

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0320b.5
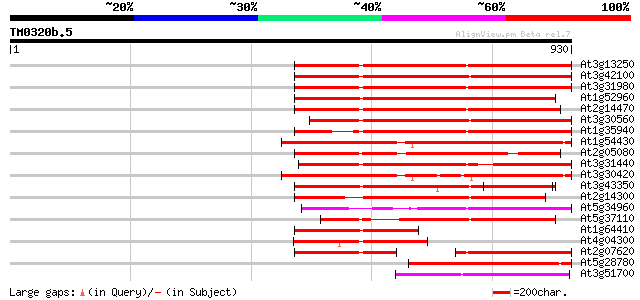
(930 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g13250 hypothetical protein 494 e-140
At3g42100 putative protein 487 e-137
At3g31980 hypothetical protein 473 e-133
At1g52960 hypothetical protein 466 e-131
At2g14470 pseudogene 453 e-127
At3g30560 hypothetical protein 447 e-125
At1g35940 hypothetical protein 433 e-121
At1g54430 hypothetical protein 420 e-117
At2g05080 putative helicase 416 e-116
At3g31440 hypothetical protein 411 e-114
At3g30420 hypothetical protein 406 e-113
At3g43350 putative protein 389 e-108
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 379 e-105
At5g34960 putative protein 329 4e-90
At5g37110 putative helicase 309 4e-84
At1g64410 unknown protein 244 2e-64
At4g04300 hypothetical protein 232 8e-61
At2g07620 putative helicase 207 2e-53
At5g28780 putative protein 195 9e-50
At3g51700 unknown protein 189 6e-48
>At3g13250 hypothetical protein
Length = 1419
Score = 494 bits (1272), Expect = e-140
Identities = 252/462 (54%), Positives = 336/462 (72%), Gaps = 8/462 (1%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W TL+AA+RS+G+I LNVASSGIASLLL GGRTAHSRFSIP+ E
Sbjct: 963 FFVYGFGGTGKTFIWKTLAAAVRSKGQICLNVASSGIASLLLEGGRTAHSRFSIPLNPDE 1022
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
S C ++ S A+++++ASLIIWDEAPM++K CFEALDK+ +DI+K +K F G
Sbjct: 1023 FSVCKIKPKSDLADLIKEASLIIWDEAPMMSKFCFEALDKSFSDIIKRV----DNKVFGG 1078
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSS-SSSL 650
KV+V GGDFRQ+LPVI R+EI+ SS N+SYLW HCKV++LT NMRL S +
Sbjct: 1079 KVMVFGGDFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLRLTKNMRLLNNDLSVDEAK 1138
Query: 651 EIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLNVVA--NLE 708
EI+EF+DWLL VGDG + ++ + II+IP LL++E+DNP+ + Y + +
Sbjct: 1139 EIQEFSDWLLAVGDGRVNEPNDGEVIIDIPEELLIQEADNPIEAISREIYGDPTKLHEIS 1198
Query: 709 NHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEF 768
+ +F++RA+LAP E V +N +M+ + EE YLS D+ SD DS + T +F
Sbjct: 1199 DPKFFQRRAILAPKNEDVNTINQYMLEHLDSEERIYLSADSIDPSDSDS-LKNPVITPDF 1257
Query: 769 LNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKT 828
LN +K SG+P+H + LKVG P+ML++N+D GLCNGTRL ++ L +I+ A ++G +
Sbjct: 1258 LNSIKVSGMPHHSLRLKVGAPVMLLRNLDPKGGLCNGTRLQITQLCSHIVEAKVITGDRI 1317
Query: 829 GKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHG 888
G+ VYIP +++TPSDT LPFK RRQFP++V F MTINKSQGQSL VGLYLP+PVF+HG
Sbjct: 1318 GQIVYIPLINITPSDTKLPFKMRRRQFPLSVAFVMTINKSQGQSLEQVGLYLPKPVFSHG 1377
Query: 889 QLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
QLYVALSRV S+ LKILI+D +G + T NVV++EVFQNI
Sbjct: 1378 QLYVALSRVTSKTGLKILILDKEGKIQKQTTNVVFKEVFQNI 1419
>At3g42100 putative protein
Length = 1752
Score = 487 bits (1254), Expect = e-137
Identities = 250/462 (54%), Positives = 333/462 (71%), Gaps = 8/462 (1%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W TL+AA+RS G+IVLNVASSGIASLLL GGRTAHSRF+IP+ E
Sbjct: 1295 FFIYGFGGTGKTFIWKTLAAAVRSRGQIVLNVASSGIASLLLEGGRTAHSRFAIPLNPDE 1354
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
S C + S A ++++ASLIIWDEAPM++K CFE+LDK+ DI+ + +K F G
Sbjct: 1355 FSVCKITPKSDLANLIKEASLIIWDEAPMMSKFCFESLDKSFYDILNNK----DNKVFGG 1410
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAG-SSSSSL 650
KVVV GGDFRQ+LPVI R EI+ SS N+SYLW HCKV+KLT NMRL G SS +
Sbjct: 1411 KVVVFGGDFRQVLPVINGAGRVEIVMSSLNASYLWDHCKVLKLTKNMRLLSGGLSSEEAK 1470
Query: 651 EIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAY--LNVVANLE 708
EI++F+DWLL VGDG I ++ +++I+IP LL++E+ NP+ + Y + + +
Sbjct: 1471 EIQQFSDWLLAVGDGRINEPNDGEALIDIPEELLIKEAGNPIEAISKEIYGDPSELHMIN 1530
Query: 709 NHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEF 768
+ +F++RA+LAPT E V +N +M+ + EE YLS D+ +D DS + T +F
Sbjct: 1531 DPKFFQRRAILAPTNEDVNTINQYMLEHLKSEERIYLSADSIDPTDSDSLANPV-ITPDF 1589
Query: 769 LNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKT 828
LN ++ +G+P+H + LKVG P+ML++N+D GLCNGTRL ++ L ++ A ++ +
Sbjct: 1590 LNSIQLTGMPHHALRLKVGAPVMLLRNLDPKGGLCNGTRLQITQLAKQVVQAKVITRDRI 1649
Query: 829 GKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHG 888
G V IP ++LTPSDT LPFK RRQFP++V FAMTINKSQGQSL VGLYLP+PVF+HG
Sbjct: 1650 GDIVLIPLINLTPSDTKLPFKMRRRQFPLSVAFAMTINKSQGQSLEQVGLYLPKPVFSHG 1709
Query: 889 QLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
QLYVALSRV S+K LKILI+D G + T NVV++EVFQNI
Sbjct: 1710 QLYVALSRVTSKKGLKILILDKDGNMQKQTTNVVFKEVFQNI 1751
>At3g31980 hypothetical protein
Length = 1099
Score = 473 bits (1216), Expect = e-133
Identities = 240/462 (51%), Positives = 327/462 (69%), Gaps = 8/462 (1%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+ KTF+W TLSAA+R G I +NVASSGIASLLL GGRTAHSRF IPI +
Sbjct: 641 FFVYGFGGTSKTFMWKTLSAAVRMRGLISVNVASSGIASLLLQGGRTAHSRFGIPINPDD 700
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
+TC++ S A ML++ASLIIWDEAPM++++CFE+LD++LND++ KPF G
Sbjct: 701 FTTCHIVPNSDLANMLKEASLIIWDEAPMMSRYCFESLDRSLNDVIGNI----DGKPFGG 756
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAG-SSSSSL 650
KVVV GGDFRQ+LPVI R+EI+ ++ NSSYLW+HCKV+ LT NMRL +
Sbjct: 757 KVVVFGGDFRQVLPVIHGAGRAEIVLAALNSSYLWEHCKVLTLTKNMRLMSNDLDKDEAE 816
Query: 651 EIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAY--LNVVANLE 708
EIKEF++WLL VGDG + ++ + +I+IP LL++++++P+ + Y L+++
Sbjct: 817 EIKEFSNWLLAVGDGRVSEPNDGEVLIDIPEELLIKDANDPIEAITKAVYGDLDLLQPNN 876
Query: 709 NHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEF 768
+ +F+QRA+L P V +N+ M+ + GE YLS D+ D S ++ T +F
Sbjct: 877 DPKFFQQRAILCPRNTDVNTINDIMLDKLNGELVTYLSADSIDPQDAAS-LNNPVLTPDF 935
Query: 769 LNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKT 828
LN +K SG+PNH + LK+G P+ML++NID GLCNGTRL V+ + +I+ A ++G +
Sbjct: 936 LNSIKLSGLPNHNLTLKIGTPVMLLRNIDPKGGLCNGTRLQVTQMGNHILEARVITGDRV 995
Query: 829 GKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHG 888
G V I + ++PSDT LPF+ RRQFPI V FAMTINKSQGQSL VG+YLP+PVF+HG
Sbjct: 996 GDKVIIIKSQISPSDTKLPFRMRRRQFPIAVAFAMTINKSQGQSLKEVGIYLPKPVFSHG 1055
Query: 889 QLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
QLYVALSRV S+K LK+LIVD +G + T NVV++E+FQNI
Sbjct: 1056 QLYVALSRVTSKKGLKVLIVDKEGNTQSQTMNVVFKEIFQNI 1097
>At1g52960 hypothetical protein
Length = 924
Score = 466 bits (1198), Expect = e-131
Identities = 231/433 (53%), Positives = 312/433 (71%), Gaps = 5/433 (1%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W LSAA+RS+G I LNVASSGIA+LLL GGRT HSRF IPI +E
Sbjct: 495 FFVYGFGGTGKTFLWKLLSAAIRSKGDISLNVASSGIAALLLDGGRTTHSRFGIPINPNE 554
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
SSTCN+ +GS E++++A+LIIWDE PM++KHCFE+LD+TL DIM DKPF G
Sbjct: 555 SSTCNISRGSDLGELVKEANLIIWDETPMMSKHCFESLDRTLRDIMNNPG----DKPFGG 610
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLE 651
K +V GGDFRQ+LPVI R EI+ ++ NSSY+W+HCKV++LT NMRL S +
Sbjct: 611 KGIVFGGDFRQVLPVINGAGREEIVFAALNSSYIWEHCKVLELTKNMRLLANISEHEKRD 670
Query: 652 IKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLNVVANLENHA 711
I+ F+ W+L VGDG I ++ ++I+IP L+ ++P+ ++ Y N ++
Sbjct: 671 IEYFSKWILDVGDGKISQPNDGIALIDIPEEFLINGDNDPVESIIEAVYGNTFMEEKDPK 730
Query: 712 YFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLND 771
+F+ RA+L PT E V +N MMSM+ GEE YLS D+ +D S + + ++++FLN
Sbjct: 731 FFQGRAILCPTNEDVNSINEHMMSMLDGEERIYLSSDSIDPADTSSA-NNDAYSADFLNS 789
Query: 772 LKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKP 831
++ SG+PNH + LKVG P+ML++N+D + GLCNGTRL V+ + +I A ++G++ GK
Sbjct: 790 VRVSGLPNHCLRLKVGCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKI 849
Query: 832 VYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLY 891
V IPR+ +TPSDT LPFK RRQFP++V FAMTINKSQGQ+L VGLYLPRPVF+HGQLY
Sbjct: 850 VLIPRMLITPSDTRLPFKMRRRQFPLSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLY 909
Query: 892 VALSRVKSRKRLK 904
VA+SRV S+ K
Sbjct: 910 VAISRVTSKTGTK 922
>At2g14470 pseudogene
Length = 1265
Score = 453 bits (1165), Expect = e-127
Identities = 231/444 (52%), Positives = 311/444 (70%), Gaps = 8/444 (1%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W TLSA +R +IVLNVASSGIASLLL GGRTAHSRF IP+ E
Sbjct: 825 FFVYGFGGTGKTFIWKTLSATIRYRDQIVLNVASSGIASLLLEGGRTAHSRFGIPLNPDE 884
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
S C ++ S A +++KASL+IWDEAPM+++ CFEALDK+ +DI+K + F G
Sbjct: 885 FSVCKIKPKSDLANLVKKASLVIWDEAPMMSRFCFEALDKSFSDIIKNT----DNTVFGG 940
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAG-SSSSSL 650
KVVV GGDFRQ+ PVI R+EI+ SS N+SYLW +CKV+KLT N RL S + +
Sbjct: 941 KVVVFGGDFRQVFPVINGAGRAEIVMSSLNASYLWDNCKVLKLTKNTRLLANNLSETEAK 1000
Query: 651 EIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLN--VVANLE 708
EI+EF+DWLL VGDG I ++ +II+IP LL+ +D P+ + N Y + ++ +
Sbjct: 1001 EIQEFSDWLLAVGDGRINESNDGVAIIDIPEDLLITNADKPIESITNEIYGDPKILHEIT 1060
Query: 709 NHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEF 768
+ +F+ RA+LA E V +N +++ + EE YLS D+ +D DS + T +F
Sbjct: 1061 DPKFFQGRAILASKNEDVNTINEYLLDQLHAEERIYLSADSIDPTDSDS-LSNPVITPDF 1119
Query: 769 LNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKT 828
LN +K G+PNH + LKVG P++L++N+D GLCNGTRL ++ L I+ A ++G +
Sbjct: 1120 LNSIKLPGLPNHSLRLKVGAPVLLLRNLDPKGGLCNGTRLQITQLCTQIVEAKVITGDRI 1179
Query: 829 GKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHG 888
G + IP ++LTP++T LPFK RRQFP++V F MTINKS+GQSL HVGLYLP+PVF+HG
Sbjct: 1180 GHIILIPTVNLTPTNTKLPFKMRRRQFPLSVAFVMTINKSEGQSLEHVGLYLPKPVFSHG 1239
Query: 889 QLYVALSRVKSRKRLKILIVDDKG 912
QLYVALSRV S+K LKILI+D G
Sbjct: 1240 QLYVALSRVTSKKGLKILILDKDG 1263
>At3g30560 hypothetical protein
Length = 1473
Score = 447 bits (1151), Expect = e-125
Identities = 227/434 (52%), Positives = 306/434 (70%), Gaps = 5/434 (1%)
Query: 497 GKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHESSTCNVRQGSHKAEMLQKASLIIWD 556
G I LNVASSGIASLLL GGRTAHSRF IP+T HE+STCN+ +GS AE++ A LIIWD
Sbjct: 1045 GDICLNVASSGIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSDLAELVTAAKLIIWD 1104
Query: 557 EAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAGKVVVLGGDFRQILPVILKGSRSEII 616
EAPM++K+CFE+LDK+L DI+ T D PF GK+++ GGDFRQILPVIL R I+
Sbjct: 1105 EAPMMSKYCFESLDKSLKDILSTP----EDMPFGGKLIIFGGDFRQILPVILAAGRELIV 1160
Query: 617 SSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLEIKEFADWLLQVGDGTIKPIDEDDSI 676
SS NSS+LW++CKV KLT NMRL Q + + EI++F+ W+L VG+G + ++ +
Sbjct: 1161 KSSLNSSHLWQYCKVFKLTKNMRLLQDIDINEAREIEDFSKWILAVGEGKLNQPNDGVTQ 1220
Query: 677 IEIPTYLLVRESDNPLLELVNFAYLNVVANLENHAYFEQRALLAPTLESVEEVNNFMMSM 736
I+I +L+ E DNP+ ++ Y + +F+ RA+L PT + V +N+ M+S
Sbjct: 1221 IQIRDDILIPEGDNPIESIIKAVYGTSFDEERDPKFFQDRAILCPTNDDVNSINDHMLSK 1280
Query: 737 IPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLNDLKCSGIPNHRIVLKVGVPIMLIQNI 796
+ GEE Y S D+ SD ++ + +T +FLN +K SG+PNH + LKVG P+ML++N+
Sbjct: 1281 LTGEEKIYRSSDSIDPSDTRADKNPV-YTPDFLNKIKISGLPNHLLWLKVGCPVMLLRNL 1339
Query: 797 DQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQFP 856
D GL NGTRL + L ++ L+G++ GK V IPR+ LTPSD LPFK RRQFP
Sbjct: 1340 DSHGGLMNGTRLQIVRLGDKLVQGRILTGTRVGKLVIIPRMPLTPSDRRLPFKMKRRQFP 1399
Query: 857 ITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSN 916
++V FAMTINKSQGQSL +VG+YLP+PVF+HGQLYVA+SRVKS+ LK+LI D KG N
Sbjct: 1400 LSVAFAMTINKSQGQSLGNVGIYLPKPVFSHGQLYVAMSRVKSKGGLKVLITDSKGKQKN 1459
Query: 917 CTRNVVYEEVFQNI 930
T NVV++E+F+N+
Sbjct: 1460 ETTNVVFKEIFRNL 1473
>At1g35940 hypothetical protein
Length = 1678
Score = 433 bits (1113), Expect = e-121
Identities = 232/462 (50%), Positives = 306/462 (66%), Gaps = 41/462 (8%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W TLSAA+R G+IVLNVASSGIASLLL GGRTAHSRF IP+ E
Sbjct: 1254 FFVYGFGGTGKTFIWKTLSAAIRCRGQIVLNVASSGIASLLLEGGRTAHSRFGIPLNHDE 1313
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
S +LDK+ +DI+K ++K F G
Sbjct: 1314 FSV---------------------------------SLDKSFSDIIKNT----NNKVFGG 1336
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAG-SSSSSL 650
KVVV GGDFRQ+LPVI R+EI+ SS N+SYLW HCKV+KLT NMRL S++ +
Sbjct: 1337 KVVVFGGDFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLANNLSATEAK 1396
Query: 651 EIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLN--VVANLE 708
EI+EF+DWLL V DG I ++ + I+IP LL+ +D P+ + N Y + ++ +
Sbjct: 1397 EIQEFSDWLLAVSDGRINEPNDGVATIDIPEDLLITNADKPIETITNEIYGDPKILHEIT 1456
Query: 709 NHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEF 768
+ +F+ RA+LAP E V +N +++ + EE YLS D+ +D DS ++ T +F
Sbjct: 1457 DPKFFQGRAILAPKNEDVNTINEYLLEQLDAEERIYLSADSIDPTDSDS-LNNPVITPDF 1515
Query: 769 LNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKT 828
LN +K G+PNH + LKVG P+ML++N+D GLCNGTRL ++ L I+ A ++G +
Sbjct: 1516 LNSIKLPGLPNHSLCLKVGAPVMLLRNLDPKGGLCNGTRLQITQLCTQIVEAKVITGDRI 1575
Query: 829 GKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHG 888
G V IP ++LTP+DT LPFK RRQFP++V FAMTINKSQGQSL H+GLYLP+PVF+HG
Sbjct: 1576 GNIVLIPTVNLTPTDTKLPFKMRRRQFPLSVAFAMTINKSQGQSLEHIGLYLPKPVFSHG 1635
Query: 889 QLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
QLYVALSRV S+K LKILI+D G + T NVV++EVFQNI
Sbjct: 1636 QLYVALSRVTSKKGLKILILDKDGKLQKQTTNVVFKEVFQNI 1677
>At1g54430 hypothetical protein
Length = 1639
Score = 420 bits (1080), Expect = e-117
Identities = 239/491 (48%), Positives = 321/491 (64%), Gaps = 21/491 (4%)
Query: 451 LNRKERIPRYWMLFRPITED--FFFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGI 508
LN ++RI +L I ++ FFLYG GG+GKTF++ T+ +ALRS GK V+ VASS I
Sbjct: 1151 LNEQQRIIYDDVLKSVINKEGKLFFLYGAGGTGKTFLYKTIISALRSNGKNVMPVASSAI 1210
Query: 509 ASLLLPGGRTAHSRFSIPITIHESSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEA 568
A+LLLPGGRTAHSRF IPI +HE S C+++ GS A +L K LIIWDEAPM ++H FEA
Sbjct: 1211 AALLLPGGRTAHSRFKIPINVHEDSICDIKIGSMLANVLSKVDLIIWDEAPMAHRHTFEA 1270
Query: 569 LDKTLNDIMKTQATFGHDKPFAGKVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKH 628
+D+TL DI+ K F GK V+LGGDFRQILPVI +G+R E +S++ N SYLW+
Sbjct: 1271 VDRTLRDILSVGDEKALTKTFGGKTVLLGGDFRQILPVIPQGTRQETVSAAINRSYLWES 1330
Query: 629 CKVMKLTTNMRLQQAGSSSSSLEIKEFADWLLQVGDGT-------IKPIDEDDSIIEIPT 681
C L+ NMR+Q EIK FA+W+LQVGDG I E+D+II I
Sbjct: 1331 CHKYLLSQNMRVQPE-------EIK-FAEWILQVGDGEAPRKTHGIDDDQEEDNII-IDK 1381
Query: 682 YLLVRESDNPLLELVNFAYLNVVANLENHAYFEQRALLAPTLESVEEVNNFMMSMIPGEE 741
LL+ E++NPL L + + ++ + A+L P E+V+E+N++++S +PG
Sbjct: 1382 NLLLPETENPLEVLCRSVFPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLLSKVPGLA 1441
Query: 742 TKYLSYDTPCRSDEDSEIDAEW-FTSEFLNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSA 800
+Y S D+ R + +E E + E+LN L+ G+P HR+ LKVGVPIML++N++Q
Sbjct: 1442 KEYFSADSIDRDEALTEEGFEMSYPMEYLNSLEFPGLPAHRLCLKVGVPIMLLRNLNQKE 1501
Query: 801 GLCNGTRLIVSALTPYIIVATALSG-SKTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITV 859
GLCNGTRLIV+ L ++ A LS +K K V IPR+ L+P D+ PF RRQFP+ +
Sbjct: 1502 GLCNGTRLIVTHLGDKVLKAEILSDTTKERKKVLIPRIILSPQDSKHPFTLRRRQFPVRM 1561
Query: 860 CFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSNCTR 919
C+AMTINKSQGQ+L+ V LYLP+PVF+HGQLYVALSRV S K L +L K T
Sbjct: 1562 CYAMTINKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLDTSKKKEGKYVT- 1620
Query: 920 NVVYEEVFQNI 930
N+VY EVF +
Sbjct: 1621 NIVYREVFNGL 1631
>At2g05080 putative helicase
Length = 1219
Score = 416 bits (1070), Expect = e-116
Identities = 216/443 (48%), Positives = 302/443 (67%), Gaps = 34/443 (7%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W TLSAALRS+G IVLNVASSGIASLLL GGRTAHSR IP+ +E
Sbjct: 770 FFVYGFGGTGKTFLWKTLSAALRSKGDIVLNVASSGIASLLLEGGRTAHSRSGIPLNPNE 829
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
+TCN++ GS +A ++++ASLIIWDEAPM+++HCFE+LD++L+DI +KPF G
Sbjct: 830 FTTCNMKAGSDRANLVKEASLIIWDEAPMMSRHCFESLDRSLSDICGN----CDNKPFGG 885
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLE 651
KVVV GGDFRQ+LPVI ++I+ ++ NSSYLW HCKV+ LT NM L
Sbjct: 886 KVVVFGGDFRQVLPVIPGADTADIVMAALNSSYLWSHCKVLTLTKNMCLFS--------- 936
Query: 652 IKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAY--LNVVANLEN 709
+W+L VGDG I ++ +++I+IP+ L+ ++ +P+ + Y + + ++
Sbjct: 937 ----EEWILAVGDGRIGEPNDGEALIDIPSEFLITKAKDPIQAICTEIYGDITKIHEQKD 992
Query: 710 HAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFL 769
+F++RA+L PT E V ++N M+ + GEE +LS D+ +D S + T EFL
Sbjct: 993 PVFFQERAILCPTNEDVNQINETMLDNLQGEELTFLSSDSLDTADIGSR-NNPVLTPEFL 1051
Query: 770 NDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTG 829
N++K G+ NH++ LK+G P+ML++NID GL NGTRL + ++P+I+ A L+G +
Sbjct: 1052 NNVKVLGLSNHKLRLKIGSPVMLLRNIDPIGGLMNGTRLQIMQMSPFILQAMILTGDR-- 1109
Query: 830 KPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQ 889
+DT LPF+ R Q P+ VCFAMTINKSQGQSL VG++LPRP F+H Q
Sbjct: 1110 ------------ADTKLPFRMRRTQLPLAVCFAMTINKSQGQSLKRVGIFLPRPCFSHSQ 1157
Query: 890 LYVALSRVKSRKRLKILIVDDKG 912
LYVA+SRV S+ LKILIV+D+G
Sbjct: 1158 LYVAISRVTSKSGLKILIVNDEG 1180
>At3g31440 hypothetical protein
Length = 536
Score = 411 bits (1056), Expect = e-114
Identities = 224/455 (49%), Positives = 300/455 (65%), Gaps = 32/455 (7%)
Query: 479 GSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHESSTCNVR 538
G+ KT +W TL AA+R +IVLN+ASSGIASLLL GGRTAHSRF I + E S C ++
Sbjct: 110 GTWKTIIWKTLFAAIRRRDQIVLNMASSGIASLLLEGGRTAHSRFGIRLNPDEFSVCKIK 169
Query: 539 QGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAGKVVVLGG 598
S A ++++ASL+I D+APM+++ CFEALDK+ +DI+K ++K F GKVVV G
Sbjct: 170 PKSDLANLVKEASLVICDKAPMMSRFCFEALDKSFSDIIKNT----YNKVFGGKVVVFSG 225
Query: 599 DFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAG-SSSSSLEIKEFAD 657
DFRQ+LPVI R+EI+ SS N+SYLW HCKV+KLT NMRL S + + EI EF+D
Sbjct: 226 DFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLANNLSETEAKEIHEFSD 285
Query: 658 WLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLN--VVANLENHAYFEQ 715
WLL VGDG I ++D +II+IP LL+ +D P+ + N Y + ++ + + +F+
Sbjct: 286 WLLAVGDGRINEPNDDVAIIDIPKDLLITNADKPIEWITNEIYGDPKILHEITDPKFFQG 345
Query: 716 RALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLNDLKCS 775
RA+LAP E V +N +++ + EE YLS D+ +D DS ++ T +FLN +K
Sbjct: 346 RAILAPKNEDVNTINEYLLEQLHAEERIYLSADSIDPTDSDS-LNNPVITPDFLNSIKLP 404
Query: 776 GIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIP 835
G GLCNG RL ++ L I+ A ++G + G V IP
Sbjct: 405 G------------------------GLCNGARLQITQLFTEIVEAKVITGDRIGHIVLIP 440
Query: 836 RLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALS 895
++LTP+DT LPFK RRQFP++V FAMTINKSQGQSL HVGLYLP+PVF+HGQLYVALS
Sbjct: 441 TVNLTPTDTKLPFKMRRRQFPLSVAFAMTINKSQGQSLEHVGLYLPKPVFSHGQLYVALS 500
Query: 896 RVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
RV S+K LKILI+D G + T N+V++EVFQNI
Sbjct: 501 RVTSKKGLKILILDKNGKLQKQTTNIVFKEVFQNI 535
>At3g30420 hypothetical protein
Length = 837
Score = 406 bits (1044), Expect = e-113
Identities = 237/498 (47%), Positives = 320/498 (63%), Gaps = 35/498 (7%)
Query: 451 LNRKERIPRYWMLFRPITED---FFFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSG 507
LN ++RI Y + + +T FFLYG GG+GKTF++ T+ +ALRS GK V+ VASS
Sbjct: 349 LNEQQRII-YDDVLKSVTNKEGKLFFLYGDGGTGKTFLYKTIISALRSNGKNVMPVASSA 407
Query: 508 IASLLLPGGRTAHSRFSIPITIHESSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFE 567
IA+LLLPGGRTAHS F IPI +HE C+++ GS A +L K LIIWDEAPM ++H FE
Sbjct: 408 IAALLLPGGRTAHSWFKIPINVHEDFICDIKIGSMLANVLSKVDLIIWDEAPMAHRHTFE 467
Query: 568 ALDKTLNDIMKTQATFGHDKPFAGKVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWK 627
A+D+TL DI+ K GK V+LGGDFRQILPVI + +R E +S++ N SYLW+
Sbjct: 468 AVDRTLRDILSVGDEKALTKTLGGKTVLLGGDFRQILPVIPQRTRQETVSAAINRSYLWE 527
Query: 628 HCKVMKLTTNMRLQQAGSSSSSLEIKEFADWLLQVGDGT-------IKPIDEDDSIIEIP 680
C L+ NMR+Q EIK FA+W+LQ+GDG I E+D+II I
Sbjct: 528 SCHKYLLSQNMRVQPE-------EIK-FAEWILQIGDGEAPRKTHGIDDDQEEDNII-ID 578
Query: 681 TYLLVRESDNP---LLELVNFAYLNVVANLENHAYFEQRALLAPTLESVEEVNNFMMSMI 737
LL+ E++NP L + V+ + N +LEN + A+L P E+V+E+N++++S +
Sbjct: 579 KNLLLPETENPLEVLCQSVSPDFTNTFQDLEN---LKGTAVLTPRNETVDEINDYLLSKV 635
Query: 738 PGEETKYLSYDTPCRSDEDSEIDAEWF----TSEFLNDLKCSGIPNHRIVLKVGVPIMLI 793
PG +Y S D+ D+D + E F E+LN L+ G+P HR+ LKVGVPIML+
Sbjct: 636 PGLAKEYFSADS---IDQDEALTEEGFEMSYPMEYLNSLEFPGLPAHRLCLKVGVPIMLL 692
Query: 794 QNIDQSAGLCNGTRLIVSALTPYIIVATALSG-SKTGKPVYIPRLSLTPSDTGLPFKFSR 852
+N++Q GLCNGTRL V+ L ++ A LS +K K V IPR+ L+P D+ PF R
Sbjct: 693 RNLNQKEGLCNGTRLTVTHLGDKVLKAEILSDTTKKRKKVLIPRIILSPQDSKHPFTLRR 752
Query: 853 RQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKG 912
RQFP+ +C+AMT+NKSQGQ+L+ V LYLP+PVF+HGQLYVALSRV S K L +L K
Sbjct: 753 RQFPVRMCYAMTVNKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLDTSKKK 812
Query: 913 VVSNCTRNVVYEEVFQNI 930
T N+VY EVF +
Sbjct: 813 EGKYVT-NIVYREVFNGL 829
>At3g43350 putative protein
Length = 830
Score = 389 bits (999), Expect = e-108
Identities = 207/434 (47%), Positives = 282/434 (64%), Gaps = 45/434 (10%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W LSAA+RS+G I LNVASSGIA+L L GGRTAHSRF IPI +E
Sbjct: 199 FFVYGFGGTGKTFLWKLLSAAVRSKGDISLNVASSGIAALRLDGGRTAHSRFDIPINPNE 258
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
SSTCN+ +GS E++++A LIIWDEAPM++KHCFE+LD+TL DI+ DKP G
Sbjct: 259 SSTCNISRGSDLGELVKEAKLIIWDEAPMMSKHCFESLDRTLKDIVNNPG----DKPLGG 314
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLE 651
KV+V GGDFRQ+LPVI R EI+ ++ NSSY+W+H KV++LT NMRL S +
Sbjct: 315 KVIVFGGDFRQVLPVINGAGREEIVFAALNSSYIWEHSKVLELTKNMRLLADISEHEKRD 374
Query: 652 IKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLNVVANLE--- 708
I++F+ W+L VGDG I ++ ++I+IP L+ ++P+ ++ Y N +
Sbjct: 375 IEDFSKWILDVGDGKISQPNDGIALIDIPEEFLINGDNDPVESIIEAVYGNTFMEEKDPK 434
Query: 709 --NHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTS 766
++ ++ RA+L PT E V +N MM M+ GEE YLS D+ +D S +A + +
Sbjct: 435 KTDYPQYQGRAILCPTNEDVNSINEHMMRMLDGEERIYLSSDSIDPADISSANNAA-YLA 493
Query: 767 EFLNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGS 826
+FLN+++ G+PNH + LKVG P+ML++N+D + GLCNGTRL V+ +T II A ++
Sbjct: 494 DFLNNVRVYGLPNHCLRLKVGCPVMLLRNMDPNKGLCNGTRLQVTQMTDTIIQARFIT-- 551
Query: 827 KTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFT 886
FAMTINKSQGQ+L VGLYLPRPVF+
Sbjct: 552 ---------------------------------AFAMTINKSQGQTLESVGLYLPRPVFS 578
Query: 887 HGQLYVALSRVKSR 900
HGQLYVA+SRV S+
Sbjct: 579 HGQLYVAISRVTSK 592
Score = 149 bits (376), Expect = 7e-36
Identities = 70/115 (60%), Positives = 92/115 (79%)
Query: 786 VGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTG 845
+G P+ML++N+D + GLCNGTRL V+ + +I A ++G++ GK V IPR+ +TPSDT
Sbjct: 594 IGCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPSDTR 653
Query: 846 LPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSR 900
LPFK R+QF ++V FAMTINKSQGQ+L VGLYLPRPVF+HGQLYVA+SRV S+
Sbjct: 654 LPFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSK 708
Score = 147 bits (372), Expect = 2e-35
Identities = 71/119 (59%), Positives = 92/119 (76%)
Query: 786 VGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTG 845
VG P+ML++N+D + GLCNGTRL V+ + +I A ++G++ GK V IPR+ +TP DT
Sbjct: 710 VGCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPLDTR 769
Query: 846 LPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLK 904
LPFK R+QF ++V FAMTINKSQGQ+L VGLYLPRPVF+HGQLYVA+SRV S+ K
Sbjct: 770 LPFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTGTK 828
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 379 bits (974), Expect = e-105
Identities = 200/417 (47%), Positives = 266/417 (62%), Gaps = 29/417 (6%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FFLY FGG+GKTF+W LSAA+R +G LNVASS IASLLL GGRTAHSRF IP+T HE
Sbjct: 843 FFLYAFGGTGKTFLWKVLSAAIRCKGDTCLNVASSSIASLLLEGGRTAHSRFGIPLTPHE 902
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
+STCN+ +GS AE++ A LIIWDE D PF
Sbjct: 903 TSTCNMERGSDLAELVTAAKLIIWDE----------------------------DMPFGR 934
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLE 651
KV++ GGDFRQIL VI R I+ SS NSSYLW+HCKV+KLT NMRL Q + + E
Sbjct: 935 KVILFGGDFRQILHVIPAAGRELIVKSSLNSSYLWQHCKVLKLTKNMRLLQDIDINEARE 994
Query: 652 IKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLNVVANLENHA 711
I++F W+L VG+G + + + I+IP +L+ E DNP+ ++ Y A +
Sbjct: 995 IEDFLKWILTVGEGKLNEPSDGVTHIQIPDDILIPEGDNPIESIIKAVYGTTFAQKRDPK 1054
Query: 712 YFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLND 771
+F+ +A+L PT + V +N+ M+S + GEE Y S ++ SD ++ + +T +FLN
Sbjct: 1055 FFQHKAILCPTNDDVNSINDHMLSKLTGEERIYRSSNSIDPSDTRADKNPI-YTPDFLNK 1113
Query: 772 LKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKP 831
+K SG+ NH + LKVG P+ML++N GL NGTRL + L ++ L+G++ GK
Sbjct: 1114 IKISGLANHLLRLKVGCPVMLLRNFYPHGGLMNGTRLQIVRLGDKLVQGRILTGTRVGKL 1173
Query: 832 VYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHG 888
V IPR+SLTPSD LPFK RR FP++V FAMTINKSQGQSL +VG+YLP+ VF+HG
Sbjct: 1174 VIIPRMSLTPSDRRLPFKMKRRHFPLSVAFAMTINKSQGQSLGNVGMYLPKAVFSHG 1230
>At5g34960 putative protein
Length = 1033
Score = 329 bits (844), Expect = 4e-90
Identities = 191/449 (42%), Positives = 264/449 (58%), Gaps = 75/449 (16%)
Query: 485 VWNTLSAALRSE-GKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHESSTCNVRQGSHK 543
V+N ++ A+ + +IVLNVASSGIASLLL GGRTAHSRF I + E S C ++ S
Sbjct: 656 VYNEITEAVFNNLDQIVLNVASSGIASLLLEGGRTAHSRFGISLNPDEFSVCKIKPKSDL 715
Query: 544 AEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAGKVVVLGGDFRQI 603
A ++++ASL+IWDEAPM++ RQ+
Sbjct: 716 ANLVKEASLVIWDEAPMMS--------------------------------------RQV 737
Query: 604 LPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLEIKEFADWLLQVG 663
L VI R+EI+ SS N+SYLW HCKV+K+ +
Sbjct: 738 LLVINGAGRAEIVMSSLNASYLWDHCKVLKIN-------------------------EPN 772
Query: 664 DGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFAYLN--VVANLENHAYFEQRALLAP 721
DG +II+IP LL+ +D P+ + N Y + ++ + + +F+ RA+LAP
Sbjct: 773 DGV--------AIIDIPEDLLITNTDKPIESITNEIYGDPKILHEITDPKFFQGRAILAP 824
Query: 722 TLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLNDLKCSGIPNHR 781
T E V +N +++ + EE YLS D+ +D +S ++ T +FLN +K +G+PNH
Sbjct: 825 TNEDVNTINEYLLEQLHAEERIYLSADSIDPTDSNS-LNNPVITPDFLNSIKLAGLPNHS 883
Query: 782 IVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTP 841
+ LKV P+ML++N+D GLCNGTRL ++ L I+ A ++G G V IP ++LTP
Sbjct: 884 LRLKVSAPVMLLRNLDPKGGLCNGTRLQITQLCTQIVEAKVITGDIIGHIVLIPTVNLTP 943
Query: 842 SDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRK 901
+DT LPFK RRQFP++V FAMTIN SQGQSL HVGLYLP+ VF+HGQLYVALSRV S+K
Sbjct: 944 TDTKLPFKMRRRQFPLSVAFAMTINTSQGQSLEHVGLYLPKAVFSHGQLYVALSRVTSKK 1003
Query: 902 RLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
LK LI+D G + T NVV++EVFQNI
Sbjct: 1004 GLKFLILDKDGKLQKQTTNVVFKEVFQNI 1032
>At5g37110 putative helicase
Length = 1307
Score = 309 bits (792), Expect = 4e-84
Identities = 167/393 (42%), Positives = 243/393 (61%), Gaps = 52/393 (13%)
Query: 515 GGRTAHSRFSIPITIHESSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLN 574
GGRTAHSRF IP+ +E +TCN++ GS +A ++++ASLIIWDEAPM++++CFE+LD++L+
Sbjct: 950 GGRTAHSRFGIPLNPNEFTTCNMKVGSDRANLVKEASLIIWDEAPMMSRYCFESLDRSLS 1009
Query: 575 DIMKTQATFGHDKPFAGKVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKL 634
DI G +KPF GKVVV GG
Sbjct: 1010 DICGN----GDNKPFGGKVVVFGGL----------------------------------- 1030
Query: 635 TTNMRLQQAGSSSSSLEIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLE 694
S S + +IKEF++W+L VGDG I ++ +++I IP+ L+ ++ +P+
Sbjct: 1031 ----------SVSEAKDIKEFSEWILAVGDGRIVEPNDGEALIVIPSEFLITKAKDPIEA 1080
Query: 695 LVNFAYLNVVANLENH--AYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCR 752
+ Y ++ E + +F+++A+L PT E V ++N M+ + GEE +LS D+
Sbjct: 1081 ICTEIYGDITKIHEQNDPIFFQEKAILCPTNEDVNQINETMLDNLQGEEFTFLSSDSLDP 1140
Query: 753 SDEDSEIDAEWFTSEFLNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSA 812
+D + + T +FLN +K S +PNH++ LK+G P+ML++NID GL NGTRL ++
Sbjct: 1141 ADIGGKNNPA-LTPDFLNSVKVSRLPNHKLRLKIGCPVMLLRNIDPIGGLMNGTRLRITQ 1199
Query: 813 LTPYIIVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQS 872
+ P+I+ A L+G + G V IPRL L PSDT LPF+ R Q P+ VCFAMTINKSQGQS
Sbjct: 1200 MGPFILQAMILTGDRAGHLVLIPRLKLAPSDTKLPFRMRRTQLPLAVCFAMTINKSQGQS 1259
Query: 873 LSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKI 905
L VG++L RP F+HGQLYVA+SRV S+ RLKI
Sbjct: 1260 LKRVGIFLLRPCFSHGQLYVAISRVTSKTRLKI 1292
>At1g64410 unknown protein
Length = 753
Score = 244 bits (622), Expect = 2e-64
Identities = 119/207 (57%), Positives = 155/207 (74%), Gaps = 4/207 (1%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W LSAA+RS+G I LNVASSGIA+LLL GGRTAHSRF IPI +E
Sbjct: 545 FFVYGFGGTGKTFLWKLLSAAIRSKGDISLNVASSGIAALLLDGGRTAHSRFGIPINPNE 604
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
SSTCN+ +G E++++A+LIIWDEA M++KHCFE+LD+TL DIM DKPF G
Sbjct: 605 SSTCNISRGLDFGELVKEANLIIWDEAHMMSKHCFESLDRTLRDIMNNPG----DKPFGG 660
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRLQQAGSSSSSLE 651
KV+V GGDFRQ+L VI R EI+ ++ NSSY+W+HCKV++LT NMRL S +
Sbjct: 661 KVIVFGGDFRQVLSVINGAGREEIVFAALNSSYIWEHCKVLELTKNMRLLANISEHEKRD 720
Query: 652 IKEFADWLLQVGDGTIKPIDEDDSIIE 678
I+ F+ W+L VGDG I ++ ++I+
Sbjct: 721 IEYFSKWILDVGDGKISQPNDGIALID 747
>At4g04300 hypothetical protein
Length = 286
Score = 232 bits (591), Expect = 8e-61
Identities = 122/238 (51%), Positives = 161/238 (67%), Gaps = 20/238 (8%)
Query: 471 FFFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIH 530
FFF+YG GG GKTF+W TL+A RS+G+ LN+ASSGIASLLL GGR AH RFSIP+
Sbjct: 25 FFFVYGSGGIGKTFIWKTLAAVGRSKGQTCLNIASSGIASLLLEGGRIAHYRFSIPLNPD 84
Query: 531 ESSTCNVRQGSHKA---------------EMLQKASLIIWDEAPMLNKHCFEALDKTLND 575
E S C ++ S A ++++KASLIIWD+APM +K CFEALDK+ +D
Sbjct: 85 EFSVCKIKPKSDLADLIKEASLIIWDKLVDLIKKASLIIWDKAPMKSKFCFEALDKSFSD 144
Query: 576 IMKTQATFGHDKPFAGKVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLT 635
I+K +K F GKV+V GGDFRQ+LPVI R+E + SS N+ Y+W HCKV+KLT
Sbjct: 145 IIKRV----DNKVFCGKVMVFGGDFRQVLPVINGAGRAETVMSSLNAVYIWDHCKVLKLT 200
Query: 636 TNMRLQQAG-SSSSSLEIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPL 692
NMRL S + EI+EF DWLL VGDG + ++ +++I+IP LL++E+D P+
Sbjct: 201 KNMRLLNNDLSVDEAKEIQEFFDWLLVVGDGRVNEPNDGEALIDIPEELLIQEADIPI 258
>At2g07620 putative helicase
Length = 1241
Score = 207 bits (528), Expect = 2e-53
Identities = 102/169 (60%), Positives = 130/169 (76%), Gaps = 4/169 (2%)
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHE 531
FF+YGFGG+GKTF+W TLSAA+R +G I +NVASSGIA LLL GGRTAHSRF IPI +
Sbjct: 870 FFVYGFGGTGKTFMWKTLSAAVRMKGLISVNVASSGIAFLLLQGGRTAHSRFGIPINPDD 929
Query: 532 SSTCNVRQGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAG 591
+TC++ S A ML++ASLIIWDEAPM++++CFE+LD++LND++ KPF G
Sbjct: 930 FTTCHIVPNSDLANMLKEASLIIWDEAPMMSRYCFESLDRSLNDVIGNV----DGKPFGG 985
Query: 592 KVVVLGGDFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKLTTNMRL 640
KVVV GGDFRQ+L VI R+EI+ ++ NSSYLW+HC V+ LT NM L
Sbjct: 986 KVVVFGGDFRQVLHVIHGAGRAEIVLAALNSSYLWEHCNVLTLTKNMSL 1034
Score = 197 bits (500), Expect = 3e-50
Identities = 101/191 (52%), Positives = 132/191 (68%), Gaps = 1/191 (0%)
Query: 740 EETKYLSYDTPCRSDEDSEIDAEWFTSEFLNDLKCSGIPNHRIVLKVGVPIMLIQNIDQS 799
E YLS D+ D S ++ FT FLN +K SG+ NH + LK+G P+ML++NID
Sbjct: 1050 ESITYLSADSIDPQDPAS-LNNPVFTPYFLNSIKLSGLSNHNLTLKIGTPVMLLKNIDPK 1108
Query: 800 AGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITV 859
GLCNGTRL V+ + +I+ A ++G + V I + ++PSDT LPF+ RRQFPI V
Sbjct: 1109 GGLCNGTRLQVTQMGNHILEARVITGDRVRDKVIIIKAQISPSDTKLPFRMRRRQFPIAV 1168
Query: 860 CFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSNCTR 919
FAM I KSQGQSL V +YLPRPVF+HGQLYVALSRV S+K LK+LIVD +G + T
Sbjct: 1169 AFAMRIKKSQGQSLKEVEIYLPRPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQTM 1228
Query: 920 NVVYEEVFQNI 930
NVV++E+FQN+
Sbjct: 1229 NVVFKEIFQNL 1239
>At5g28780 putative protein
Length = 337
Score = 195 bits (496), Expect = 9e-50
Identities = 109/270 (40%), Positives = 163/270 (60%), Gaps = 3/270 (1%)
Query: 662 VGDGTIKPIDEDDS-IIEIPTYLLVRESDNPLLELVNFAYLNVVANLENHAYFEQRALLA 720
VG+G + DD ++E+ T ++ + N L ++ AY+ + N Y +R +L
Sbjct: 63 VGNGNAPTCETDDGEVVEMDTSFFLKHNGNRLQQVTKGAYVQFSVSQPNFQYLTERGILT 122
Query: 721 PTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLNDLKCSGIPNH 780
P E V+E+N +M+S + G+ +YLS + ++D + ++LN L+ +P H
Sbjct: 123 PHNEYVDEINAYMLSQVGGDSKEYLSSYSIGKADTIGADYEALYHVKYLNSLEFPSLPKH 182
Query: 781 RIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLT 840
+I LK GVPIM ++N +Q GLCNGTRLIV+ L +I A ++G+ GK V IPR L+
Sbjct: 183 KISLKKGVPIMQMRNFNQKEGLCNGTRLIVTNLGEQVIEAQIVTGTHAGKMVSIPRFILS 242
Query: 841 PSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSR 900
P + PF R+QFP+ VC+AMTI K+QGQSL LYLP PVF+H QLYVALSRV S
Sbjct: 243 PPQSEHPFTLRRQQFPMRVCYAMTIIKNQGQSLKSDVLYLPNPVFSHVQLYVALSRVTSP 302
Query: 901 KRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
L IL DD+ ++ +N+VY+E + ++
Sbjct: 303 IGLTILHGDDQ--KNDEVKNIVYKEFYNDL 330
>At3g51700 unknown protein
Length = 344
Score = 189 bits (480), Expect = 6e-48
Identities = 104/289 (35%), Positives = 168/289 (57%), Gaps = 2/289 (0%)
Query: 640 LQQAGSSSSSLEIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLLELVNFA 699
L +G+ + + F W+ +G I ++ ++ I+I LL+ E +P+ +V+
Sbjct: 45 LDSSGTKIHTTIDEAFTKWITNIGGENINKPNDGETKIDIHEDLLITECKDPIKTIVDEV 104
Query: 700 YLNVVANLENHAYFEQRALLAPTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEI 759
Y N ++++RA+L T + +E+N++M+S + GEETK DT S
Sbjct: 105 YGESFTESYNPDFYQERAILCHTNDVADEINDYMLSQLQGEETKCYGADT-IYPTHASPN 163
Query: 760 DAEWFTSEFLNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIV 819
D + EFLN +K G P+ ++ LKVG P+ML++++ L GTRL ++ + +++
Sbjct: 164 DKMLYPLEFLNSIKIPGFPDFKLRLKVGAPVMLLRDLAPYGWLRKGTRLQITRVETFVLE 223
Query: 820 ATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLY 879
A ++G+ G+ V IPR+ + P K RRQFP+ + FAMTI++SQ Q+LS VG+Y
Sbjct: 224 AMIITGNNHGEKVLIPRIPSDLREAKFPIKMRRRQFPVKLAFAMTIDESQRQTLSKVGIY 283
Query: 880 LPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVV-SNCTRNVVYEEVF 927
LPR + HGQ YVA+S+VKSR LK+LI D G T+NVV++E+F
Sbjct: 284 LPRQLLFHGQRYVAISKVKSRAGLKVLITDKDGKPDQEETKNVVFKELF 332
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.344 0.151 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,695,747
Number of Sequences: 26719
Number of extensions: 689253
Number of successful extensions: 3208
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 3085
Number of HSP's gapped (non-prelim): 59
length of query: 930
length of database: 11,318,596
effective HSP length: 109
effective length of query: 821
effective length of database: 8,406,225
effective search space: 6901510725
effective search space used: 6901510725
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0320b.5


