

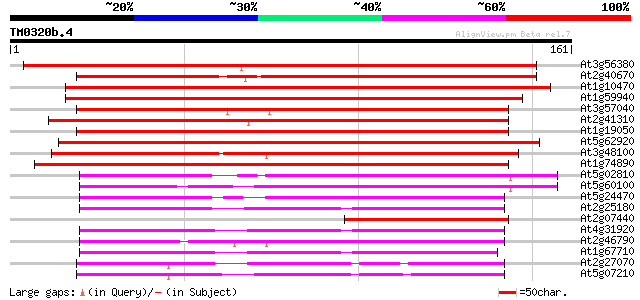
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0320b.4
(161 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g56380 response regulator-like protein 201 1e-52
At2g40670 putative two-component response regulator protein 189 7e-49
At1g10470 putative response regulator 3 172 9e-44
At1g59940 putative protein 170 4e-43
At3g57040 responce reactor 4 168 1e-42
At2g41310 putative two-component response regulator 3 protein 166 5e-42
At1g19050 putative protein 165 1e-41
At5g62920 response regulator 6 (ARR6) 164 2e-41
At3g48100 response reactor 2 (ATRR2) 164 3e-41
At1g74890 putative response regulator (two component phosphotran... 162 7e-41
At5g02810 unknown protein 71 2e-13
At5g60100 pseudo-response regulator 3 (APRR3) 70 4e-13
At5g24470 pseudo-response regulator 5 (APRR5) 68 2e-12
At2g25180 putative two-component response regulator protein 66 9e-12
At2g07440 putative two-component response regulator protein 62 1e-10
At4g31920 predicted protein 61 3e-10
At2g46790 hypothetical protein 60 5e-10
At1g67710 response regulator 11 (ARR11) 59 1e-09
At2g27070 putative two-component response regulator protein 57 3e-09
At5g07210 putative protein 56 1e-08
>At3g56380 response regulator-like protein
Length = 153
Score = 201 bits (511), Expect = 1e-52
Identities = 104/148 (70%), Positives = 121/148 (81%), Gaps = 1/148 (0%)
Query: 5 GSSSSNWVMESGDIPHVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGE 64
GS S + + + HVLAVDDNLIDRKLVE++L+ SSCKVTTAENG RALE+LGL +
Sbjct: 6 GSGSDSCLSSMEEELHVLAVDDNLIDRKLVERILKISSCKVTTAENGLRALEYLGLGDPQ 65
Query: 65 Q-NTLNGRSKVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLE 123
Q ++L KVNLIITDYCMPGMTG+ELLKK+KESS +KEVPVVI+SSENIPTRINKCL
Sbjct: 66 QTDSLTNVMKVNLIITDYCMPGMTGFELLKKVKESSNLKEVPVVILSSENIPTRINKCLA 125
Query: 124 EGAQMFILKPLKQSDIKKLKCKLSNCSS 151
GAQMF+ KPLK SD++KLKC L NC S
Sbjct: 126 SGAQMFMQKPLKLSDVEKLKCHLLNCRS 153
>At2g40670 putative two-component response regulator protein
Length = 164
Score = 189 bits (479), Expect = 7e-49
Identities = 101/138 (73%), Positives = 114/138 (82%), Gaps = 9/138 (6%)
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQN------TLNGRSK 73
HVLAVDDNLIDRKLVE+LL+ S CKVTTAEN RALE+LGL G+QN T N K
Sbjct: 30 HVLAVDDNLIDRKLVERLLKISCCKVTTAENALRALEYLGL--GDQNQHIDALTCNVM-K 86
Query: 74 VNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKP 133
V+LIITDYCMPGMTG+ELLKK+KESS ++EVPVVIMSSENIPTRINKCL GAQMF+ KP
Sbjct: 87 VSLIITDYCMPGMTGFELLKKVKESSNLREVPVVIMSSENIPTRINKCLASGAQMFMQKP 146
Query: 134 LKQSDIKKLKCKLSNCSS 151
LK +D++KLKC L NC S
Sbjct: 147 LKLADVEKLKCHLMNCRS 164
>At1g10470 putative response regulator 3
Length = 259
Score = 172 bits (435), Expect = 9e-44
Identities = 88/139 (63%), Positives = 111/139 (79%)
Query: 17 DIPHVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNL 76
D HVLAVDD+L+DR ++E+LLR +SCKVT ++G RALEFLGL + + + R KV+L
Sbjct: 32 DEVHVLAVDDSLVDRIVIERLLRITSCKVTAVDSGWRALEFLGLDNEKASAEFDRLKVDL 91
Query: 77 IITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQ 136
IITDYCMPGMTGYELLKKIKESS +EVPVVIMSSEN+ TRI++CLEEGAQ F+LKP+K
Sbjct: 92 IITDYCMPGMTGYELLKKIKESSNFREVPVVIMSSENVLTRIDRCLEEGAQDFLLKPVKL 151
Query: 137 SDIKKLKCKLSNCSSLGVG 155
+D+K+L+ L+ L G
Sbjct: 152 ADVKRLRSHLTKDVKLSNG 170
>At1g59940 putative protein
Length = 231
Score = 170 bits (430), Expect = 4e-43
Identities = 85/131 (64%), Positives = 108/131 (81%)
Query: 17 DIPHVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNL 76
D HVLAVDD+L+DR ++E+LLR +SCKVT ++G RALEFLGL + R KV+L
Sbjct: 31 DQVHVLAVDDSLVDRIVIERLLRITSCKVTAVDSGWRALEFLGLDDDKAAVEFDRLKVDL 90
Query: 77 IITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQ 136
IITDYCMPGMTGYELLKKIKES++ KEVPVVIMSSEN+ TRI++CLEEGA+ F+LKP+K
Sbjct: 91 IITDYCMPGMTGYELLKKIKESTSFKEVPVVIMSSENVMTRIDRCLEEGAEDFLLKPVKL 150
Query: 137 SDIKKLKCKLS 147
+D+K+L+ L+
Sbjct: 151 ADVKRLRSYLT 161
>At3g57040 responce reactor 4
Length = 234
Score = 168 bits (426), Expect = 1e-42
Identities = 85/134 (63%), Positives = 107/134 (79%), Gaps = 10/134 (7%)
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLT-SGEQNTLNGRSK----- 73
HVLAVDD+L DRKL+E+LL+ SSC+VTT ++G +ALEFLGL S + N N SK
Sbjct: 10 HVLAVDDSLFDRKLIERLLQKSSCQVTTVDSGSKALEFLGLRQSTDSNDPNAFSKAPVNH 69
Query: 74 ----VNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMF 129
VNLIITDYCMPGMTGY+LLKK+KESSA +++PVVIMSSEN+P RI++CLEEGA+ F
Sbjct: 70 QVVEVNLIITDYCMPGMTGYDLLKKVKESSAFRDIPVVIMSSENVPARISRCLEEGAEEF 129
Query: 130 ILKPLKQSDIKKLK 143
LKP++ +D+ KLK
Sbjct: 130 FLKPVRLADLNKLK 143
>At2g41310 putative two-component response regulator 3 protein
Length = 225
Score = 166 bits (420), Expect = 5e-42
Identities = 81/140 (57%), Positives = 109/140 (77%), Gaps = 8/140 (5%)
Query: 12 VMESGDIPHVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNT---- 67
VME+ HVLAVDD+L DRK++E+LL+ SSC+VTT ++G +ALEFLGL + +
Sbjct: 2 VMETESKFHVLAVDDSLFDRKMIERLLQKSSCQVTTVDSGSKALEFLGLRVDDNDPNALS 61
Query: 68 ----LNGRSKVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLE 123
++ ++NLIITDYCMPGMTGY+LLKK+KES+A + +PVVIMSSEN+P RI++CLE
Sbjct: 62 TSPQIHQEVEINLIITDYCMPGMTGYDLLKKVKESAAFRSIPVVIMSSENVPARISRCLE 121
Query: 124 EGAQMFILKPLKQSDIKKLK 143
EGA+ F LKP+K +D+ KLK
Sbjct: 122 EGAEEFFLKPVKLADLTKLK 141
>At1g19050 putative protein
Length = 206
Score = 165 bits (417), Expect = 1e-41
Identities = 80/124 (64%), Positives = 105/124 (84%)
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIIT 79
HVLAVDD+++DRK++E+LLR SSCKVTT E+G RAL++LGL G+ + KVNLI+T
Sbjct: 25 HVLAVDDSIVDRKVIERLLRISSCKVTTVESGTRALQYLGLDGGKGASNLKDLKVNLIVT 84
Query: 80 DYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDI 139
DY MPG++GY+LLKKIKESSA +EVPVVIMSSENI RI +CL+EGA+ F+LKP+K +D+
Sbjct: 85 DYSMPGLSGYDLLKKIKESSAFREVPVVIMSSENILPRIQECLKEGAEEFLLKPVKLADV 144
Query: 140 KKLK 143
K++K
Sbjct: 145 KRIK 148
>At5g62920 response regulator 6 (ARR6)
Length = 186
Score = 164 bits (415), Expect = 2e-41
Identities = 86/138 (62%), Positives = 106/138 (76%)
Query: 15 SGDIPHVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKV 74
S D HVLAVDD+ +DRK +E+LLR SSCKVT ++ RAL++LGL E++ KV
Sbjct: 21 SPDPLHVLAVDDSHVDRKFIERLLRVSSCKVTVVDSATRALQYLGLDVEEKSVGFEDLKV 80
Query: 75 NLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPL 134
NLI+TDY MPGMTGYELLKKIKESSA +EVPVVIMSSENI RI++CLEEGA+ F+LKP+
Sbjct: 81 NLIMTDYSMPGMTGYELLKKIKESSAFREVPVVIMSSENILPRIDRCLEEGAEDFLLKPV 140
Query: 135 KQSDIKKLKCKLSNCSSL 152
K SD+K+L+ L L
Sbjct: 141 KLSDVKRLRDSLMKVEDL 158
>At3g48100 response reactor 2 (ATRR2)
Length = 184
Score = 164 bits (414), Expect = 3e-41
Identities = 84/136 (61%), Positives = 109/136 (79%), Gaps = 3/136 (2%)
Query: 13 MESGDIPHVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRS 72
+ S + HVLAVDD+++DRK +E+LLR SSCKVT ++ RAL++LGL GE N+ G
Sbjct: 19 LASPKLLHVLAVDDSMVDRKFIERLLRVSSCKVTVVDSATRALQYLGL-DGENNSSVGFE 77
Query: 73 --KVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFI 130
K+NLI+TDY MPGMTGYELLKKIKESSA +E+PVVIMSSENI RI++CLEEGA+ F+
Sbjct: 78 DLKINLIMTDYSMPGMTGYELLKKIKESSAFREIPVVIMSSENILPRIDRCLEEGAEDFL 137
Query: 131 LKPLKQSDIKKLKCKL 146
LKP+K +D+K+L+ L
Sbjct: 138 LKPVKLADVKRLRDSL 153
>At1g74890 putative response regulator (two component
phosphotransfer signaling)
Length = 206
Score = 162 bits (410), Expect = 7e-41
Identities = 81/136 (59%), Positives = 107/136 (78%)
Query: 8 SSNWVMESGDIPHVLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNT 67
SS+ S HVLAVDD+ +DRK++E+LL+ S+CKVTT E+G RAL++LGL ++
Sbjct: 7 SSSLSSSSSPELHVLAVDDSFVDRKVIERLLKISACKVTTVESGTRALQYLGLDGDNGSS 66
Query: 68 LNGRSKVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQ 127
KVNLI+TDY MPG+TGYELLKKIKESSA++E+PVVIMSSENI RI +C+ EGA+
Sbjct: 67 GLKDLKVNLIVTDYSMPGLTGYELLKKIKESSALREIPVVIMSSENIQPRIEQCMIEGAE 126
Query: 128 MFILKPLKQSDIKKLK 143
F+LKP+K +D+K+LK
Sbjct: 127 EFLLKPVKLADVKRLK 142
>At5g02810 unknown protein
Length = 727
Score = 71.2 bits (173), Expect = 2e-13
Identities = 46/143 (32%), Positives = 80/143 (55%), Gaps = 15/143 (10%)
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL V+++ R +V LLRN S +V A NG +A + L LN +++++T+
Sbjct: 80 VLLVENDDCTRYIVTALLRNCSYEVVEASNGIQAWKVL-------EDLNNH--IDIVLTE 130
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
MP ++G LL KI + + +PV++MSS + + KCL +GA F++KP++++++K
Sbjct: 131 VIMPYLSGIGLLCKILNHKSRRNIPVIMMSSHDSMGLVFKCLSKGAVDFLVKPIRKNELK 190
Query: 141 KL------KCKLSNCSSLGVGMH 157
L +C+ S+ S G H
Sbjct: 191 ILWQHVWRRCQSSSGSGSESGTH 213
>At5g60100 pseudo-response regulator 3 (APRR3)
Length = 495
Score = 70.5 bits (171), Expect = 4e-13
Identities = 46/143 (32%), Positives = 77/143 (53%), Gaps = 15/143 (10%)
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL V+++ R +V LL+N S +VT P LE + E +S ++L++T+
Sbjct: 66 VLLVENDDSTRHIVTALLKNCSYEVTAV---PDVLEAWRILEDE------KSCIDLVLTE 116
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
MP +G LL KI +K +PV++MSS + + KCL GA F++KP++++++K
Sbjct: 117 VDMPVHSGTGLLSKIMSHKTLKNIPVIMMSSHDSMVLVFKCLSNGAVDFLVKPIRKNELK 176
Query: 141 KL------KCKLSNCSSLGVGMH 157
L +C S+ S G+H
Sbjct: 177 NLWQHVWRRCHSSSGSGSESGIH 199
>At5g24470 pseudo-response regulator 5 (APRR5)
Length = 667
Score = 68.2 bits (165), Expect = 2e-12
Identities = 38/122 (31%), Positives = 72/122 (58%), Gaps = 9/122 (7%)
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL V+ + R+++ LLR S +V +G +A E L G+ + V+LI+T+
Sbjct: 161 VLLVEADDSTRQIIAALLRKCSYRVAAVPDGLKAWEML---KGKPES------VDLILTE 211
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
+P ++GY LL I E K +PV++MS+++ + KC+ +GA +++KPL++++++
Sbjct: 212 VDLPSISGYALLTLIMEHDICKNIPVIMMSTQDSVNTVYKCMLKGAADYLVKPLRRNELR 271
Query: 141 KL 142
L
Sbjct: 272 NL 273
>At2g25180 putative two-component response regulator protein
Length = 573
Score = 65.9 bits (159), Expect = 9e-12
Identities = 39/122 (31%), Positives = 69/122 (55%), Gaps = 12/122 (9%)
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VLAVDD+ K++E LLR+ VTT +ALE L ++K +L+I+D
Sbjct: 19 VLAVDDDQTCLKILESLLRHCQYHVTTTNQAQKALELL---------RENKNKFDLVISD 69
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
MP M G++LL+ + ++PV+++S+ + P + K + GA ++LKP++ ++K
Sbjct: 70 VDMPDMDGFKLLELV---GLEMDLPVIMLSAHSDPKYVMKGVTHGACDYLLKPVRIEELK 126
Query: 141 KL 142
+
Sbjct: 127 NI 128
>At2g07440 putative two-component response regulator protein
Length = 136
Score = 62.0 bits (149), Expect = 1e-10
Identities = 29/47 (61%), Positives = 38/47 (80%)
Query: 97 ESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIKKLK 143
ES+A + + VVIMSSEN+PTRI++CLE+GA+ F LKP+K D KLK
Sbjct: 9 ESAAFRSIHVVIMSSENVPTRISRCLEKGAEEFFLKPVKLVDHTKLK 55
>At4g31920 predicted protein
Length = 552
Score = 60.8 bits (146), Expect = 3e-10
Identities = 36/122 (29%), Positives = 68/122 (55%), Gaps = 12/122 (9%)
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VLAVDD+ ++++ LL+ VTT ALE L ++K +L+I+D
Sbjct: 19 VLAVDDDQTCLRILQTLLQRCQYHVTTTNQAQTALELLR---------ENKNKFDLVISD 69
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
MP M G++LL+ + ++PV+++S+ + P + K ++ GA ++LKP++ ++K
Sbjct: 70 VDMPDMDGFKLLELV---GLEMDLPVIMLSAHSDPKYVMKGVKHGACDYLLKPVRIEELK 126
Query: 141 KL 142
+
Sbjct: 127 NI 128
>At2g46790 hypothetical protein
Length = 454
Score = 60.1 bits (144), Expect = 5e-10
Identities = 36/125 (28%), Positives = 70/125 (55%), Gaps = 5/125 (4%)
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSG--EQNTLNGRS-KVNLI 77
VL V+ + R+++ LLR K P F+ ++ G L +S ++LI
Sbjct: 39 VLLVESDYSTRQIITALLRKCCYKGRLVF--PFISLFVAVSDGLAAWEVLKEKSHNIDLI 96
Query: 78 ITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQS 137
+T+ +P ++G+ LL + E A K +PV++MSS++ + KC+ GA +++KP++++
Sbjct: 97 LTELDLPSISGFALLALVMEHEACKNIPVIMMSSQDSIKMVLKCMLRGAADYLIKPMRKN 156
Query: 138 DIKKL 142
++K L
Sbjct: 157 ELKNL 161
>At1g67710 response regulator 11 (ARR11)
Length = 521
Score = 58.5 bits (140), Expect = 1e-09
Identities = 36/120 (30%), Positives = 66/120 (55%), Gaps = 12/120 (10%)
Query: 21 VLAVDDNLIDRKLVEKLLRNSSCKVTTAENGPRALEFLGLTSGEQNTLNGRSKVNLIITD 80
VL VDD+ K++EK+L+ S +VTT AL L + +++I+D
Sbjct: 13 VLVVDDDPTWLKILEKMLKKCSYEVTTCGLAREALRLLR---------ERKDGYDIVISD 63
Query: 81 YCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQSDIK 140
MP M G++LL+ + ++PV++MS + +R+ K ++ GA ++LKP++ ++K
Sbjct: 64 VNMPDMDGFKLLEHV---GLELDLPVIMMSVDGETSRVMKGVQHGACDYLLKPIRMKELK 120
>At2g27070 putative two-component response regulator protein
Length = 575
Score = 57.4 bits (137), Expect = 3e-09
Identities = 41/126 (32%), Positives = 65/126 (51%), Gaps = 16/126 (12%)
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCK---VTTAENGPRALEFLGLTSGEQNTLNGRSKVNL 76
+V+ VDDN + + ++L S + V + +AL L N R ++L
Sbjct: 17 NVMVVDDNRVFLDIWSRMLEKSKYREITVIAVDYPKKALSTLK---------NQRDNIDL 67
Query: 77 IITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFILKPLKQ 136
IITDY MPGM G +L K+I + + V++MSS+ P + + L GA FI KP+
Sbjct: 68 IITDYYMPGMNGLQLKKQITQE--FGNLSVLVMSSD--PNKEEESLSCGAMGFIPKPIAP 123
Query: 137 SDIKKL 142
+D+ K+
Sbjct: 124 TDLPKI 129
>At5g07210 putative protein
Length = 621
Score = 55.8 bits (133), Expect = 1e-08
Identities = 39/131 (29%), Positives = 69/131 (51%), Gaps = 21/131 (16%)
Query: 20 HVLAVDDNLIDRKLVEKLLRNSSCK--------VTTAENGPRALEFLGLTSGEQNTLNGR 71
+V+ VDD+ + ++ ++L++S + V ++ +AL L + R
Sbjct: 17 NVMVVDDDHVFLDIMSRMLQHSKYRDPSVMEIAVIAVDDPKKALSTLKIQ---------R 67
Query: 72 SKVNLIITDYCMPGMTGYELLKKIKESSAMKEVPVVIMSSENIPTRINKCLEEGAQMFIL 131
++LIITDY MPGM G +L K+I + +PV++MSS+ + + L GA FI
Sbjct: 68 DNIDLIITDYYMPGMNGLQLKKQITQE--FGNLPVLVMSSDT--NKEEESLSCGAMGFIP 123
Query: 132 KPLKQSDIKKL 142
KP+ +D+ K+
Sbjct: 124 KPIHPTDLTKI 134
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,416,477
Number of Sequences: 26719
Number of extensions: 126125
Number of successful extensions: 501
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 433
Number of HSP's gapped (non-prelim): 54
length of query: 161
length of database: 11,318,596
effective HSP length: 91
effective length of query: 70
effective length of database: 8,887,167
effective search space: 622101690
effective search space used: 622101690
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0320b.4


