

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.7
(448 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
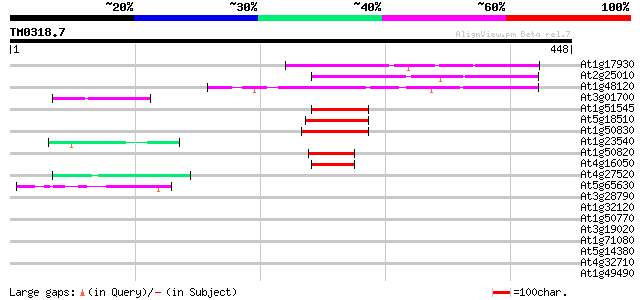
Score E
Sequences producing significant alignments: (bits) Value
At1g17930 unknown protein 104 9e-23
At2g25010 unknown protein 95 7e-20
At1g48120 serine/threonine phosphatase PP7, putative 83 4e-16
At3g01700 arabinogalactan-protein AGP11 47 2e-05
At1g51545 unknown protein 47 2e-05
At5g18510 putative protein 47 3e-05
At1g50830 hypothetical protein 45 8e-05
At1g23540 putative serine/threonine protein kinase 44 2e-04
At1g50820 hypothetical protein 43 3e-04
At4g16050 hypothetical protein 42 5e-04
At4g27520 early nodulin-like 2 predicted GPI-anchored protein 42 7e-04
At5g65630 unknown protein 42 9e-04
At3g28790 unknown protein 41 0.001
At1g32120 hypothetical protein 40 0.002
At1g50770 hypothetical protein 40 0.003
At3g19020 hypothetical protein 40 0.003
At1g71080 unknown protein 40 0.003
At5g14380 agp6 39 0.006
At4g32710 putative protein kinase 39 0.006
At1g49490 hypothetical protein 39 0.008
>At1g17930 unknown protein
Length = 478
Score = 104 bits (260), Expect = 9e-23
Identities = 69/205 (33%), Positives = 110/205 (53%), Gaps = 8/205 (3%)
Query: 221 LIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMG 280
L+E+ G + LI ALVERW ET++FH P GEMTITLD+VS +L L +
Sbjct: 45 LVEKAGFGWFRLVGSISLNNSLISALVERWRRETNTFHFPCGEMTITLDEVSLILGLAVD 104
Query: 281 SRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNR--SQTIRFGVLQTRYEAALAEHR 338
+ + + ++ + +C +L+G + + E NR ++ ++ + A + E
Sbjct: 105 GKPVVGVKEKDEDPSQVCLRLLG---KLPKGELSGNRVTAKWLKESFAECPKGATMKEIE 161
Query: 339 YEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQ 398
Y R +L+ +G+T+FA+ + Y I + ED + EYAWGA ALA LY Q+
Sbjct: 162 YH--TRAYLIYIVGSTIFATTDPSKISVDYLI-LFEDFEKAGEYAWGAAALAFLYRQIGN 218
Query: 399 ASRRGTAQMGGFTSLLLGWVYEYLS 423
AS+R + +GG +LL W Y +L+
Sbjct: 219 ASQRSQSIIGGCLTLLQCWSYFHLN 243
>At2g25010 unknown protein
Length = 509
Score = 95.1 bits (235), Expect = 7e-20
Identities = 59/184 (32%), Positives = 96/184 (52%), Gaps = 7/184 (3%)
Query: 242 LILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERDECAALCAQL 301
LI ALVERW ET++FH+P GEMTITLD+V+ +L L + + + +C +L
Sbjct: 75 LISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVGDEVAMDMCGRL 134
Query: 302 MGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDA---ARIWLVNQLGATLFAS 358
+G E + +R ++ L+ + + ++ R +L+ +G+T+FA+
Sbjct: 135 LGKLPSAANKEVNCSR---VKLNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTIFAT 191
Query: 359 KSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRRGTAQMGGFTSLLLGWV 418
G +V ++ + ED + YAWGA ALA LY L AS + + + G +LL W
Sbjct: 192 TDGD-KVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCWS 250
Query: 419 YEYL 422
Y +L
Sbjct: 251 YFHL 254
>At1g48120 serine/threonine phosphatase PP7, putative
Length = 1338
Score = 82.8 bits (203), Expect = 4e-16
Identities = 74/269 (27%), Positives = 122/269 (44%), Gaps = 28/269 (10%)
Query: 159 GPVELSLLTYYADHKAPWAWHALLRTDERYVDRRQL--KVATAGGKVWNLAYDGDSDSHR 216
GPV+ S+L + +H++ W E V R+L + G + W L
Sbjct: 14 GPVDQSILVWQHEHRSAAIW-------EDEVPPRELTCRHKLLGMRDWPL--------DP 58
Query: 217 RVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLH 276
V + + + GL+ + ++ D LI ALVERW ET +FH+P GE+T+TL DV+ LL
Sbjct: 59 LVCQKLIEFGLYGVYKVAFIQLDYALITALVERWRPETHTFHLPAGEITVTLQDVNILLG 118
Query: 277 LPMGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALA- 335
L + T + A LC L+G G + + L+ + A
Sbjct: 119 LRVDGPAVT--GSTKYNWADLCEDLLGHRPGPKDL-----HGSHVSLAWLRENFRNLPAD 171
Query: 336 --EHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIALATLY 393
E + R +++ + L+ KS + + ++ +L D V + +WG+ LA LY
Sbjct: 172 PDEVTLKCHTRAFVLALMSGFLYGDKS-KHDVALTFLPLLRDFDEVAKLSWGSATLALLY 230
Query: 394 DQLSQASRRGTAQMGGFTSLLLGWVYEYL 422
+L +AS+R + + G LL W +E L
Sbjct: 231 RELCRASKRTVSTICGPLVLLQLWAWERL 259
>At3g01700 arabinogalactan-protein AGP11
Length = 136
Score = 47.0 bits (110), Expect = 2e-05
Identities = 26/78 (33%), Positives = 41/78 (52%), Gaps = 2/78 (2%)
Query: 35 ATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSG 94
+++ A +AS+PV P+P + PS + E PTV SPP+P E+ G S G S G
Sbjct: 46 SSAAAPKASSPVAEEPTPEDDYSAAS--PSDSAEAPTVSSPPAPTPEADGPSSDGPSSDG 103
Query: 95 EESSGEESSGEESSGEAS 112
++ SG ++ + S
Sbjct: 104 PAAAESPKSGATTNVKLS 121
>At1g51545 unknown protein
Length = 629
Score = 47.0 bits (110), Expect = 2e-05
Identities = 25/46 (54%), Positives = 31/46 (67%), Gaps = 1/46 (2%)
Query: 242 LILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPM-GSRFYTP 286
L+LALVE+W ET SF P+GE TITL+DV LL + GS + P
Sbjct: 31 LLLALVEKWCPETKSFLFPWGEATITLEDVLVLLGFSVQGSPVFAP 76
>At5g18510 putative protein
Length = 702
Score = 46.6 bits (109), Expect = 3e-05
Identities = 23/51 (45%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query: 237 VTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLP-MGSRFYTP 286
+ D IL++ E+W ET SF P+GE TITL+DV LL +GS ++P
Sbjct: 92 IKDTSSILSIAEKWCSETKSFIFPWGEATITLEDVMVLLGFSVLGSPVFSP 142
>At1g50830 hypothetical protein
Length = 768
Score = 45.1 bits (105), Expect = 8e-05
Identities = 25/55 (45%), Positives = 35/55 (63%), Gaps = 2/55 (3%)
Query: 234 SYPVT-DAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLP-MGSRFYTP 286
+Y +T + LIL++ E+W ET SF P+GE TITL+DV LL +GS + P
Sbjct: 110 TYSITKNPSLILSVSEKWCPETKSFVFPWGEATITLEDVMVLLGFSVLGSPVFAP 164
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 43.5 bits (101), Expect = 2e-04
Identities = 34/107 (31%), Positives = 40/107 (36%), Gaps = 19/107 (17%)
Query: 32 DHAATSQAVEASAPVVS---PPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESS 88
D ++T E S P PP P I PL D PP P+ P V S PSP +S E S
Sbjct: 45 DSSSTPPLSEPSTPPPDSQLPPLPSILPPLTDSPPPPSDSSPPVDSTPSPPPPTSNESPS 104
Query: 89 GEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
E S E D+ PPP+ D PP
Sbjct: 105 PPEDS----------------ETPPAPPNESNDNNPPPSQDLQSPPP 135
Score = 35.4 bits (80), Expect = 0.064
Identities = 34/120 (28%), Positives = 42/120 (34%), Gaps = 8/120 (6%)
Query: 44 APVVSPPSPMIEVPLVDYPPSPTVEIPTV-VSPPSPMVESSGEESSGEESSGEESSGEES 102
+P SPP+P + PS +P V SPPSP +SS E S+ S
Sbjct: 7 SPSSSPPAPPADTAPPPETPSENSALPPVDSSPPSPPADSSSTPPLSEPSTPPPDSQLPP 66
Query: 103 SGEESSGEASSGMGGSDED-------SIPPPTVDDDVLPPEQGEQGAQGGEEDLIQRLPP 155
S SD S PPPT ++ PPE E E PP
Sbjct: 67 LPSILPPLTDSPPPPSDSSPPVDSTPSPPPPTSNESPSPPEDSETPPAPPNESNDNNPPP 126
Score = 29.3 bits (64), Expect = 4.6
Identities = 28/119 (23%), Positives = 43/119 (35%), Gaps = 24/119 (20%)
Query: 41 EASAPVVSPPSPMI-----EVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGE 95
+ +P S PSP + E P + PP+P PT P SP+ ++
Sbjct: 129 DLQSPPPSSPSPNVGPTNPESPPLQSPPAPPASDPTNSPPASPLDPTNPPPI-------- 180
Query: 96 ESSGEESSGEESSGEASSGMGGSDEDSIPP------PTVDDDVLPPEQGEQGAQGGEED 148
+ SG +S A+ S ++PP P V + P +G G D
Sbjct: 181 -----QPSGPATSPPANPNAPPSPFPTVPPKTPSSGPVVSPSLTSPSKGTPTPNQGNGD 234
>At1g50820 hypothetical protein
Length = 528
Score = 43.1 bits (100), Expect = 3e-04
Identities = 19/37 (51%), Positives = 25/37 (67%)
Query: 239 DAGLILALVERWHEETSSFHMPFGEMTITLDDVSALL 275
D L+L L E+W +T +F P+GE TITL+DV LL
Sbjct: 94 DTDLVLGLAEKWCPDTKTFIFPWGEATITLEDVMVLL 130
>At4g16050 hypothetical protein
Length = 900
Score = 42.4 bits (98), Expect = 5e-04
Identities = 19/34 (55%), Positives = 26/34 (75%)
Query: 242 LILALVERWHEETSSFHMPFGEMTITLDDVSALL 275
LIL+L + W ET++F P+GE TITL+DV+ LL
Sbjct: 363 LILSLAQNWCPETNTFVFPWGEATITLEDVNVLL 396
>At4g27520 early nodulin-like 2 predicted GPI-anchored protein
Length = 349
Score = 42.0 bits (97), Expect = 7e-04
Identities = 32/111 (28%), Positives = 45/111 (39%), Gaps = 5/111 (4%)
Query: 35 ATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMV-ESSGEESSGEESS 93
+TS +SAP+ SPP+PM PPS + SPP M +SS S+ S
Sbjct: 223 STSPVSPSSAPMTSPPAPMAPKSSSTIPPSSA----PMTSPPGSMAPKSSSPVSNSPTVS 278
Query: 94 GEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGEQGAQG 144
+ G +S S + S MG S + + P Q + A G
Sbjct: 279 PSLAPGGSTSSSPSDSPSGSAMGPSGDGPSAAGDISTPAGAPGQKKSSANG 329
Score = 28.5 bits (62), Expect = 7.9
Identities = 31/107 (28%), Positives = 41/107 (37%), Gaps = 13/107 (12%)
Query: 44 APVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGE---ESSGEESSGEESSGE 100
+P SPP+P P P SP+ + SPP+PM S SS +S S
Sbjct: 212 SPTTSPPAP----PKSTSPVSPS--SAPMTSPPAPMAPKSSSTIPPSSAPMTSPPGSMAP 265
Query: 101 ESSGEESSG---EASSGMGGSDEDSIPPPTVDDDVLPPEQGEQGAQG 144
+SS S+ S GGS S P + + P A G
Sbjct: 266 KSSSPVSNSPTVSPSLAPGGSTSSS-PSDSPSGSAMGPSGDGPSAAG 311
>At5g65630 unknown protein
Length = 590
Score = 41.6 bits (96), Expect = 9e-04
Identities = 39/130 (30%), Positives = 54/130 (41%), Gaps = 39/130 (30%)
Query: 6 RSHASSEDVGATEDRHRRLHASSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVDYPPSP 65
R+ AS ++G+ E R +RRGD A + V+ ++P+ DYP
Sbjct: 488 RNMASVAEMGSAEKR-------TRRGD--AGEEDVDIGE----------DIPIEDYP--- 525
Query: 66 TVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGG------SD 119
S +E G + SSG SSG SS SS + SG GG SD
Sbjct: 526 -----------SVEIERDGTAVAAAASSGSSSSGSSSSSGGSSSSSDSGSGGSSSGSDSD 574
Query: 120 EDSIPPPTVD 129
DS+ P V+
Sbjct: 575 ADSVQSPFVE 584
Score = 28.9 bits (63), Expect = 6.0
Identities = 33/118 (27%), Positives = 48/118 (39%), Gaps = 13/118 (11%)
Query: 38 QAVEAS---APVVSPPSPMIEVPLVD-YPPSPTVEIPTVVSPPSPMVESSGEESSGEESS 93
Q VE S +P PP P+I+ L PP P +EI V +PP S G + +
Sbjct: 337 QLVEPSRVQSPSPPPPPPVIQPELPQPQPPPPQLEI-EVEAPPDVSEVSKGRKGKLPKPK 395
Query: 94 GEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDD--DVLPPEQGEQGAQGGEEDL 149
++ + + EE S GM D +PP + +L G G E +L
Sbjct: 396 AKDPNKRLMTMEEKS---KLGMNLQD---LPPEKLGQLLQILRKRNGHLAQDGDEIEL 447
>At3g28790 unknown protein
Length = 608
Score = 41.2 bits (95), Expect = 0.001
Identities = 38/162 (23%), Positives = 59/162 (35%), Gaps = 19/162 (11%)
Query: 3 NRKRSHASSEDV-GATEDRHRRLHASSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVDY 61
++ S A +++V G + + S G + S S P S P+P P
Sbjct: 243 SKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPSTPTPSTPTPSTPT 302
Query: 62 PPSPTVEIPTVVSPPSPMVESSGEESSG--EESSGEESSGEESSG---------EESSGE 110
P +PT P +P + G ES+ +ES+ + S +SG + SSG+
Sbjct: 303 PSTPTPSTPAPSTPAAGKTSEKGSESASMKKESNSKSESESAASGSVSKTKETNKGSSGD 362
Query: 111 -------ASSGMGGSDEDSIPPPTVDDDVLPPEQGEQGAQGG 145
SSG P P+ D +G A G
Sbjct: 363 TYKDTTGTSSGSPSGSPSGSPTPSTSTDGKASSKGSASASAG 404
>At1g32120 hypothetical protein
Length = 1206
Score = 40.4 bits (93), Expect = 0.002
Identities = 18/33 (54%), Positives = 26/33 (78%)
Query: 242 LILALVERWHEETSSFHMPFGEMTITLDDVSAL 274
LI+ALVE+W ET++F P+GE T+TL+D+ L
Sbjct: 121 LIVALVEKWCIETNTFVFPWGEATLTLEDMIVL 153
>At1g50770 hypothetical protein
Length = 632
Score = 40.0 bits (92), Expect = 0.003
Identities = 17/34 (50%), Positives = 24/34 (70%)
Query: 242 LILALVERWHEETSSFHMPFGEMTITLDDVSALL 275
L+L + E+W +T +F P+GE TITL+DV LL
Sbjct: 100 LVLGIAEKWCPDTKTFVFPWGETTITLEDVMLLL 133
>At3g19020 hypothetical protein
Length = 951
Score = 39.7 bits (91), Expect = 0.003
Identities = 27/105 (25%), Positives = 43/105 (40%), Gaps = 14/105 (13%)
Query: 44 APVVSPPSPMIEVPLVDYPPSPTVEIPTVVSP----PSPMVESSGEESSGEESSGEESSG 99
+P+ SPP P+ PP P +P SP P+P SGE S+ ++ +S
Sbjct: 801 SPIYSPPPPVFSP-----PPKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSED 855
Query: 100 EESSGEESSGEASSGMGGSDEDS-----IPPPTVDDDVLPPEQGE 139
E+ + + DS P P+ + +V P+Q E
Sbjct: 856 IEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEPEVEAPKQSE 900
Score = 33.9 bits (76), Expect = 0.19
Identities = 38/139 (27%), Positives = 53/139 (37%), Gaps = 29/139 (20%)
Query: 26 ASSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPS-PMVESSG 84
+SS G+ + QA + + PS P+ P+P SP S P VE+
Sbjct: 835 SSSESGEISTPVQAPTPDSEDIEAPSDSNHSPVFKSSPAP--------SPDSEPEVEAPV 886
Query: 85 EESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDD-----VLPPEQGE 139
S E + ++S SS SS + D PP+ D+D +LPP G
Sbjct: 887 PSSEPEVEAPKQSEATPSSSPPSSNPS--------PDVTAPPSEDNDDGDNFILPPNIGH 938
Query: 140 QGAQGGEEDLIQRLPPFPG 158
Q A P FPG
Sbjct: 939 QYASPPP-------PMFPG 950
Score = 32.0 bits (71), Expect = 0.71
Identities = 18/38 (47%), Positives = 19/38 (49%), Gaps = 3/38 (7%)
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEI---PTVVSPPSPM 79
PV SPP PM P Y P P V P V SPP P+
Sbjct: 654 PVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPV 691
Score = 30.0 bits (66), Expect = 2.7
Identities = 27/114 (23%), Positives = 42/114 (36%), Gaps = 10/114 (8%)
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEES-S 103
P P I P ++ P P E PSP E+ E S + ES +ES
Sbjct: 410 PKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPESPKQESPK 469
Query: 104 GEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGEQGAQGGEEDLIQRLPPFP 157
E+ + S S + P PEQ + + +++ ++ PP P
Sbjct: 470 TEQPKPKPESPKQESPKQEAP---------KPEQPKPKPESPKQESSKQEPPKP 514
Score = 29.6 bits (65), Expect = 3.5
Identities = 18/39 (46%), Positives = 19/39 (48%), Gaps = 4/39 (10%)
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVV-SPPSPMVES 82
PV SPP P P+ PP P P V SPP P V S
Sbjct: 733 PVFSPPPP---APIYSPPPPPVHSPPPPVHSPPPPPVHS 768
Score = 29.6 bits (65), Expect = 3.5
Identities = 25/93 (26%), Positives = 30/93 (31%), Gaps = 4/93 (4%)
Query: 49 PPSPMIEVPLVDYPP----SPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSG 104
P SP E P + P SP E P +P + E E S E EES
Sbjct: 460 PESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPK 519
Query: 105 EESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQ 137
E S + P P PP+Q
Sbjct: 520 PEPPKPEESPKPQPPKQETPKPEESPKPQPPKQ 552
Score = 28.9 bits (63), Expect = 6.0
Identities = 16/42 (38%), Positives = 19/42 (45%), Gaps = 3/42 (7%)
Query: 44 APVVSPPSPMIEVP---LVDYPPSPTVEIPTVVSPPSPMVES 82
AP+ SPP P + P + PP P P V P P V S
Sbjct: 741 APIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHS 782
Score = 28.9 bits (63), Expect = 6.0
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 3/37 (8%)
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEI---PTVVSPPSP 78
PV SPP P+ P + P P V+ P V SPP P
Sbjct: 704 PVHSPPPPVHSPPPPVHSPPPPVQSPPPPPVFSPPPP 740
Score = 28.9 bits (63), Expect = 6.0
Identities = 18/40 (45%), Positives = 20/40 (50%), Gaps = 2/40 (5%)
Query: 45 PVVSPPSPMIEVPL-VDYPPSPTVEIPTVV-SPPSPMVES 82
PV SPP P+ P + PP P P V SPP P V S
Sbjct: 647 PVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHS 686
Score = 28.9 bits (63), Expect = 6.0
Identities = 18/52 (34%), Positives = 20/52 (37%), Gaps = 2/52 (3%)
Query: 33 HAATSQAVEASAPVVSPPSPMIEVPL--VDYPPSPTVEIPTVVSPPSPMVES 82
H+ PV SPP P + P V PP P P V P P V S
Sbjct: 663 HSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHS 714
Score = 28.5 bits (62), Expect = 7.9
Identities = 17/39 (43%), Positives = 19/39 (48%), Gaps = 1/39 (2%)
Query: 45 PVVSPPSPMIEVPL-VDYPPSPTVEIPTVVSPPSPMVES 82
PV SPP P+ P V PP P P V P P V+S
Sbjct: 690 PVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQS 728
Score = 28.5 bits (62), Expect = 7.9
Identities = 17/40 (42%), Positives = 18/40 (44%), Gaps = 2/40 (5%)
Query: 45 PVVSPPSPMIEVPL--VDYPPSPTVEIPTVVSPPSPMVES 82
PV SPP P + P V PP P P V P P V S
Sbjct: 757 PVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHS 796
>At1g71080 unknown protein
Length = 326
Score = 39.7 bits (91), Expect = 0.003
Identities = 21/43 (48%), Positives = 26/43 (59%), Gaps = 3/43 (6%)
Query: 82 SSGEESSGEESSGEE---SSGEESSGEESSGEASSGMGGSDED 121
SS SSG +S G SSG S G+ SSG +S G GGSD++
Sbjct: 278 SSSGSSSGSDSDGRSKSVSSGSGSGGQSSSGSSSRGSGGSDDE 320
>At5g14380 agp6
Length = 150
Score = 38.9 bits (89), Expect = 0.006
Identities = 24/67 (35%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Query: 34 AATSQAVEASAPVVSPPSPMIEVPLVDYP---PSPTVEIPTVVSPPSPMVESSGEESSGE 90
AA ++ AS+P S P+ VP DY PS + E PTV SPP+P +S+
Sbjct: 57 AAAPKSSSASSPKASSPAAEGPVPEDDYSASSPSDSAEAPTVSSPPAPTPDSTSAADGPS 116
Query: 91 ESSGEES 97
+ ES
Sbjct: 117 DGPTAES 123
Score = 31.2 bits (69), Expect = 1.2
Identities = 22/62 (35%), Positives = 26/62 (41%), Gaps = 6/62 (9%)
Query: 27 SSRRGDHAATSQAVEASAPV----VSPPSPMIEVPLVDYPPSPTVEIPTVVSPPS--PMV 80
SS A+S A E P S PS E P V PP+PT + + PS P
Sbjct: 62 SSSASSPKASSPAAEGPVPEDDYSASSPSDSAEAPTVSSPPAPTPDSTSAADGPSDGPTA 121
Query: 81 ES 82
ES
Sbjct: 122 ES 123
>At4g32710 putative protein kinase
Length = 731
Score = 38.9 bits (89), Expect = 0.006
Identities = 22/41 (53%), Positives = 26/41 (62%), Gaps = 6/41 (14%)
Query: 45 PVVSPPSPMIE---VPLVDYPPSPTVEIPTVVSPPSPMVES 82
PV SPPSP IE PL++ PP P +E P SPPSP V +
Sbjct: 67 PVESPPSPPIESPPPPLLESPPPPPLESP---SPPSPHVSA 104
Score = 32.7 bits (73), Expect = 0.42
Identities = 22/49 (44%), Positives = 26/49 (52%), Gaps = 10/49 (20%)
Query: 43 SAPVVSPPSPMIEVPL-VDYPPSPTVEIPT---VVSPPSPMVESSGEES 87
SAP SPP PL V+ PPSP +E P + SPP P +ES S
Sbjct: 57 SAPTASPP------PLPVESPPSPPIESPPPPLLESPPPPPLESPSPPS 99
Score = 30.4 bits (67), Expect = 2.1
Identities = 15/42 (35%), Positives = 21/42 (49%), Gaps = 2/42 (4%)
Query: 43 SAPVVSPPSPMIEVPLVDYPPSPTVEIPT--VVSPPSPMVES 82
S+P P P + P PP P P+ + SPP P++ES
Sbjct: 45 SSPPPLPSPPPLSAPTASPPPLPVESPPSPPIESPPPPLLES 86
Score = 30.0 bits (66), Expect = 2.7
Identities = 22/69 (31%), Positives = 26/69 (36%), Gaps = 3/69 (4%)
Query: 43 SAPVVSPPSPMIEV---PLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSG 99
SAP SPP P + P PPS TV +SPP + S S S
Sbjct: 103 SAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISPPPRSLPSESTPPVNTASPPPPSPP 162
Query: 100 EESSGEESS 108
SG + S
Sbjct: 163 RRRSGPKPS 171
>At1g49490 hypothetical protein
Length = 847
Score = 38.5 bits (88), Expect = 0.008
Identities = 42/135 (31%), Positives = 53/135 (39%), Gaps = 22/135 (16%)
Query: 27 SSRRGDHAATSQAVEASAPVVSPPSP----MIEVPLVDYPPSPT---------VEIPTVV 73
SS A V+A PV +P + + P + SP+ V PT
Sbjct: 706 SSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPN 765
Query: 74 SPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDD-- 131
S P S E S E S E + E + SS +SS S + SIPPP +DD
Sbjct: 766 SKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPSSVPSSSP---STDTSIPPPENNDDDD 822
Query: 132 ----VLPPEQGEQGA 142
VLPP G Q A
Sbjct: 823 DGDFVLPPHIGFQYA 837
Score = 33.5 bits (75), Expect = 0.25
Identities = 22/91 (24%), Positives = 32/91 (34%), Gaps = 4/91 (4%)
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSG 104
PV SPP P P P + PT P+P S E + S E + S
Sbjct: 628 PVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTP----SSETTQVPTPSSESDQSQILSP 683
Query: 105 EESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
++ S S+ +P P+ + P
Sbjct: 684 VQAPTPVQSSTPSSEPTQVPTPSSSESYQAP 714
Score = 31.6 bits (70), Expect = 0.93
Identities = 20/82 (24%), Positives = 31/82 (37%), Gaps = 3/82 (3%)
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSG 104
PV SPP P P+ PP P V SPP P ++ + G + + +
Sbjct: 605 PVFSPPPPS---PVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTP 661
Query: 105 EESSGEASSGMGGSDEDSIPPP 126
+ + + SD+ I P
Sbjct: 662 SSETTQVPTPSSESDQSQILSP 683
Score = 30.4 bits (67), Expect = 2.1
Identities = 16/35 (45%), Positives = 17/35 (47%), Gaps = 1/35 (2%)
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVE-IPTVVSPPSP 78
PV SPP P P+ PP P P V SPP P
Sbjct: 578 PVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPP 612
Score = 29.3 bits (64), Expect = 4.6
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 2/37 (5%)
Query: 45 PVVSPPSPMIEVPLVD--YPPSPTVEIPTVVSPPSPM 79
PV SPP P+ P Y P P V P SPP P+
Sbjct: 555 PVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPV 591
Score = 29.3 bits (64), Expect = 4.6
Identities = 29/130 (22%), Positives = 42/130 (32%), Gaps = 19/130 (14%)
Query: 50 PSPMIEVPLVDYP----PSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEES--- 102
PSP + P + P P E+P P P + E+ EES E
Sbjct: 431 PSPKPQPPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEESPKPEQPQI 490
Query: 103 -SGEESSGEASSGMGGSDED-----------SIPPPTVDDDVLPPEQGEQGAQGGEEDLI 150
+ + G + +D S PPP V+D +PP Q + +
Sbjct: 491 PEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPSPIY 550
Query: 151 QRLPPFPGGP 160
PP P
Sbjct: 551 SPPPPVHSPP 560
Score = 29.3 bits (64), Expect = 4.6
Identities = 35/141 (24%), Positives = 50/141 (34%), Gaps = 21/141 (14%)
Query: 31 GDHAATSQAVEASAPVVSPPSPMI---------EVPLVDYPPSPTVEIPTVVSPPSPMVE 81
G + + S P S P P++ E P PSP + P SP E
Sbjct: 390 GGSSPSPNPPRTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPSPKPQPPKHESPKPEEPE 449
Query: 82 SSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGEQG 141
+ E +ES + S E S + + ++ IP PT V PP +
Sbjct: 450 NKHELPKQKESPKPQPSKPEDSPKPEQPKPEES-PKPEQPQIPEPT--KPVSPPNE---- 502
Query: 142 AQGGEEDLIQRLPPFPGGPVE 162
AQG D P+ PV+
Sbjct: 503 AQGPTPD-----DPYDASPVK 518
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,757,874
Number of Sequences: 26719
Number of extensions: 509806
Number of successful extensions: 3861
Number of sequences better than 10.0: 254
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 168
Number of HSP's that attempted gapping in prelim test: 2675
Number of HSP's gapped (non-prelim): 893
length of query: 448
length of database: 11,318,596
effective HSP length: 102
effective length of query: 346
effective length of database: 8,593,258
effective search space: 2973267268
effective search space used: 2973267268
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0318.7


