

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.6
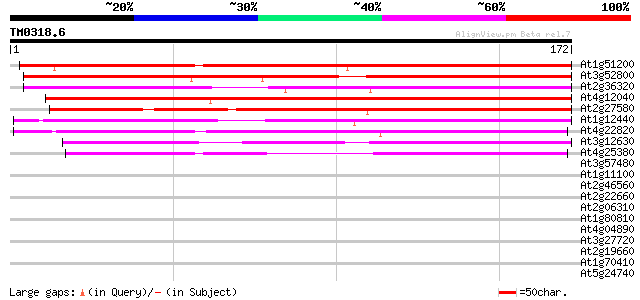
(172 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g51200 unknown protein 191 1e-49
At3g52800 zinc finger like protein 152 1e-37
At2g36320 unknown protein 142 9e-35
At4g12040 unknown protein 141 2e-34
At2g27580 unknown protein 140 3e-34
At1g12440 unknown protein 140 4e-34
At4g22820 predicted protein 138 1e-33
At3g12630 unknown protein 121 2e-28
At4g25380 putative protein 94 3e-20
At3g57480 unknown protein 31 0.29
At1g11100 transcription factor RUSH-1alpha isolog 30 0.49
At2g46560 hypothetical protein 30 0.65
At2g22660 unknown protein 29 1.4
At2g06310 putative Athila retroelement ORF1 protein 29 1.4
At1g80810 unknown protein 28 3.2
At4g04890 putative homeotic protein 27 4.2
At3g27720 hypothetical protein 27 4.2
At2g19660 unknown protein 27 5.5
At1g70410 unknown protein 27 5.5
At5g24740 VPS13 - like protein 27 7.1
>At1g51200 unknown protein
Length = 173
Score = 191 bits (486), Expect = 1e-49
Identities = 97/173 (56%), Positives = 118/173 (68%), Gaps = 6/173 (3%)
Query: 4 HDETGCQAP-ERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSC 62
HD+TGCQ+P E P LC NNCGFFG AATMNMCSKC+KDML +QEQ A++V S
Sbjct: 3 HDKTGCQSPPEGPKLCTNNCGFFGSAATMNMCSKCHKDMLFQQEQGAKFASAVSG--TSS 60
Query: 63 SNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESL---EVKAKTGPSRCGTCRKR 119
S+ K+ TA V++ + VE V+ Q S+ E + E GPSRC TC KR
Sbjct: 61 SSNIIKETFTAALVDIETKSVEPMTVSVQPSSVQVVAEVVAPEEAAKPKGPSRCTTCNKR 120
Query: 120 VGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
VGLTGF C+CG++FC HRY+D HDC F+Y A QEAIAKANPV+KA+KLDKI
Sbjct: 121 VGLTGFKCRCGSLFCGTHRYADVHDCSFNYHAAAQEAIAKANPVVKAEKLDKI 173
>At3g52800 zinc finger like protein
Length = 170
Score = 152 bits (383), Expect = 1e-37
Identities = 76/176 (43%), Positives = 109/176 (61%), Gaps = 16/176 (9%)
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATS-VENIVNSCS 63
+E CQ PE LCVNNCGF G +ATMN+CS CY D+ LKQ+Q + S VE+ ++
Sbjct: 3 EEHRCQTPESNRLCVNNCGFLGSSATMNLCSNCYGDLCLKQQQQSSSIKSTVESSLSVSP 62
Query: 64 NGNGKQAITADAV-------NVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTC 116
+ I++ + +V++E E KAV + + + + + P+RC TC
Sbjct: 63 PSSSSSEISSPIIPPLLKNPSVKLEVPEKKAVISLPTTEQNQQQR--------PNRCTTC 114
Query: 117 RKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
RKRVGLTGF C+CG +FC +HRY + H C +D+++ G+E IAKANP++KA KL KI
Sbjct: 115 RKRVGLTGFKCRCGTMFCGVHRYPEIHGCSYDFKSAGREEIAKANPLVKAAKLQKI 170
>At2g36320 unknown protein
Length = 161
Score = 142 bits (358), Expect = 9e-35
Identities = 74/176 (42%), Positives = 99/176 (56%), Gaps = 25/176 (14%)
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E C+ PE LCVNNCGFFG +ATMN+CS CY D+ LKQ+Q ++VE+ ++
Sbjct: 3 EEHRCETPEGHRLCVNNCGFFGSSATMNLCSNCYGDLCLKQQQQASMKSTVESSLSPV-- 60
Query: 65 GNGKQAITADAVNVRVEPV-EVKAVTAQISADSSSGESLEVKAKTG-------PSRCGTC 116
+ PV E A +I + E ++ T P+RC C
Sbjct: 61 ---------------IAPVLENYAAELEIPTTKKTEEKKPIQIPTEQPSPPQRPNRCTVC 105
Query: 117 RKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
RKRVGLTGF C+CG FC HRY + H C FD+++ G+E IAKANP++ A KL KI
Sbjct: 106 RKRVGLTGFMCRCGTTFCGSHRYPEVHGCTFDFKSAGREEIAKANPLVIAAKLQKI 161
>At4g12040 unknown protein
Length = 175
Score = 141 bits (355), Expect = 2e-34
Identities = 66/163 (40%), Positives = 99/163 (60%), Gaps = 2/163 (1%)
Query: 12 PERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN--SCSNGNGKQ 69
P P LC N CGFFG + MN+CSKCY+ + +++Q +A +V+N + SCS Q
Sbjct: 13 PTEPKLCDNGCGFFGSPSNMNLCSKCYRSLRAEEDQTAVAKAAVKNSLKLPSCSIIAPGQ 72
Query: 70 AITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKC 129
+ +E V V A + + + E+ + +RC +C K+VG+ GF CKC
Sbjct: 73 KHPLEIKPAHLETVVVTAEPSSVPVAAEQDEAEPSRPVRPNNRCFSCNKKVGVMGFKCKC 132
Query: 130 GNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
G+ FC HRY +KH+C FD++ VG++AIAKANP++KADK+ +I
Sbjct: 133 GSTFCGSHRYPEKHECSFDFKEVGRDAIAKANPLVKADKVQRI 175
>At2g27580 unknown protein
Length = 163
Score = 140 bits (354), Expect = 3e-34
Identities = 67/161 (41%), Positives = 100/161 (61%), Gaps = 6/161 (3%)
Query: 13 ERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSNGNGKQAIT 72
+ P LC NNCGFFG AT N+CSKC++D+ Q Q++ ++T+ + S + A +
Sbjct: 8 QEPRLCANNCGFFGSTATQNLCSKCFRDL---QHQEQNSSTAKHALTQSLAAVGA--AAS 62
Query: 73 ADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKT-GPSRCGTCRKRVGLTGFSCKCGN 131
+ P + K + S ++ E E P RC TCR+RVG+TGF C+CG
Sbjct: 63 SSVSPPPPPPADSKEIVEAKSEKRAAAEPEEADGPPQDPKRCLTCRRRVGITGFRCRCGF 122
Query: 132 VFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
VFC HRY+++H+C FD++ +G++ IAKANP++KADKL+KI
Sbjct: 123 VFCGTHRYAEQHECSFDFKRMGKDKIAKANPIVKADKLEKI 163
>At1g12440 unknown protein
Length = 168
Score = 140 bits (352), Expect = 4e-34
Identities = 70/180 (38%), Positives = 100/180 (54%), Gaps = 24/180 (13%)
Query: 2 ESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNS 61
E +D T +P P LCV CGFFG + MN+CSKCY+D+ +EQ A +VE +N
Sbjct: 4 EQNDSTSF-SPSEPKLCVKGCGFFGSPSNMNLCSKCYRDIRATEEQTASAKAAVEKSLNP 62
Query: 62 CSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEV---------KAKTGPSR 112
+ +P + + +T + SS S K+ +R
Sbjct: 63 NKP--------------KTQPQQSQEITQGVLGSGSSSSSTRGGDSAAAPLDPPKSTATR 108
Query: 113 CGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
C +C K+VG+TGF C+CG+ FC HRY + H+C FD++ V +EAIAKANPV+KADK+D+I
Sbjct: 109 CLSCNKKVGVTGFKCRCGSTFCGTHRYPESHECQFDFKGVAREAIAKANPVVKADKVDRI 168
>At4g22820 predicted protein
Length = 176
Score = 138 bits (348), Expect = 1e-33
Identities = 73/173 (42%), Positives = 99/173 (57%), Gaps = 7/173 (4%)
Query: 2 ESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNS 61
+S T QA E P LCV CGFFG + M++CSKCY+ + ++ Q +A +VE S
Sbjct: 7 DSTSFTQSQASE-PKLCVKGCGFFGSPSNMDLCSKCYRGICAEEAQTAVAKAAVEK---S 62
Query: 62 CSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSR---CGTCRK 118
+ A+ V VEP KA +SA+ SS E + P+R C C K
Sbjct: 63 FKPSPPRSLFIAEPPAVVVEPKPEKAAVVVVSAEPSSSAVPEANEPSRPARTNRCLCCNK 122
Query: 119 RVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDK 171
+VG+ GF CKCG+ FC HRY + HDC FD++ VG+ IAKANPV+KADK+ +
Sbjct: 123 KVGIMGFKCKCGSTFCGEHRYPETHDCSFDFKEVGRGEIAKANPVVKADKIQR 175
>At3g12630 unknown protein
Length = 160
Score = 121 bits (304), Expect = 2e-28
Identities = 61/156 (39%), Positives = 86/156 (55%), Gaps = 20/156 (12%)
Query: 17 LCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSNGNGKQAITADAV 76
LC NNCG AT NMC KC+ L+ + + S+ +A +V
Sbjct: 25 LCTNNCGVTANPATNNMCQKCFNASLVSAAAGVVESGSILKR-------------SARSV 71
Query: 77 NVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCGNVFCAM 136
N+R P +V +I A + + +RC CRK+VGLTGF C+CG +FC+
Sbjct: 72 NLRSSPAKVVIRPREIDAVKKRDQQIV-------NRCSGCRKKVGLTGFRCRCGELFCSE 124
Query: 137 HRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
HRYSD+HDC +DY+ G+EAIA+ NPV+KA K+ K+
Sbjct: 125 HRYSDRHDCSYDYKTAGREAIARENPVVKAAKMVKV 160
>At4g25380 putative protein
Length = 130
Score = 94.4 bits (233), Expect = 3e-20
Identities = 47/154 (30%), Positives = 69/154 (44%), Gaps = 34/154 (22%)
Query: 18 CVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSNGNGKQAITADAVN 77
C CG +G N+CS CYK +L+ E + C A+ + V
Sbjct: 10 CEGGCGLYGTRVNNNLCSLCYKKSVLQHSPALRFEPETEQ--SQCCPPTNSPAVEEEPVK 67
Query: 78 VRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCGNVFCAMH 137
R RCG C+++VG+ GF C+CG++FC H
Sbjct: 68 KR--------------------------------RCGICKRKVGMLGFKCRCGHMFCGSH 95
Query: 138 RYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDK 171
RY ++H CPFDY+ G+ A+A P+I+ADKL +
Sbjct: 96 RYPEEHSCPFDYKQSGRLALATQLPLIRADKLQR 129
>At3g57480 unknown protein
Length = 249
Score = 31.2 bits (69), Expect = 0.29
Identities = 13/30 (43%), Positives = 18/30 (59%), Gaps = 1/30 (3%)
Query: 118 KRVGLTGFSC-KCGNVFCAMHRYSDKHDCP 146
K++ F+C +C V+C HR KHDCP
Sbjct: 19 KQIDFLPFTCDRCLQVYCLDHRSYMKHDCP 48
>At1g11100 transcription factor RUSH-1alpha isolog
Length = 1227
Score = 30.4 bits (67), Expect = 0.49
Identities = 16/47 (34%), Positives = 19/47 (40%), Gaps = 4/47 (8%)
Query: 113 CGTCRKRVGLTGFSCKCGNVFC----AMHRYSDKHDCPFDYRAVGQE 155
CG C R+ CG+VFC D + CP Y VG E
Sbjct: 919 CGICNVRLSTHAVVSLCGHVFCNQCICECLTRDNNQCPLSYCKVGLE 965
>At2g46560 hypothetical protein
Length = 2480
Score = 30.0 bits (66), Expect = 0.65
Identities = 17/64 (26%), Positives = 32/64 (49%), Gaps = 6/64 (9%)
Query: 35 SKCYKDMLLKQE------QDKLAATSVENIVNSCSNGNGKQAITADAVNVRVEPVEVKAV 88
S Y++++ K E QD ++ V ++ N SN + K A+TA+ + P E+
Sbjct: 1986 SDMYREVIRKNELYVPCNQDGRSSNEVASLANHASNSSPKAAVTANENSAFQNPKEIHKR 2045
Query: 89 TAQI 92
T ++
Sbjct: 2046 TGEL 2049
>At2g22660 unknown protein
Length = 819
Score = 28.9 bits (63), Expect = 1.4
Identities = 31/108 (28%), Positives = 46/108 (41%), Gaps = 28/108 (25%)
Query: 39 KDMLLKQEQDKLAATSVENIVNSCSNGNGKQAITA-------------DAVNVRVEPVEV 85
KD+ + +Q KL A + N+ + +N + AITA + N+ V
Sbjct: 664 KDIKEESKQTKLVAATENNVNANSTNVETQTAITAPKKGSGCGGGCSGECGNM------V 717
Query: 86 KAVTAQISADSSSGESLE-VKAKTGPSRCGTCRKRVGLTGFSCKCGNV 132
KA A S SGE + VK+ S CG +G S +CGN+
Sbjct: 718 KAANASGCGSSCSGECGDMVKSAANASGCG--------SGCSGECGNM 757
>At2g06310 putative Athila retroelement ORF1 protein
Length = 733
Score = 28.9 bits (63), Expect = 1.4
Identities = 25/68 (36%), Positives = 32/68 (46%), Gaps = 7/68 (10%)
Query: 111 SRCGTCRKRVGLTGFSCKCGNVFC-AMHRYSD---KHDCPFDYRAVGQEAIAKANPVIKA 166
S T R R ++GF K G FC A R+ D DC R+ ++ K IKA
Sbjct: 127 SNARTARLRNEISGFLQKAGESFCEAWERFKDGNYNEDCDRTVRSTA-DSDDKHRKEIKA 185
Query: 167 --DKLDKI 172
DKLD+I
Sbjct: 186 LNDKLDRI 193
>At1g80810 unknown protein
Length = 826
Score = 27.7 bits (60), Expect = 3.2
Identities = 15/40 (37%), Positives = 22/40 (54%)
Query: 86 KAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGF 125
K + +Q++A + ES E K+ P+R T RK V GF
Sbjct: 515 KKMVSQVAARQLANESEEETPKSHPTRRRTVRKEVESDGF 554
>At4g04890 putative homeotic protein
Length = 743
Score = 27.3 bits (59), Expect = 4.2
Identities = 28/102 (27%), Positives = 47/102 (45%), Gaps = 13/102 (12%)
Query: 24 FFGRAATMNMCSKCYKDMLLKQEQDKLAATS---VENIVN-SCSNGNGKQAI---TADAV 76
F + M S+ +++ +LK + DKL A + E + N +C N G AI + D
Sbjct: 110 FQNKRTQMKAQSERHENQILKSDNDKLRAENNRYKEALSNATCPNCGGPAAIGEMSFDEQ 169
Query: 77 NVRVEPVEVKAVTAQISADSSS------GESLEVKAKTGPSR 112
++R+E ++ +ISA ++ G S A PSR
Sbjct: 170 HLRIENARLREEIDRISAIAAKYVGKPLGSSFAPLAIHAPSR 211
>At3g27720 hypothetical protein
Length = 493
Score = 27.3 bits (59), Expect = 4.2
Identities = 13/40 (32%), Positives = 18/40 (44%)
Query: 95 DSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCGNVFC 134
+S + + V K P +KR G +CKCG FC
Sbjct: 227 ESETVNWMTVNTKLCPKCSKPIQKRDGCNHMTCKCGQHFC 266
>At2g19660 unknown protein
Length = 438
Score = 26.9 bits (58), Expect = 5.5
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 3/26 (11%)
Query: 109 GPSRCGTCRKRVG--LTGFSC-KCGN 131
G + CG CRK+V G+SC KC N
Sbjct: 80 GQNTCGVCRKKVDGFYGGYSCSKCSN 105
>At1g70410 unknown protein
Length = 280
Score = 26.9 bits (58), Expect = 5.5
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 9/77 (11%)
Query: 36 KCYKDMLLKQ--EQDKLAATSVENIVNSCSNGNGKQAITADAVNVRVEPVEVKAVTAQI- 92
KC+ K E+D++A S E + N + AD NV ++KA+TA++
Sbjct: 7 KCFMFCCAKTSPEKDEMATESYEAAIKGL---NDLLSTKADLGNVAA--AKIKALTAELK 61
Query: 93 SADSSSGESLEVKAKTG 109
DSS+ +++E + KTG
Sbjct: 62 ELDSSNSDAIE-RIKTG 77
>At5g24740 VPS13 - like protein
Length = 3306
Score = 26.6 bits (57), Expect = 7.1
Identities = 21/75 (28%), Positives = 32/75 (42%), Gaps = 7/75 (9%)
Query: 44 KQEQDKLAATSVENIVNSCSNGNGKQAITADAVNV---RVEPVEVKAVTAQISADSSSGE 100
K D S N++ S S+G+G A T+D N+ R P ++ ++S S
Sbjct: 752 KDAADSHILCSSGNLLKSSSHGHGMDAHTSDETNIIDYRTTPEAAISLNCKVSQTSYKIS 811
Query: 101 SLEVKAKTGPSRCGT 115
S + T PS T
Sbjct: 812 S----SYTNPSSLAT 822
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,759,805
Number of Sequences: 26719
Number of extensions: 135022
Number of successful extensions: 492
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 460
Number of HSP's gapped (non-prelim): 37
length of query: 172
length of database: 11,318,596
effective HSP length: 92
effective length of query: 80
effective length of database: 8,860,448
effective search space: 708835840
effective search space used: 708835840
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0318.6


