

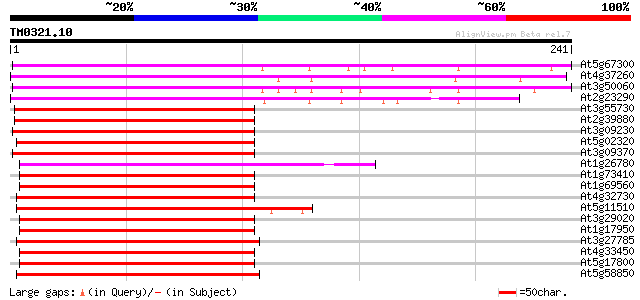
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.10
(241 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g67300 myb-related protein, 33.3K (pir||S71284) 264 3e-71
At4g37260 myb-related protein 248 3e-66
At3g50060 R2R3-MYB transcription factor 242 1e-64
At2g23290 putative MYB family transcription factor 238 2e-63
At3g55730 MYB transcription factor - like protein 159 1e-39
At2g39880 putative MYB family transcription factor 157 5e-39
At3g09230 unknown protein 153 7e-38
At5g02320 myb -like protein 136 1e-32
At3g09370 MYB family like transcription factor 135 1e-32
At1g26780 putative transcription factor MYB117 (MYB117) 133 1e-31
At1g73410 putative protein 130 5e-31
At1g69560 myb-related transcription factor, putative 130 5e-31
At4g32730 putative myb-protein 129 1e-30
At5g11510 putative c-myb-like transcription factor MYB3R-4 (MYB3R4) 128 2e-30
At3g29020 putative transcription factor (MYB110) 128 2e-30
At1g17950 unknown protein 127 7e-30
At3g27785 putative transcription factor MYB118 (MYB118) 125 2e-29
At4g33450 putative transcription factor (MYB69) 125 2e-29
At5g17800 MYB56 R2R3-MYB factor family member 124 3e-29
At5g58850 putative transcription factor MYB119 (MYB119) 121 4e-28
>At5g67300 myb-related protein, 33.3K (pir||S71284)
Length = 305
Score = 264 bits (675), Expect = 3e-71
Identities = 151/303 (49%), Positives = 181/303 (58%), Gaps = 63/303 (20%)
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
DR+KGPWSPEEDE L RLV +GP+NWT ISKSIPGRSGKSCRLRWCNQLSP+VEHRPF+
Sbjct: 3 DRIKGPWSPEEDEQLRRLVVKYGPRNWTVISKSIPGRSGKSCRLRWCNQLSPQVEHRPFS 62
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCS-----AVDTAVDHR 116
EED I RAHA+FGNKWATIAR+LNGRTDNAVKNHWNSTLKRKC D + DHR
Sbjct: 63 AEEDETIARAHAQFGNKWATIARLLNGRTDNAVKNHWNSTLKRKCGGYDHRGYDGSEDHR 122
Query: 117 PLKRSASVGAG------CLNPSSPTGSAMSDPGP-PAAISTD--APIAVPAAVVVP---- 163
P+KRS S G+ ++P SPTGS +SD P S + P+ P AVV+P
Sbjct: 123 PVKRSVSAGSPPVVTGLYMSPGSPTGSDVSDSSTIPILPSVELFKPVPRPGAVVLPLPIE 182
Query: 164 ----ATDPATSLSLSLPGFDSSGSGSGSKQLF---------------------------- 191
+ DP TSLSLSLPG D S + S +
Sbjct: 183 TSSSSDDPPTSLSLSLPGADVSEESNRSHESTNINNTTSSRHNHNNTVSFMPFSGGFRGA 242
Query: 192 -----------GAEFLAVMQEMIRKEVRSYMSGMEEKNGACMPAEAIGNAV--MKRMGIS 238
G EF+AV+QEMI+ EVRSYM+ M+ NG I N + M ++G+
Sbjct: 243 IEEMGKSFPGNGGEFMAVVQEMIKAEVRSYMTEMQRNNGGGFVGGFIDNGMIPMSQIGVG 302
Query: 239 NVK 241
++
Sbjct: 303 RIE 305
>At4g37260 myb-related protein
Length = 320
Score = 248 bits (632), Expect = 3e-66
Identities = 143/304 (47%), Positives = 169/304 (55%), Gaps = 65/304 (21%)
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
M+R+KGPWSPEED+ L RLVQ HGP+NW+ ISKSIPGRSGKSCRLRWCNQLSPEVEHR F
Sbjct: 9 MERIKGPWSPEEDDLLQRLVQKHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPEVEHRAF 68
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVD------ 114
+ EED IIRAHARFGNKWATI+R+LNGRTDNA+KNHWNSTLKRKCS + D
Sbjct: 69 SQEEDETIIRAHARFGNKWATISRLLNGRTDNAIKNHWNSTLKRKCSVEGQSCDFGGNGG 128
Query: 115 -------HRPLKRSASVGAGC-----LNPSSPTGSAMSDPGPPAAISTDAPIAVPAAVVV 162
+PLKR+AS G G ++P SP+GS +S+ A +
Sbjct: 129 YDGNLGEEQPLKRTASGGGGVSTGLYMSPGSPSGSDVSEQSSGGAHVFKPTVRSEVTASS 188
Query: 163 PATDPATSLSLSLPGFDSSGSGSGSKQL-------------------------------- 190
DP T LSLSLP D + + QL
Sbjct: 189 SGEDPPTYLSLSLPWTDETVRVNEPVQLNQNTVMDGGYTAELFPVRKEEQVEVEEEEAKG 248
Query: 191 ----FGAEFLAVMQEMIRKEVRSYMSGMEEKN-----------GACMPAEAIGNAVMKRM 235
FG EF+ V+QEMIR EVRSYM+ ++ N G+CMP V R
Sbjct: 249 ISGGFGGEFMTVVQEMIRTEVRSYMADLQRGNVGGSSSGGGGGGSCMPQSVNSRRVGFRE 308
Query: 236 GISN 239
I N
Sbjct: 309 FIVN 312
>At3g50060 R2R3-MYB transcription factor
Length = 301
Score = 242 bits (617), Expect = 1e-64
Identities = 144/297 (48%), Positives = 179/297 (59%), Gaps = 57/297 (19%)
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
DRVKGPWS EEDE L R+V+ +GP+NW+AISKSIPGRSGKSCRLRWCNQLSPEVEHRPF+
Sbjct: 3 DRVKGPWSQEEDEQLRRMVEKYGPRNWSAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFS 62
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCS---AVDTAVD---- 114
PEED I+ A A+FGNKWATIAR+LNGRTDNAVKNHWNSTLKRKCS AV T +
Sbjct: 63 PEEDETIVTARAQFGNKWATIARLLNGRTDNAVKNHWNSTLKRKCSGGVAVTTVTETEED 122
Query: 115 -HRPLKRS--------ASVGAGC-LNPSSPTGSAMSD----PGPPAAIS----------- 149
RP KR A V G ++P SP G +SD P P + ++
Sbjct: 123 QDRPKKRRSVSFDSAFAPVDTGLYMSPESPNGIDVSDSSTIPSPSSPVAQLFKPMPISGG 182
Query: 150 -TDAPIAVPAAVVVPATDPATSLSLSLPGFD---SSGSGSGSKQLF-------------- 191
T P +P + + DP TSLSLSLPG + SS + + + +F
Sbjct: 183 FTVVPQPLPVEMSSSSEDPPTSLSLSLPGAENTSSSHNNNNNALMFPRFESQMKINVEER 242
Query: 192 ----GAEFLAVMQEMIRKEVRSYMSGMEEKNGACMPA---EAIGNAVMKRMGISNVK 241
EF+ V+QEMI+ EVRSYM+ M++ +G + E+ GN + G+ K
Sbjct: 243 GEGRRGEFMTVVQEMIKAEVRSYMAEMQKTSGGFVVGGLYESGGNGGFRDCGVITPK 299
>At2g23290 putative MYB family transcription factor
Length = 309
Score = 238 bits (608), Expect = 2e-63
Identities = 138/272 (50%), Positives = 164/272 (59%), Gaps = 56/272 (20%)
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPWSPEED+ L LVQ HGP+NW+ ISKSIPGRSGKSCRLRWCNQLSPEVEHR F
Sbjct: 9 MDRIKGPWSPEEDDLLQSLVQKHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPEVEHRGF 68
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSA------------ 108
T EED II AHARFGNKWATIAR+LNGRTDNA+KNHWNSTLKRKCS
Sbjct: 69 TAEEDDTIILAHARFGNKWATIARLLNGRTDNAIKNHWNSTLKRKCSGGGGGGEEGQSCD 128
Query: 109 -------VDTAVDHRPLKRSASVGAG--CLNPSSPTGSAMSD----PGPPAAISTDAPIA 155
D +PLKR AS G G + SPTGS +S+ G +S+ +
Sbjct: 129 FGGNGGYDGNLTDEKPLKRRASGGGGVVVVTALSPTGSDVSEQSQSSGSVLPVSSSCHVF 188
Query: 156 VPAA----VVVPAT-------DPATSLSLSLPGFDSSGSGSGSKQLF------------- 191
P A VV+ ++ DP T L LSLP + S + +LF
Sbjct: 189 KPTARAGGVVIESSSPEEEEKDPMTCLRLSLPWVNES---TTPPELFPVKREEEEEKERE 245
Query: 192 ----GAEFLAVMQEMIRKEVRSYMSGMEEKNG 219
G +F+ V+QEMI+ EVRSYM+ ++ NG
Sbjct: 246 ISGLGGDFMTVVQEMIKTEVRSYMADLQLGNG 277
>At3g55730 MYB transcription factor - like protein
Length = 399
Score = 159 bits (403), Expect = 1e-39
Identities = 70/103 (67%), Positives = 85/103 (81%)
Query: 3 RVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTP 62
+VKGPWS EED LT+LV+ GP+NW+ I++ IPGRSGKSCRLRWCNQL P ++ +PF+
Sbjct: 54 KVKGPWSTEEDAVLTKLVRKLGPRNWSLIARGIPGRSGKSCRLRWCNQLDPCLKRKPFSD 113
Query: 63 EEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
EED II AHA GNKWA IA++L GRTDNA+KNHWNSTL+RK
Sbjct: 114 EEDRMIISAHAVHGNKWAVIAKLLTGRTDNAIKNHWNSTLRRK 156
>At2g39880 putative MYB family transcription factor
Length = 367
Score = 157 bits (397), Expect = 5e-39
Identities = 68/103 (66%), Positives = 85/103 (82%)
Query: 3 RVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTP 62
+VKGPW PE+DEALTRLV++ GP+NW IS+ IPGRSGKSCRLRWCNQL P ++ +PF+
Sbjct: 48 KVKGPWLPEQDEALTRLVKMCGPRNWNLISRGIPGRSGKSCRLRWCNQLDPILKRKPFSD 107
Query: 63 EEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
EE+ I+ A A GNKW+ IA++L GRTDNA+KNHWNS L+RK
Sbjct: 108 EEEHMIMSAQAVLGNKWSVIAKLLPGRTDNAIKNHWNSNLRRK 150
>At3g09230 unknown protein
Length = 393
Score = 153 bits (387), Expect = 7e-38
Identities = 69/104 (66%), Positives = 83/104 (79%)
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
DRVKGPWS EED+ L+ LV+ G +NW+ I++SIPGRSGKSCRLRWCNQL+P + FT
Sbjct: 52 DRVKGPWSKEEDDVLSELVKRLGARNWSFIARSIPGRSGKSCRLRWCNQLNPNLIRNSFT 111
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
ED AII AHA GNKWA IA++L GRTDNA+KNHWNS L+R+
Sbjct: 112 EVEDQAIIAAHAIHGNKWAVIAKLLPGRTDNAIKNHWNSALRRR 155
>At5g02320 myb -like protein
Length = 529
Score = 136 bits (342), Expect = 1e-32
Identities = 57/102 (55%), Positives = 78/102 (75%)
Query: 4 VKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPE 63
VKGPW+ EED+ + LV+ +GP W+ I+KS+PGR GK CR RW N L+P + +T E
Sbjct: 107 VKGPWTQEEDDKIVELVKKYGPAKWSVIAKSLPGRIGKQCRERWHNHLNPGIRKDAWTVE 166
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
E+SA++ +H +GNKWA IA++L GRTDNA+KNHWNS+LK+K
Sbjct: 167 EESALMNSHRMYGNKWAEIAKVLPGRTDNAIKNHWNSSLKKK 208
Score = 37.0 bits (84), Expect = 0.009
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVE 56
K W+ EE+ AL +++G K W I+K +PGR+ + + W + L ++E
Sbjct: 160 KDAWTVEEESALMNSHRMYGNK-WAEIAKVLPGRTDNAIKNHWNSSLKKKLE 210
>At3g09370 MYB family like transcription factor
Length = 505
Score = 135 bits (341), Expect = 1e-32
Identities = 57/104 (54%), Positives = 78/104 (74%)
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
D +KGPW+ EEDE + LV+ +GP W+ I++S+PGR GK CR RW N L+P++ +T
Sbjct: 127 DLIKGPWTHEEDEKIVELVEKYGPAKWSIIAQSLPGRIGKQCRERWHNHLNPDINKDAWT 186
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
EE+ A++ AH GNKWA IA++L GRTDNA+KNHWNS+LK+K
Sbjct: 187 TEEEVALMNAHRSHGNKWAEIAKVLPGRTDNAIKNHWNSSLKKK 230
Score = 37.0 bits (84), Expect = 0.009
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVE 56
K W+ EE+ AL + HG K W I+K +PGR+ + + W + L + E
Sbjct: 182 KDAWTTEEEVALMNAHRSHGNK-WAEIAKVLPGRTDNAIKNHWNSSLKKKSE 232
>At1g26780 putative transcription factor MYB117 (MYB117)
Length = 280
Score = 133 bits (334), Expect = 1e-31
Identities = 66/153 (43%), Positives = 89/153 (58%), Gaps = 4/153 (2%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W P ED L LV ++GP+NW I++ + GRSGKSCRLRW NQL P + R FT EE
Sbjct: 98 RGHWRPAEDVKLKELVSIYGPQNWNLIAEKLQGRSGKSCRLRWFNQLDPRINRRAFTEEE 157
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHRPLKRSASV 124
+ +++AH +GNKWA IAR+ GRTDN+VKNHW+ + RK +A R L + +
Sbjct: 158 EERLMQAHRLYGNKWAMIARLFPGRTDNSVKNHWHVVMARKYREHSSAYRRRKLMSNNPL 217
Query: 125 GAGCLNPSSPTGSAMSDPGPPAAISTDAPIAVP 157
N P +P + IST+ A P
Sbjct: 218 KPHLTNNHHPN----PNPNYHSFISTNHYFAQP 246
>At1g73410 putative protein
Length = 243
Score = 130 bits (328), Expect = 5e-31
Identities = 56/101 (55%), Positives = 71/101 (69%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W P EDE L LV+ +GP NW AI+ +PGRSGKSCRLRW NQL P + PFT EE
Sbjct: 6 RGHWRPAEDEKLKDLVEQYGPHNWNAIALKLPGRSGKSCRLRWFNQLDPRINRNPFTEEE 65
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
+ ++ AH GN+W+ IAR+ GRTDNAVKNHW+ + R+
Sbjct: 66 EERLLAAHRIHGNRWSIIARLFPGRTDNAVKNHWHVIMARR 106
>At1g69560 myb-related transcription factor, putative
Length = 330
Score = 130 bits (328), Expect = 5e-31
Identities = 56/101 (55%), Positives = 72/101 (70%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W P ED L LV V+GP+NW I++ + GRSGKSCRLRW NQL P + R FT EE
Sbjct: 107 RGHWRPAEDTKLKELVAVYGPQNWNLIAEKLQGRSGKSCRLRWFNQLDPRINRRAFTEEE 166
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
+ +++AH +GNKWA IAR+ GRTDN+VKNHW+ + RK
Sbjct: 167 EERLMQAHRLYGNKWAMIARLFPGRTDNSVKNHWHVIMARK 207
>At4g32730 putative myb-protein
Length = 776
Score = 129 bits (324), Expect = 1e-30
Identities = 54/102 (52%), Positives = 74/102 (71%)
Query: 4 VKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPE 63
VKGPWS EED + LV+ +GPK W+ IS+ +PGR GK CR RW N L+P + +T E
Sbjct: 86 VKGPWSKEEDNTIIDLVEKYGPKKWSTISQHLPGRIGKQCRERWHNHLNPGINKNAWTQE 145
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
E+ +IRAH +GNKWA + + L GR+DN++KNHWNS++K+K
Sbjct: 146 EELTLIRAHQIYGNKWAELMKFLPGRSDNSIKNHWNSSVKKK 187
Score = 38.1 bits (87), Expect = 0.004
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVE 56
K W+ EE+ L R Q++G K W + K +PGRS S + W + + +++
Sbjct: 139 KNAWTQEEELTLIRAHQIYGNK-WAELMKFLPGRSDNSIKNHWNSSVKKKLD 189
>At5g11510 putative c-myb-like transcription factor MYB3R-4 (MYB3R4)
Length = 961
Score = 128 bits (322), Expect = 2e-30
Identities = 59/145 (40%), Positives = 91/145 (62%), Gaps = 18/145 (12%)
Query: 4 VKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPE 63
VKGPW+ EEDE + +L++ +GPK W+ I++ +PGR GK CR RW N L+P + +T E
Sbjct: 80 VKGPWTKEEDEMIVQLIEKYGPKKWSTIARFLPGRIGKQCRERWHNHLNPAINKEAWTQE 139
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDT--------AVDH 115
E+ +IRAH +GN+WA + + L GR+DN +KNHW+S++K+K + + A+
Sbjct: 140 EELLLIRAHQIYGNRWAELTKFLPGRSDNGIKNHWHSSVKKKLDSYMSSGLLDQYQAMPL 199
Query: 116 RPLKRSASV----------GAGCLN 130
P +RS+++ G GCLN
Sbjct: 200 APYERSSTLQSTFMQSNIDGNGCLN 224
>At3g29020 putative transcription factor (MYB110)
Length = 233
Score = 128 bits (322), Expect = 2e-30
Identities = 55/101 (54%), Positives = 72/101 (70%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W ED L LV V+GP+NW I++S+ GR+GKSCRLRW NQL P + R F+ EE
Sbjct: 65 RGHWRISEDTQLMELVSVYGPQNWNHIAESMQGRTGKSCRLRWFNQLDPRINKRAFSDEE 124
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
+ ++ AH FGNKWA IA++ NGRTDNA+KNHW+ + RK
Sbjct: 125 EERLLAAHRAFGNKWAMIAKLFNGRTDNALKNHWHVLMARK 165
>At1g17950 unknown protein
Length = 249
Score = 127 bits (318), Expect = 7e-30
Identities = 54/101 (53%), Positives = 69/101 (67%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W P EDE L LV+ GP NW AI++ + GRSGKSCRLRW NQL P + PFT EE
Sbjct: 5 RGHWRPAEDEKLRELVEQFGPHNWNAIAQKLSGRSGKSCRLRWFNQLDPRINRNPFTEEE 64
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
+ ++ +H GN+W+ IAR GRTDNAVKNHW+ + R+
Sbjct: 65 EERLLASHRIHGNRWSVIARFFPGRTDNAVKNHWHVIMARR 105
>At3g27785 putative transcription factor MYB118 (MYB118)
Length = 437
Score = 125 bits (315), Expect = 2e-29
Identities = 54/104 (51%), Positives = 74/104 (70%)
Query: 4 VKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPE 63
+KG W+PEED+ L +LV +HG K W+ I+K + GR GK CR RW N L P+++ +T E
Sbjct: 188 IKGQWTPEEDKLLVQLVDLHGTKKWSQIAKMLQGRVGKQCRERWHNHLRPDIKKDGWTEE 247
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCS 107
ED +I+AH GN+WA IAR L GRT+N +KNHWN+T +R+ S
Sbjct: 248 EDIILIKAHKEIGNRWAEIARKLPGRTENTIKNHWNATKRRQHS 291
>At4g33450 putative transcription factor (MYB69)
Length = 250
Score = 125 bits (314), Expect = 2e-29
Identities = 54/101 (53%), Positives = 75/101 (73%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W P ED+ L +LV+ +GPKNW I++ + GRSGKSCRLRW NQL P + +PFT EE
Sbjct: 19 RGHWRPVEDDNLRQLVEQYGPKNWNFIAQHLYGRSGKSCRLRWYNQLDPNITKKPFTEEE 78
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
+ +++AH GN+WA+IAR+ GRTDNAVKNH++ + R+
Sbjct: 79 EERLLKAHRIQGNRWASIARLFPGRTDNAVKNHFHVIMARR 119
>At5g17800 MYB56 R2R3-MYB factor family member
Length = 323
Score = 124 bits (312), Expect = 3e-29
Identities = 55/101 (54%), Positives = 68/101 (66%)
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W P ED L LV GP+NW IS + GRSGKSCRLRW NQL P + R FT EE
Sbjct: 93 RGHWRPTEDAKLKELVAQFGPQNWNLISNHLLGRSGKSCRLRWFNQLDPRINKRAFTEEE 152
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
+ ++ AH +GNKWA I+R+ GRTDNAVKNHW+ + R+
Sbjct: 153 EFRLLAAHRAYGNKWALISRLFPGRTDNAVKNHWHVIMARR 193
>At5g58850 putative transcription factor MYB119 (MYB119)
Length = 430
Score = 121 bits (303), Expect = 4e-28
Identities = 49/104 (47%), Positives = 74/104 (71%)
Query: 4 VKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPE 63
+KG W+ EED L RLV+ HG + W IS+ + GR+GK CR RW N L P+++ ++ E
Sbjct: 104 IKGQWTAEEDRKLIRLVRQHGERKWAMISEKLEGRAGKQCRERWHNHLRPDIKKDGWSEE 163
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCS 107
E+ ++ +H R GNKWA IA+++ GRT+N++KNHWN+T +R+ S
Sbjct: 164 EERVLVESHMRIGNKWAEIAKLIPGRTENSIKNHWNATKRRQNS 207
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.129 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,848,295
Number of Sequences: 26719
Number of extensions: 255824
Number of successful extensions: 1175
Number of sequences better than 10.0: 192
Number of HSP's better than 10.0 without gapping: 167
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 688
Number of HSP's gapped (non-prelim): 289
length of query: 241
length of database: 11,318,596
effective HSP length: 96
effective length of query: 145
effective length of database: 8,753,572
effective search space: 1269267940
effective search space used: 1269267940
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0321.10


