CR+W++ GIPC H+ + + E+YV Y
Sbjct: 334 EIYVNDVGYFVDMDKKTCGCRKWEMVGIPCVHTPCVIIGRKEKVEDYVSDYY 385
>At2g13970 Mutator-like transposase
Length = 597
Score = 88.2 bits (217), Expect = 3e-17
Identities = 60/224 (26%), Positives = 98/224 (42%), Gaps = 33/224 (14%)
Query: 831 GLVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKA 890
GL + +IP +HR +HI N++K EL+ WK +
Sbjct: 190 GLGKAIHTVIPQAEHRQYARHIMDNWKKNSHDMELQRLFWKTNPLT-------------- 235
Query: 891 FSEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKF 950
W+R+ + + C+ +NN+ E+FN + R KP++ +LE ++
Sbjct: 236 ----------------WSRAFFRIGSCCNDNLNNLSESFNRTIRNARRKPLLDMLEDIRR 279
Query: 951 YINNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDS 1010
R K+ + R K S +E + A C W N +V D DS
Sbjct: 280 QCMVRNEKRFVIAYRLK-SRFTKRAHMEIEKMIDGAAVCDR-WMARHNRIEVRVGLD-DS 336
Query: 1011 YVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
+ VD+ +RTC CR+WQ+TGIPC H+ + + R+ E++V Y
Sbjct: 337 FSVDMNDRTCGCRKWQMTGIPCIHAASVIIGNRQKVEDFVSDWY 380
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 87.8 bits (216), Expect = 3e-17
Identities = 57/224 (25%), Positives = 106/224 (46%), Gaps = 7/224 (3%)
Query: 832 LVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAF 891
+V ++ P H C++++ NFR + ++L W A A T E++ + M
Sbjct: 294 VVEAVETHFPTAFHGFCLRYVSENFRDTFKNTKLVNIFWSAVYALTPAEFETKSNEMIEI 353
Query: 892 SEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFY 951
S++ WA AY R + E LE + PII ++E ++
Sbjct: 354 SQDVVQWFELYLPHLWA-VAYFQGVRYGHFGLGITEVLYNWALECHELPIIQMMEHIRHQ 412
Query: 952 INNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAAD-KCVAVWNGSDNFRHFQVKRDSDS 1010
I++ +R+L + W SI+ P ++R+ ++ AD +C V ++ F++ +
Sbjct: 413 ISSWFDNRRELSMGWN-SILVPSAERRI--TEAVADARCYQVLRANEV--EFEIVSTERT 467
Query: 1011 YVVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
+VD+ R C+CRRWQ+ G+PC+H+ A + RN + +P +
Sbjct: 468 NIVDIRTRDCSCRRWQIYGLPCAHAAAALISCGRNVHLFAEPCF 511
Score = 42.4 bits (98), Expect = 0.002
Identities = 21/71 (29%), Positives = 36/71 (50%), Gaps = 1/71 (1%)
Query: 515 LGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCVK-GCPFNLRISKTNSRHYF 573
+G+ F + +K+ A+ DL+IVK+D+ R + C K GCP+ + +K F
Sbjct: 27 VGKEFPDVETCRRTLKDLAIALHFDLRIVKSDRSRFIAKCSKEGCPWRIHAAKCPGVQTF 86
Query: 574 LLVTYEAAHNC 584
+ T + H C
Sbjct: 87 TVRTLNSEHTC 97
>At1g06740 mudrA-like protein
Length = 726
Score = 85.1 bits (209), Expect = 2e-16
Identities = 52/223 (23%), Positives = 104/223 (46%), Gaps = 4/223 (1%)
Query: 832 LVPTLQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAF 891
+V ++ P H C+ ++ F++++ S L W+AA TV E+K ++ ++
Sbjct: 427 VVDGVEANFPAAFHGFCLHYLTERFQREFQSSVLVDLFWEAAHCLTVLEFKSKINKIEQI 486
Query: 892 SEEAYNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFY 951
S EA + +WA S + L N + E+ + V + PII +E + +
Sbjct: 487 SPEASLWIQNKSPARWASSYFEGTRFGQLTANVITESLSNWVEDTSGLPIIQTMECIHRH 546
Query: 952 INNRIVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSY 1011
+ N + ++R+ L W ++ +Q L +++ V N ++ F+V +
Sbjct: 547 LINMLKERRETSLHWSNVLVPSAEKQMLAAIEQSRAHRVYRANEAE----FEVMTCEGNV 602
Query: 1012 VVDLEERTCACRRWQLTGIPCSHSIACMWFARRNPENYVDPSY 1054
VV++E +C C RWQ+ G+PCSH++ + + Y + +
Sbjct: 603 VVNIENCSCLCGRWQVYGLPCSHAVGALLSCEEDVYRYTESCF 645
Score = 42.7 bits (99), Expect = 0.001
Identities = 21/78 (26%), Positives = 37/78 (46%), Gaps = 1/78 (1%)
Query: 508 GDGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNC-VKGCPFNLRISK 566
G V+G F A+K A+ + +++ +K+DK R C KGCP+ + +K
Sbjct: 151 GTDHEMVVGMEFSDAYACRRAIKNAAISLRFEMRTIKSDKTRFTAKCNSKGCPWRIHCAK 210
Query: 567 TNSRHYFLLVTYEAAHNC 584
++ F + T +H C
Sbjct: 211 VSNAPTFTIRTIHGSHTC 228
>At3g47280 putative protein
Length = 739
Score = 80.9 bits (198), Expect = 4e-15
Identities = 51/211 (24%), Positives = 101/211 (47%), Gaps = 4/211 (1%)
Query: 841 PDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEAYNDMM 900
P H LC H+ N + GS L + A+R T E+ + ++ +
Sbjct: 445 PLAHHGLCTFHLQKNLETHFRGSSLIPVYYAASRIYTKTEFDSLFWEITNSDKKLAQYLC 504
Query: 901 RLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNRIVKQR 960
+ ++W+R AYS R ++ +N+ E+ N + ++R+ P++ L E ++ I R +R
Sbjct: 505 EVDVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYPVVCLFESIRS-IMTRWFNER 562
Query: 961 QLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSYVVDLEERTC 1020
+ S + + ++++ S + + + + V N F+VK D+ +++V+L++RTC
Sbjct: 563 REESSQHPSAVTINVGKKMKASYDTSTRWLEVCQV--NQEEFEVKGDTKTHLVNLDKRTC 620
Query: 1021 ACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
C + + PC+H IA N +VD
Sbjct: 621 TCCMFDIDKFPCAHGIASAKHINLNENMFVD 651
Score = 48.1 bits (113), Expect = 3e-05
Identities = 20/77 (25%), Positives = 43/77 (54%), Gaps = 1/77 (1%)
Query: 509 DGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCV-KGCPFNLRISKT 567
D + +G++F KD + +++FA++H VK+D R V++C+ + C + LR ++
Sbjct: 167 DADSLFIGKIFKDKDEMVFTLRKFAVKHSFKFHTVKSDLTRYVLHCIDENCSWRLRATRA 226
Query: 568 NSRHYFLLVTYEAAHNC 584
+++ Y + H+C
Sbjct: 227 GGSESYVIRKYVSHHSC 243
>At2g15210 putaive transposase of transposon FARE2.7 (cds1)
Length = 580
Score = 80.9 bits (198), Expect = 4e-15
Identities = 52/211 (24%), Positives = 101/211 (47%), Gaps = 4/211 (1%)
Query: 841 PDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEAYNDMM 900
P H LC H+ N + GS L + A+R T E+ + ++ +
Sbjct: 286 PLAHHGLCTFHLQKNLETHFRGSSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLW 345
Query: 901 RLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNRIVKQR 960
+ ++W+R AYS R ++ +N+ E+ N + ++R+ PI+ L E ++ I R +R
Sbjct: 346 EVHVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFESIRS-IMTRWFNER 403
Query: 961 QLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSYVVDLEERTC 1020
+ S + + ++++ S + + + + V N F+VK D+ +++V+L++RTC
Sbjct: 404 REESSQHPSAVTINVGKKMKASYDTSTRWLEVCQV--NQEEFEVKGDTKTHLVNLDKRTC 461
Query: 1021 ACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
C + + PC+H IA N +VD
Sbjct: 462 TCCMFDIDKFPCAHGIASANHINLNENMFVD 492
Score = 45.8 bits (107), Expect = 1e-04
Identities = 20/77 (25%), Positives = 42/77 (53%), Gaps = 1/77 (1%)
Query: 509 DGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCV-KGCPFNLRISKT 567
D + +G+ F KD + ++ FA++H + VK+D R V++C+ + C + LR ++
Sbjct: 44 DADSLFIGKNFKDKDEMVFTLRMFAVKHNFEFHTVKSDLTRYVLHCIDENCSWRLRATRA 103
Query: 568 NSRHYFLLVTYEAAHNC 584
+++ Y + H+C
Sbjct: 104 GGSESYVIRKYVSHHSC 120
>At2g15150 putative transposase of FARE2.2 (CDS1)
Length = 607
Score = 80.9 bits (198), Expect = 4e-15
Identities = 52/211 (24%), Positives = 101/211 (47%), Gaps = 4/211 (1%)
Query: 841 PDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEAYNDMM 900
P H LC H+ N + GS L + A+R T E+ + ++ +
Sbjct: 322 PLAHHGLCTFHLQKNLETHFRGSSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLW 381
Query: 901 RLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNRIVKQR 960
+ ++W+R AYS R ++ +N+ E+ N + ++R+ PI+ L E ++ I R +R
Sbjct: 382 EVHVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFESIRS-IMTRWFNER 439
Query: 961 QLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSYVVDLEERTC 1020
+ S + + ++++ S + + + + V N F+VK D+ +++V+L++RTC
Sbjct: 440 REESSQHPSAVTINVGKKMKASYDTSTRWLEVCQV--NQEEFEVKGDTKTHLVNLDKRTC 497
Query: 1021 ACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
C + + PC+H IA N +VD
Sbjct: 498 TCCMFDIDKFPCAHGIASANHINLNENMFVD 528
Score = 47.4 bits (111), Expect = 5e-05
Identities = 20/77 (25%), Positives = 42/77 (53%), Gaps = 1/77 (1%)
Query: 509 DGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCV-KGCPFNLRISKT 567
D + +G++F KD + ++ FA++H + VK+D R V++C+ + C + LR ++
Sbjct: 44 DADSLFIGKIFKDKDEMVFTLRMFAVKHNFEFHTVKSDLTRYVLHCIDENCSWRLRATRA 103
Query: 568 NSRHYFLLVTYEAAHNC 584
+ + Y + H+C
Sbjct: 104 GGSESYAIRKYVSHHSC 120
>At2g06450 putative transposase of FARE2.11
Length = 580
Score = 80.5 bits (197), Expect = 5e-15
Identities = 51/211 (24%), Positives = 101/211 (47%), Gaps = 4/211 (1%)
Query: 841 PDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEAYNDMM 900
P H LC H+ N + GS L + A+R T E+ + ++ +
Sbjct: 286 PLAHHGLCTFHLQKNLETHFRGSSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLW 345
Query: 901 RLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNRIVKQR 960
+ ++W+R+ YS R ++ +N+ E+ N + ++R+ PI+ L E ++ I R +R
Sbjct: 346 EVDVRKWSRT-YSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFESIRS-IMTRWFNER 403
Query: 961 QLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSYVVDLEERTC 1020
+ S + + ++++ S + + + + V N F+VK D+ +++V+L++RTC
Sbjct: 404 REESSQHPSAVTINVGKKMKASYDTSTRWLEVCQV--NQEEFEVKGDTKTHLVNLDKRTC 461
Query: 1021 ACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
C + + PC+H IA N +VD
Sbjct: 462 TCCMFDIDKFPCAHGIASAKHINLNENMFVD 492
Score = 47.0 bits (110), Expect = 6e-05
Identities = 20/77 (25%), Positives = 42/77 (53%), Gaps = 1/77 (1%)
Query: 509 DGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCV-KGCPFNLRISKT 567
D + +G++F KD + ++ FA++H VK+D R V++C+ + C + LR ++
Sbjct: 44 DADSLFIGKIFKDKDEMVFTLRMFAVKHNFKFHTVKSDLTRYVLHCIDENCSWRLRATRA 103
Query: 568 NSRHYFLLVTYEAAHNC 584
+++ Y + H+C
Sbjct: 104 GGSESYVIRKYVSHHSC 120
>At4g03980 putative transposon protein
Length = 580
Score = 79.0 bits (193), Expect = 2e-14
Identities = 53/216 (24%), Positives = 102/216 (46%), Gaps = 4/216 (1%)
Query: 836 LQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEA 895
L E P H LC H+ N + GS L + A+R T E+ + ++
Sbjct: 281 LSENYPLAHHGLCTFHLQKNLETHFRGSSLIPVNYAASRVYTKTEFDSLFWKITNSDKKL 340
Query: 896 YNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNR 955
+ + ++W+R AYS R ++ +N+ E+ N + ++R+ PI+ L E ++ I R
Sbjct: 341 AQYLWEVDVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFESIRS-IMTR 398
Query: 956 IVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSYVVDL 1015
+R+ S + + ++++ S + + + + V N F+VK D+ +++V+L
Sbjct: 399 WFNERREESSQHPSAVTINVGKKMKASYDTSTRWLEVCQV--NQEEFEVKGDTKTHLVNL 456
Query: 1016 EERTCACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
++RTC C + + P +H IA N +VD
Sbjct: 457 DKRTCTCCMFDIDKFPYAHGIASAKHINLNKNMFVD 492
Score = 47.4 bits (111), Expect = 5e-05
Identities = 21/77 (27%), Positives = 42/77 (54%), Gaps = 1/77 (1%)
Query: 509 DGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCV-KGCPFNLRISKT 567
D + +G+ F KD + ++ FA++H + VK+D R V++C+ + C + LR +K
Sbjct: 44 DADSLFIGKKFKDKDEMVFTLRMFAVKHSFEFHTVKSDLTRYVLHCIDENCSWRLRATKA 103
Query: 568 NSRHYFLLVTYEAAHNC 584
+++ Y + H+C
Sbjct: 104 GGSESYVIRKYVSHHSC 120
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 79.0 bits (193), Expect = 2e-14
Identities = 51/211 (24%), Positives = 101/211 (47%), Gaps = 4/211 (1%)
Query: 841 PDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEAYNDMM 900
P + LC H+ N + GS L + A+R T E+ + ++ +
Sbjct: 322 PLAHNGLCTFHLQKNLETHFRGSSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLW 381
Query: 901 RLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNRIVKQR 960
+ ++W+R AYS R ++ +N+ E+ N + ++R+ PI+ L E ++ I R +R
Sbjct: 382 EVDVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFESIRS-IMTRWFNER 439
Query: 961 QLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSYVVDLEERTC 1020
+ S + + ++++ S + + + + V N F+VK D+ +++V+L++RTC
Sbjct: 440 REESSQHPSAVTINVGKKMKASYDTSTRWLEVCQV--NQEEFEVKGDTKTHLVNLDKRTC 497
Query: 1021 ACRRWQLTGIPCSHSIACMWFARRNPENYVD 1051
C + + PC+H IA N +VD
Sbjct: 498 TCCMFDIDKFPCAHGIASAKHINLNENMFVD 528
Score = 42.4 bits (98), Expect = 0.002
Identities = 18/77 (23%), Positives = 41/77 (52%), Gaps = 1/77 (1%)
Query: 509 DGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDKKRVVVNCV-KGCPFNLRISKT 567
D + +G+ F KD + ++ A++H + VK+D + V++C+ + C + LR ++
Sbjct: 44 DADSLFIGKFFKDKDEMVFTLRMSAVKHSFEFHTVKSDLTKYVLHCIDENCSWRLRATRA 103
Query: 568 NSRHYFLLVTYEAAHNC 584
+++ Y + H+C
Sbjct: 104 GGSESYVIRKYVSHHSC 120
>At1g34620 Mutator-like protein
Length = 1034
Score = 79.0 bits (193), Expect = 2e-14
Identities = 51/202 (25%), Positives = 101/202 (49%), Gaps = 4/202 (1%)
Query: 836 LQELIPDVDHRLCVKHIYGNFRKKYPGSELKVALWKAARASTVQEWKQEMELMKAFSEEA 895
L ++ +H C H++ N R Y L + +AA+A V ++ + ++ +
Sbjct: 638 LSKVYTQANHAACTVHLWRNIRHLYKPKSLCRLMSEAAQAFHVTDFNRIFLKIQKLNPGC 697
Query: 896 YNDMMRLPAKQWARSAYSTDTRCDLQVNNMCEAFNMAVLEHRDKPIISLLEGLKFYINNR 955
++ L +W R +S R ++ +N+CE++N + E R+ P+I +LE ++ + +
Sbjct: 698 AAYLVDLGFSEWTR-VHSKGRRFNIMDSNICESWNNVIREAREYPLICMLEYIRTTLMDW 756
Query: 956 IVKQRQLMLRWKTSIICPLIQQRLEFSKEAADKCVAVWNGSDNFRHFQVKRDSDSYVVDL 1015
+R T++ P +Q+R+E + ++A NF +R + ++V L
Sbjct: 757 FATRRAQAEDCPTTL-APRVQERVEENYQSAMSMSV--KPICNFEFQVQERTGECFIVKL 813
Query: 1016 EERTCACRRWQLTGIPCSHSIA 1037
+E TC+C +Q GIPC+H+IA
Sbjct: 814 DESTCSCLEFQGLGIPCAHAIA 835
Score = 34.3 bits (77), Expect = 0.43
Identities = 25/98 (25%), Positives = 45/98 (45%), Gaps = 5/98 (5%)
Query: 488 SEGEGEGRARRYPRFRPAEDGDGVNFVLGQVFDSKDIFIHAVKEFALQHKKDLKIVKNDK 547
++G+GEG + F +GD +G+VF SK + A+ K + ++
Sbjct: 344 NDGKGEGVDSAF--FDVKYEGD--KLFVGRVFKSKSDCKIKIAIHAINRKFHFRTARSTP 399
Query: 548 KRVVVNCV-KGCPFNLRISKTNSRHYFLLVTYEAAHNC 584
K +V+ C+ K CP+ + SK ++ F + H C
Sbjct: 400 KFMVLKCISKTCPWRVYASKVDTSDSFQVRQANQRHTC 437
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.338 0.149 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,590,180
Number of Sequences: 26719
Number of extensions: 1179191
Number of successful extensions: 12653
Number of sequences better than 10.0: 437
Number of HSP's better than 10.0 without gapping: 138
Number of HSP's successfully gapped in prelim test: 313
Number of HSP's that attempted gapping in prelim test: 9111
Number of HSP's gapped (non-prelim): 1794
length of query: 1171
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1061
effective length of database: 8,379,506
effective search space: 8890655866
effective search space used: 8890655866
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0321.4


