

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0316.4
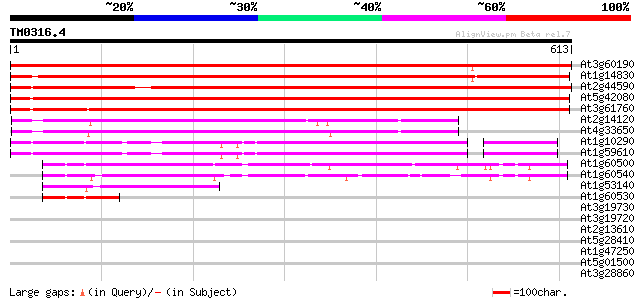
(613 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g60190 dynamin-like protein 4 (ADL4) 1022 0.0
At1g14830 dynamin-like protein 962 0.0
At2g44590 dynamin-like protein D 894 0.0
At5g42080 dynamin-like protein (pir||S59558) 868 0.0
At3g61760 dynamin-like protein B 847 0.0
At2g14120 dynamin like protein 2b (ADL2b) 317 2e-86
At4g33650 dynamin like protein 2a (ADL2a) 300 2e-81
At1g10290 putative phragmoplastin 235 6e-62
At1g59610 dynamin-like protein CF1 228 8e-60
At1g60500 169 3e-42
At1g60540 168 9e-42
At1g53140 dynamin-like protein 94 2e-19
At1g60530 unknown protein 60 5e-09
At3g19730 dynamin-like protein 41 0.002
At3g19720 hypothetical protein 39 0.011
At2g13610 putative ABC transporter 32 1.0
At5g28410 putative protein 32 1.4
At1g47250 20S proteasome subunit PAF2 32 1.4
At5g01500 unknown protein 30 3.0
At3g28860 P-glycoprotein, putative 30 3.9
>At3g60190 dynamin-like protein 4 (ADL4)
Length = 624
Score = 1022 bits (2643), Expect = 0.0
Identities = 500/621 (80%), Positives = 574/621 (91%), Gaps = 8/621 (1%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLIGLVNRIQRACTVLGD+G N+F+SLWEALPTVAVVGGQSSGKSSVLESIVGR
Sbjct: 4 MESLIGLVNRIQRACTVLGDYGGGTGSNAFNSLWEALPTVAVVGGQSSGKSSVLESIVGR 63
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK + G++EYAEFLH+P+++FTDF+LVR+EIQDETDR+TGK
Sbjct: 64 DFLPRGSGIVTRRPLVLQLHKTDDGTEEYAEFLHLPKKQFTDFALVRREIQDETDRITGK 123
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQISP+PIHLSIYSPNVVNLTL+DLPGLTKVAVEGQPE+I ++IE+MVRTYV+KPN II
Sbjct: 124 NKQISPVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPETIAEDIESMVRTYVDKPNCII 183
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLA++VDPTGERTFGVLTKLDLMDKGTNAL+VLEGRSYRLQHPW
Sbjct: 184 LAISPANQDIATSDAIKLAKDVDPTGERTFGVLTKLDLMDKGTNALEVLEGRSYRLQHPW 243
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKNVDM++ARRKEREYF SPDYGHLA+KMGSEYLAKLLS+HLESVIR
Sbjct: 244 VGIVNRSQADINKNVDMMLARRKEREYFDTSPDYGHLASKMGSEYLAKLLSKHLESVIRT 303
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSI SLINKS+EELE E+D +GRP+A+DAGAQLYTILE+CRAF++IFKEHLDGGRPGG
Sbjct: 304 RIPSILSLINKSIEELERELDRMGRPVAVDAGAQLYTILEMCRAFDKIFKEHLDGGRPGG 363
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFDNQLPAAL+KLPFDRHLSLQ+V+K+VSEADGYQPHLIAPEQGYRRLI+ AL Y
Sbjct: 364 DRIYGVFDNQLPAALKKLPFDRHLSLQSVKKIVSEADGYQPHLIAPEQGYRRLIEGALGY 423
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASVDAV++VLKELVRKSI+ET+ELKRFP+ Q LA AAN +LE+FREESKK+ +
Sbjct: 424 FRGPAEASVDAVHYVLKELVRKSISETEELKRFPSLQVELAAAANSSLEKFREESKKSVI 483
Query: 481 RLVDMESSYLTVDFFRRLPQEVEK--------AGGPATPNVDRYAEGHFRRIASNVSSYI 532
RLVDMES+YLT +FFR+LPQE+E+ P++ +D+Y +GHFRRIASNVS+Y+
Sbjct: 484 RLVDMESAYLTAEFFRKLPQEIERPVTNSKNQTASPSSATLDQYGDGHFRRIASNVSAYV 543
Query: 533 GLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQC 592
+V+DTLRNTIPKA V+CQVRQAK +LLN+FY+QI K+EGKQL ++LDEDPALM+RR +C
Sbjct: 544 NMVSDTLRNTIPKACVYCQVRQAKLALLNYFYSQISKREGKQLGQLLDEDPALMDRRLEC 603
Query: 593 AKRLELYKAARDEIDSVSWVR 613
AKRLELYK ARDEID+V+WVR
Sbjct: 604 AKRLELYKKARDEIDAVAWVR 624
>At1g14830 dynamin-like protein
Length = 614
Score = 962 bits (2488), Expect = 0.0
Identities = 481/616 (78%), Positives = 551/616 (89%), Gaps = 11/616 (1%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M+SLIGL+N+IQRACTVLGDHG SLWEALPTVAVVGGQSSGKSSVLES+VGR
Sbjct: 4 MKSLIGLINKIQRACTVLGDHGGEG-----MSLWEALPTVAVVGGQSSGKSSVLESVVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+ EYAEFLH P+++F DF+ VR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTEDGTTEYAEFLHAPKKRFADFAAVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+KQIS IPI LSIYSPNVVNLTL+DLPGLTKVAV+GQPESIVQ+IE MVR+YVEKPN II
Sbjct: 119 SKQISNIPIQLSIYSPNVVNLTLIDLPGLTKVAVDGQPESIVQDIENMVRSYVEKPNCII 178
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDAIKLAREVDPTGERTFGV TKLD+MDKGT+ LDVLEGRSYRLQHPW
Sbjct: 179 LAISPANQDIATSDAIKLAREVDPTGERTFGVATKLDIMDKGTDCLDVLEGRSYRLQHPW 238
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINK VDMI ARRKE+EYF SP+YGHLA++MGSEYLAKLLSQHLE+VIR
Sbjct: 239 VGIVNRSQADINKRVDMIAARRKEQEYFETSPEYGHLASRMGSEYLAKLLSQHLETVIRQ 298
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
+IPSI +LINKS++E+ +E+D +GRPIA+D+GAQLYTILELCRAF+R+FKEHLDGGRPGG
Sbjct: 299 KIPSIVALINKSIDEINAELDRIGRPIAVDSGAQLYTILELCRAFDRVFKEHLDGGRPGG 358
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
DRIY VFD+QLPAAL+KLPFDRHLS +NV+KVVSEADGYQPHLIAPEQGYRRLID ++SY
Sbjct: 359 DRIYGVFDHQLPAALKKLPFDRHLSTKNVQKVVSEADGYQPHLIAPEQGYRRLIDGSISY 418
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
F+GPAEA+VDAV+FVLKELVRKSI+ET+ELKRFPT + +A AANEALERFR+ES+KT L
Sbjct: 419 FKGPAEATVDAVHFVLKELVRKSISETEELKRFPTLASDIAAAANEALERFRDESRKTVL 478
Query: 481 RLVDMESSYLTVDFFRRLPQEVEK-----AGGPATPNVDRYAEGHFRRIASNVSSYIGLV 535
RLVDMESSYLTV+FFR+L E EK PA PN D Y++ HFR+I SNVS+YI +V
Sbjct: 479 RLVDMESSYLTVEFFRKLHLEPEKEKPNPRNAPA-PNADPYSDNHFRKIGSNVSAYINMV 537
Query: 536 ADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKR 595
DTLRN++PKAVV+CQVR+AK+SLLN FY Q+G+KE ++L MLDEDP LMERR AKR
Sbjct: 538 CDTLRNSLPKAVVYCQVREAKRSLLNFFYAQVGRKEKEKLGAMLDEDPQLMERRGTLAKR 597
Query: 596 LELYKAARDEIDSVSW 611
LELYK ARD+ID+V+W
Sbjct: 598 LELYKQARDDIDAVAW 613
>At2g44590 dynamin-like protein D
Length = 596
Score = 894 bits (2310), Expect = 0.0
Identities = 451/615 (73%), Positives = 531/615 (86%), Gaps = 21/615 (3%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI L+N IQRACTV+GDHG D N+ SSLWEALP+VAVVGGQSSGKSSVLESIVGR
Sbjct: 1 MESLIVLINTIQRACTVVGDHGG--DSNALSSLWEALPSVAVVGGQSSGKSSVLESIVGR 58
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQLHK E G+++ AEFLH+ +KFT+FSLVR+EI+DETDR+TGK
Sbjct: 59 DFLPRGSGIVTRRPLVLQLHKTENGTEDNAEFLHLTNKKFTNFSLVRKEIEDETDRITGK 118
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
KQIS IPIHLSI+SPN EGQPE+IV++IE+MVR+YVEKPN +I
Sbjct: 119 NKQISSIPIHLSIFSPN-----------------EGQPETIVEDIESMVRSYVEKPNCLI 161
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQDIATSDA+KLA+EVDP G+RTFGVLTKLDLMDKGTNALDV+ GRSY+L++PW
Sbjct: 162 LAISPANQDIATSDAMKLAKEVDPIGDRTFGVLTKLDLMDKGTNALDVINGRSYKLKYPW 221
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VGIVNRSQADINKNVDM++ARRKEREYF SPDYGHLA +MGSEYLAKLLS+ LESVIR+
Sbjct: 222 VGIVNRSQADINKNVDMMVARRKEREYFETSPDYGHLATRMGSEYLAKLLSKLLESVIRS 281
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIPSI SLIN ++EELE E+D LGRPIA+DAGAQLYTIL +CRAFE+IFKEHLDGGRPGG
Sbjct: 282 RIPSILSLINNNIEELERELDQLGRPIAIDAGAQLYTILGMCRAFEKIFKEHLDGGRPGG 341
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
RIY +FD LP A++KLPFDRHLSLQ+V+++VSE+DGYQPHLIAPE GYRRLI+ +L++
Sbjct: 342 ARIYGIFDYNLPTAIKKLPFDRHLSLQSVKRIVSESDGYQPHLIAPELGYRRLIEGSLNH 401
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
FRGPAEASV+A++ +LKELVRK+IAET+ELKRFP+ Q L AAN +L++FREES K+ L
Sbjct: 402 FRGPAEASVNAIHLILKELVRKAIAETEELKRFPSLQIELVAAANSSLDKFREESMKSVL 461
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAG-GPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
RLVDMESSYLTVDFFR+L E + T +D+Y +GHFR+IASNV++YI +VA+TL
Sbjct: 462 RLVDMESSYLTVDFFRKLHVESQNMSLSSPTSAIDQYGDGHFRKIASNVAAYIKMVAETL 521
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGK-KEGKQLSEMLDEDPALMERRQQCAKRLEL 598
NTIPKAVV CQVRQAK SLLN+FY QI + ++GK+L ++LDE+PALMERR QCAKRLEL
Sbjct: 522 VNTIPKAVVHCQVRQAKLSLLNYFYAQISQSQQGKRLGQLLDENPALMERRMQCAKRLEL 581
Query: 599 YKAARDEIDSVSWVR 613
YK ARDEID+ WVR
Sbjct: 582 YKKARDEIDAAVWVR 596
>At5g42080 dynamin-like protein (pir||S59558)
Length = 610
Score = 868 bits (2242), Expect = 0.0
Identities = 416/611 (68%), Positives = 532/611 (86%), Gaps = 3/611 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME+LI LVN+IQRACT LGDHG D ++ +LW++LP +AVVGGQSSGKSSVLESIVG+
Sbjct: 1 MENLISLVNKIQRACTALGDHG---DSSALPTLWDSLPAIAVVGGQSSGKSSVLESIVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL K++ G++EYAEFLH+PR+KFTDF+ VR+EIQDETDR TG+
Sbjct: 58 DFLPRGSGIVTRRPLVLQLQKIDDGTREYAEFLHLPRKKFTDFAAVRKEIQDETDRETGR 117
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+K IS +PIHLSIYSPNVVNLTL+DLPGLTKVAV+GQ +SIV++IE MVR+Y+EKPN II
Sbjct: 118 SKAISSVPIHLSIYSPNVVNLTLIDLPGLTKVAVDGQSDSIVKDIENMVRSYIEKPNCII 177
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQD+ATSDAIK++REVDP+G+RTFGVLTK+DLMDKGT+A+++LEGRS++L++PW
Sbjct: 178 LAISPANQDLATSDAIKISREVDPSGDRTFGVLTKIDLMDKGTDAVEILEGRSFKLKYPW 237
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VG+VNRSQADINKNVDMI AR++EREYF+N+ +Y HLANKMGSE+LAK+LS+HLE VI++
Sbjct: 238 VGVVNRSQADINKNVDMIAARKREREYFSNTTEYRHLANKMGSEHLAKMLSKHLERVIKS 297
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIP I SLINK++ ELE+E+ LG+PIA DAG +LY+I+E+CR F++IFKEHLDG R GG
Sbjct: 298 RIPGIQSLINKTVLELETELSRLGKPIAADAGGKLYSIMEICRLFDQIFKEHLDGVRAGG 357
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
+++Y+VFDNQLPAAL++L FD+ L++ N+RK+V+EADGYQPHLIAPEQGYRRLI+S++
Sbjct: 358 EKVYNVFDNQLPAALKRLQFDKQLAMDNIRKLVTEADGYQPHLIAPEQGYRRLIESSIVS 417
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
RGPAEASVD V+ +LK+LV KS+ ET ELK++P + + AA E+L++ RE SKK TL
Sbjct: 418 IRGPAEASVDTVHAILKDLVHKSVNETVELKQYPALRVEVTNAAIESLDKMREGSKKATL 477
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLR 540
+LVDME SYLTVDFFR+LPQ+VEK G P DRY + + RRI SNV SY+ +V LR
Sbjct: 478 QLVDMECSYLTVDFFRKLPQDVEKGGNPTHSIFDRYNDSYLRRIGSNVLSYVNMVCAGLR 537
Query: 541 NTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
N+IPK++V+CQVR+AK+SLL+HF+ ++G + K+LS +L+EDPA+MERR +KRLELY+
Sbjct: 538 NSIPKSIVYCQVREAKRSLLDHFFAELGTMDMKRLSSLLNEDPAIMERRSAISKRLELYR 597
Query: 601 AARDEIDSVSW 611
AA+ EID+V+W
Sbjct: 598 AAQSEIDAVAW 608
>At3g61760 dynamin-like protein B
Length = 610
Score = 847 bits (2189), Expect = 0.0
Identities = 407/612 (66%), Positives = 525/612 (85%), Gaps = 5/612 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
MESLI LVN+IQRACT LGDHG +G+S +LW++LP +AVVGGQSSGKSSVLES+VG+
Sbjct: 1 MESLIALVNKIQRACTALGDHG---EGSSLPTLWDSLPAIAVVGGQSSGKSSVLESVVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRG+GIVTRRPLVLQLH+++ G +EYAEF+H+P++KFTDF+ VRQEI DETDR TG+
Sbjct: 58 DFLPRGAGIVTRRPLVLQLHRIDEG-KEYAEFMHLPKKKFTDFAAVRQEISDETDRETGR 116
Query: 121 T-KQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSI 179
+ K IS +PIHLSI+SPNVVNLTLVDLPGLTKVAV+GQPESIVQ+IE MVR+++EKPN I
Sbjct: 117 SSKVISTVPIHLSIFSPNVVNLTLVDLPGLTKVAVDGQPESIVQDIENMVRSFIEKPNCI 176
Query: 180 ILAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHP 239
ILAISPANQD+ATSDAIK++REVDP G+RTFGVLTK+DLMD+GTNA+D+LEGR Y+L++P
Sbjct: 177 ILAISPANQDLATSDAIKISREVDPKGDRTFGVLTKIDLMDQGTNAVDILEGRGYKLRYP 236
Query: 240 WVGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIR 299
WVG+VNRSQADINK+VDMI ARR+ER+YF SP+Y HL +MGSEYL K+LS+HLE VI+
Sbjct: 237 WVGVVNRSQADINKSVDMIAARRRERDYFQTSPEYRHLTERMGSEYLGKMLSKHLEVVIK 296
Query: 300 ARIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPG 359
+RIP + SLI K++ ELE+E+ LG+P+A DAG +LY I+E+CRAF++ FKEHLDG R G
Sbjct: 297 SRIPGLQSLITKTISELETELSRLGKPVAADAGGKLYMIMEICRAFDQTFKEHLDGTRSG 356
Query: 360 GDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALS 419
G++I VFDNQ PAA+++L FD+HLS+ NVRK+++EADGYQPHLIAPEQGYRRLI+S L
Sbjct: 357 GEKINSVFDNQFPAAIKRLQFDKHLSMDNVRKLITEADGYQPHLIAPEQGYRRLIESCLV 416
Query: 420 YFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTT 479
RGPAEA+VDAV+ +LK+L+ KS+ ET ELK++PT + ++ AA ++L+R R+ES+K T
Sbjct: 417 SIRGPAEAAVDAVHSILKDLIHKSMGETSELKQYPTLRVEVSGAAVDSLDRMRDESRKAT 476
Query: 480 LRLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
L LVDMES YLTV+FFR+LPQ+ EK G P DRY + + RRI SNV SY+ +V L
Sbjct: 477 LLLVDMESGYLTVEFFRKLPQDSEKGGNPTHSIFDRYNDAYLRRIGSNVLSYVNMVCAGL 536
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELY 599
RN+IPK++V+CQVR+AK+SLL+ F+T++G+KE +LS++LDEDPA+ +RR AKRLELY
Sbjct: 537 RNSIPKSIVYCQVREAKRSLLDIFFTELGQKEMSKLSKLLDEDPAVQQRRTSIAKRLELY 596
Query: 600 KAARDEIDSVSW 611
++A+ +I++V+W
Sbjct: 597 RSAQTDIEAVAW 608
>At2g14120 dynamin like protein 2b (ADL2b)
Length = 780
Score = 317 bits (811), Expect = 2e-86
Identities = 202/503 (40%), Positives = 287/503 (56%), Gaps = 30/503 (5%)
Query: 3 SLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGRDF 62
S+I +VN++Q LG A LP VAVVG QSSGKSSVLE++VGRDF
Sbjct: 20 SVIPIVNKLQDIFAQLGSQSTIA-----------LPQVAVVGSQSSGKSSVLEALVGRDF 68
Query: 63 LPRGSGIVTRRPLVLQLHKLETGS-----QEYAEFLHM-PRRKFTDFSLVRQEIQDETDR 116
LPRG+ I TRRPL LQL + + S +E+ EFLH P R+ DFS +R+EI+ ET+R
Sbjct: 69 LPRGNDICTRRPLRLQLVQTKPSSDGGSDEEWGEFLHHDPVRRIYDFSEIRREIEAETNR 128
Query: 117 VTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKP 176
V+G+ K +S IPI L I+SPNV++++LVDLPG+TKV V QP I I TM+ TY+++P
Sbjct: 129 VSGENKGVSDIPIGLKIFSPNVLDISLVDLPGITKVPVGDQPSDIEARIRTMILTYIKEP 188
Query: 177 NSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRL 236
+ +ILA+SPAN D+A SDA+++A DP G RT GV+TKLD+MD+GT+A + L G++ L
Sbjct: 189 SCLILAVSPANTDLANSDALQIAGNADPDGHRTIGVITKLDIMDRGTDARNHLLGKTIPL 248
Query: 237 QHPWVGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLES 296
+ +VG+VNRSQ DI N + A E ++F + P Y L +++G LAK L+Q L
Sbjct: 249 RLGYVGVVNRSQEDILMNRSIKDALVAEEKFFRSRPVYSGLTDRLGVPQLAKKLNQVLVQ 308
Query: 297 VIRARIPSITSLINKSLEELESEMDHLGRPIALDAGAQ----LYTILELCRAF----ERI 348
I+A +PS+ S IN +L E + G I G Q L I + C A+ E
Sbjct: 309 HIKALLPSLKSRINNALFATAKEYESYG-DITESRGGQGALLLSFITKYCEAYSSTLEGK 367
Query: 349 FKEHLDGGRPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQ 408
KE GG RI ++F + +L ++ L+ ++R + A G + L P+
Sbjct: 368 SKEMSTSELSGGARILYIFQSVFVKSLEEVDPCEDLTADDIRTAIQNATGPRSALFVPDV 427
Query: 409 GYRRLIDSALSYFRGPAEASVDAVNFVLKELVRKS-IAETKELKRFPTFQAALATAANEA 467
+ L+ +S P S+ F+ ELV+ S KEL+RFP Q +
Sbjct: 428 PFEVLVRRQISRLLDP---SLQCARFIFDELVKISHQCMMKELQRFPVLQKRMDEVIGNF 484
Query: 468 LERFREESKKTTLRLVDMESSYL 490
L E S+ L++ME Y+
Sbjct: 485 LREGLEPSQAMIRDLIEMEMDYI 507
Score = 33.1 bits (74), Expect = 0.47
Identities = 19/82 (23%), Positives = 41/82 (49%), Gaps = 2/82 (2%)
Query: 528 VSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALME 587
+ SY +V + + +PKA++ V K+ L N F ++ ++ + E+L E L
Sbjct: 661 LKSYYDIVRKNVEDLVPKAIMHFLVNYTKRELHNVFIEKLYRE--NLIEELLKEPDELAI 718
Query: 588 RRQQCAKRLELYKAARDEIDSV 609
+R++ + L + + A +D +
Sbjct: 719 KRKRTQETLRILQQANRTLDEL 740
>At4g33650 dynamin like protein 2a (ADL2a)
Length = 808
Score = 300 bits (767), Expect = 2e-81
Identities = 189/501 (37%), Positives = 278/501 (54%), Gaps = 27/501 (5%)
Query: 3 SLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGRDF 62
S+I +VN++Q LG A LP V VVG QSSGKSSVLE++VGRDF
Sbjct: 36 SVIPIVNKLQDIFAQLGSQSTIA-----------LPQVVVVGSQSSGKSSVLEALVGRDF 84
Query: 63 LPRGSGIVTRRPLVLQLHKLET-----GSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRV 117
LPRG+ I TRRPLVLQL + ++ E+ EF H+P +F DFS +R+EI+ ET+R+
Sbjct: 85 LPRGNDICTRRPLVLQLLQTKSRANGGSDDEWGEFRHLPETRFYDFSEIRREIEAETNRL 144
Query: 118 TGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPN 177
G+ K ++ I L I SPNV+N+TLVDLPG+TKV V QP I I TM+ +Y+++
Sbjct: 145 VGENKGVADTQIRLKISSPNVLNITLVDLPGITKVPVGDQPSDIEARIRTMILSYIKQDT 204
Query: 178 SIILAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQ 237
+ILA++PAN D+A SDA+++A VDP G RT GV+TKLD+MDKGT+A +L G L+
Sbjct: 205 CLILAVTPANTDLANSDALQIASIVDPDGHRTIGVITKLDIMDKGTDARKLLLGNVVPLR 264
Query: 238 HPWVGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESV 297
+VG+VNR Q DI N + A E ++F + P Y LA+++G LAK L+Q L
Sbjct: 265 LGYVGVVNRCQEDILLNRTVKEALLAEEKFFRSHPVYHGLADRLGVPQLAKKLNQILVQH 324
Query: 298 IRARIPSITSLINKSLEELESEMDHLGRPIALDA--GAQLYTIL-ELCRAFERIF----K 350
I+ +P + S I+ +L E G A GA L L + C A+ + +
Sbjct: 325 IKVLLPDLKSRISNALVATAKEHQSYGELTESRAGQGALLLNFLSKYCEAYSSLLEGKSE 384
Query: 351 EHLDGGRPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGY 410
E GG RI+++F + +L ++ L+ ++R + A G + L P+ +
Sbjct: 385 EMSTSELSGGARIHYIFQSIFVKSLEEVDPCEDLTDDDIRTAIQNATGPRSALFVPDVPF 444
Query: 411 RRLIDSALSYFRGPAEASVDAVNFVLKELVRKS-IAETKELKRFPTFQAALATAANEALE 469
L+ +S P S+ F+ +EL++ S EL+RFP + + + L
Sbjct: 445 EVLVRRQISRLLDP---SLQCARFIFEELIKISHRCMMNELQRFPVLRKRMDEVIGDFLR 501
Query: 470 RFREESKKTTLRLVDMESSYL 490
E S+ ++DME Y+
Sbjct: 502 EGLEPSEAMIGDIIDMEMDYI 522
Score = 33.9 bits (76), Expect = 0.27
Identities = 25/106 (23%), Positives = 48/106 (44%), Gaps = 9/106 (8%)
Query: 511 PNVDRYAEGHFRRIASNVS-------SYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHF 563
P V R E H + A + SY +V + +++PKA++ V K+ L N F
Sbjct: 653 PAVLRPTETHSEQEAVEIQITKLLLRSYYDIVRKNIEDSVPKAIMHFLVNHTKRELHNVF 712
Query: 564 YTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSV 609
++ ++ EML E + +R++ + L + + A +D +
Sbjct: 713 IKKLYRE--NLFEEMLQEPDEIAVKRKRTQETLHVLQQAYRTLDEL 756
>At1g10290 putative phragmoplastin
Length = 914
Score = 235 bits (599), Expect = 6e-62
Identities = 165/516 (31%), Positives = 263/516 (49%), Gaps = 35/516 (6%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
++ L L + +++A ++L D D S S V +G +GKS+VL S++G
Sbjct: 4 IDELSQLSDSMKQAASLLADEDP--DETSSSKRPATFLNVVALGNVGAGKSAVLNSLIGH 61
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRR-KFTDFSLVRQEIQDETDR-VT 118
LP G TR P++++L + E+ A L + + + S +R +QD + +
Sbjct: 62 PVLPTGENGATRAPIIIELSR-ESSLSSKAIILQIDNKSQQVSASALRHSLQDRLSKGAS 120
Query: 119 GKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNS 178
GK + I+L + + L LVDLPGL + V+ E+M+ Y + ++
Sbjct: 121 GKNRD----EINLKLRTSTAPPLKLVDLPGLDQRIVD----------ESMIAEYAQHNDA 166
Query: 179 IILAISPANQ--DIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLE------ 230
I+L I PA+Q +I++S A+K+A+E DP RT G++ K+D + + AL ++
Sbjct: 167 ILLVIVPASQASEISSSRALKIAKEYDPESTRTIGIIGKIDQAAENSKALAAVQALLSNQ 226
Query: 231 GRSYRLQHPWVGIVNRS------QADINKNVDMIIARRKEREYFANSPDYGHLANKMGSE 284
G PWV ++ +S Q+ +N + A R E E S G +K+G
Sbjct: 227 GPPKTTDIPWVAVIGQSVSIASAQSGSGEN-SLETAWRAESESL-KSILTGAPQSKLGRI 284
Query: 285 YLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRA 344
L L+ + S ++ R+PS+ S + + ++ E+ LG + A LELCR
Sbjct: 285 ALVDTLASQIRSRMKLRLPSVLSGLQGKSQIVQDELARLGEQLVNSAEGTRAIALELCRE 344
Query: 345 FERIFKEHLDGGRPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLI 404
FE F HL GG G ++ F+ P +++LP DRH L NV++VV EADGYQP+LI
Sbjct: 345 FEDKFLLHLAGGEGSGWKVVASFEGNFPNRIKQLPLDRHFDLNNVKRVVLEADGYQPYLI 404
Query: 405 APEQGYRRLIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAA 464
+PE+G R LI L + PA VD V+ VL ++V S T L R+P F+ + A
Sbjct: 405 SPEKGLRSLIKIVLELAKDPARLCVDEVHRVLVDIVSASANATPGLGRYPPFKREVVAIA 464
Query: 465 NEALERFREESKKTTLRLVDMESSYLTVDFFRRLPQ 500
+ AL+ F+ E+KK + LVDME +++ F RL Q
Sbjct: 465 SAALDGFKNEAKKMVVALVDMERAFVPPQHFIRLVQ 500
Score = 55.5 bits (132), Expect = 9e-08
Identities = 26/81 (32%), Positives = 45/81 (55%)
Query: 518 EGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSE 577
E R ++ V Y+ V ++L +PKAVV CQV +AK+ +LN Y+ I +++
Sbjct: 727 EEELRWMSQEVRGYVEAVLNSLAANVPKAVVLCQVEKAKEDMLNQLYSSISAIGNERIES 786
Query: 578 MLDEDPALMERRQQCAKRLEL 598
++ ED + RR++ K+ L
Sbjct: 787 LIQEDQNVKRRRERYQKQSSL 807
>At1g59610 dynamin-like protein CF1
Length = 920
Score = 228 bits (581), Expect = 8e-60
Identities = 157/515 (30%), Positives = 263/515 (50%), Gaps = 33/515 (6%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
++ L L + +++A ++L D D S S V +G +GKS+VL S++G
Sbjct: 4 IDELSQLSDSMRQAASLLADEDP--DETSSSRRPATSLNVVALGNVGAGKSAVLNSLIGH 61
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDR-VTG 119
LP G TR P+++ L + E+ S + + + S +R +QD + +G
Sbjct: 62 PVLPTGENGATRAPIIIDLSREESLSSKAIILQIDNKNQQVSASALRHSLQDRLSKGASG 121
Query: 120 KTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSI 179
+ + I+L + + L L+DLPGL + V+ ++M+ + + ++I
Sbjct: 122 RGRD----EIYLKLRTSTAPPLKLIDLPGLDQRIVD----------DSMIGEHAQHNDAI 167
Query: 180 ILAISPANQ--DIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLE------G 231
+L + PA+Q +I++S A+K+A+E DP RT G+++K+D + +L ++ G
Sbjct: 168 LLVVVPASQASEISSSRALKIAKEYDPESTRTVGIISKIDQAAENPKSLAAVQALLSNQG 227
Query: 232 RSYRLQHPWVGIVNRS------QADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEY 285
PWV ++ +S Q+ ++N + A R E E S G +K+G
Sbjct: 228 PPKTTDIPWVALIGQSVSIASAQSGGSEN-SLETAWRAESESL-KSILTGAPQSKLGRIA 285
Query: 286 LAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAF 345
L L+ + S ++ R+P+I + + + ++ E+ LG + A LELCR F
Sbjct: 286 LVDTLASQIRSRMKLRLPNILTGLQGKSQIVQDELARLGEQLVSSAEGTRAIALELCREF 345
Query: 346 ERIFKEHLDGGRPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIA 405
E F HL GG G ++ F+ P ++KLP DRH L NV+++V EADGYQP+LI+
Sbjct: 346 EDKFLLHLAGGEGSGWKVVASFEGNFPNRIKKLPLDRHFDLNNVKRIVLEADGYQPYLIS 405
Query: 406 PEQGYRRLIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAAN 465
PE+G R LI + L + PA VD V+ VL ++V S T L R+P F+ + A+
Sbjct: 406 PEKGLRSLIKTVLELAKDPARLCVDEVHRVLVDIVSASANATPGLGRYPPFKREVVAIAS 465
Query: 466 EALERFREESKKTTLRLVDMESSYLTVDFFRRLPQ 500
AL+ F+ E+KK + LVDME +++ F RL Q
Sbjct: 466 AALDGFKNEAKKMVVALVDMERAFVPPQHFIRLVQ 500
Score = 53.5 bits (127), Expect = 3e-07
Identities = 25/81 (30%), Positives = 44/81 (53%)
Query: 518 EGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSE 577
E R ++ V Y+ V ++L +PKAVV CQV ++K+ +LN Y+ I +++
Sbjct: 734 EEELRWMSQEVRGYVEAVLNSLAANVPKAVVLCQVEKSKEDMLNQLYSSISAIGNERIES 793
Query: 578 MLDEDPALMERRQQCAKRLEL 598
++ ED + RR + K+ L
Sbjct: 794 LIQEDQNVKRRRDRYQKQSSL 814
>At1g60500
Length = 669
Score = 169 bits (429), Expect = 3e-42
Identities = 156/618 (25%), Positives = 280/618 (45%), Gaps = 63/618 (10%)
Query: 37 LPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMP 96
LPT+ VVG QSSGKSSVLES+ G LPRG GI TR PLV++L + + S E +L
Sbjct: 65 LPTIVVVGDQSSGKSSVLESLAGIS-LPRGQGICTRVPLVMRLQR--SSSPEPEIWLEYN 121
Query: 97 RRKF-TDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVE 155
+ TD + + I+ TD + G K +S P+ L + V +LT+VDLPG+T+V V
Sbjct: 122 DKVVPTDEEHIAEAIRAATDVIAGSGKGVSDAPLTLHVKKAGVPDLTMVDLPGITRVPVN 181
Query: 156 GQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLTK 215
GQPE+I ++I M+ Y+E SIIL + A D T ++I+++R+VD TG+RT V+TK
Sbjct: 182 GQPENIYEQISGMIMEYIEPQESIILNVLSATVDFTTCESIRMSRKVDKTGQRTLAVVTK 241
Query: 216 LDLMDKGTNALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSPDYG 275
D+ +G L + + +V + NR + + AR +E F P
Sbjct: 242 ADMAPEG--LLQKVTADDVSIVLGYVCVRNRIGEETYEE-----ARMQEELLFRTHPVLS 294
Query: 276 HL-ANKMGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALDAGAQ 334
+ + +G LA+ L S+I +P I S IN+ L+ E++ L +A G
Sbjct: 295 LIDEDIVGIPVLAQKLMLIQSSMIARCLPKIVSKINQKLDTAVLELNKLPMVMA-STGEA 353
Query: 335 LYTILELCRAFERI---------FKEHLDGGRPGGDRIYHVFDNQLPAALRKLPFDRHLS 385
L ++++ + + F E+ D +Q +L+ P +
Sbjct: 354 LMALMDIIGSAKESLLRILVQGDFSEYPDDQNMHCTARLADMLSQFSDSLQAKPKEVAEF 413
Query: 386 LQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSYFRGPAEASVDAVNFVLKELVRKSIA 445
L + K++ E P + ++ + + ++ + +++++ S
Sbjct: 414 LMDEIKILDECKCVGLPNFIPRSAFLAILSQHVDGIQDKPVEFINKIWDYIEDVL--SSV 471
Query: 446 ETKELKRFPTFQAALATAANEALERFREESKKTTLRLVDMES--------SYLTVDFFRR 497
K FP Q+++ A + + +E+S + +V+ME Y+T +
Sbjct: 472 TAKRSDNFPQIQSSIKRAGRNLISKIKEQSVNRVMEIVEMEKLTDYTCNPEYMTSWTQKT 531
Query: 498 LPQEVEKAGGPATPNVDRYAE---------GHFRR-----------IASNVSSYIGLVAD 537
QE N+ Y H R+ + ++SY +V
Sbjct: 532 SAQESFIDAVVKNENIPDYFSVTGFGNVKISHLRKYHAHLLIPAFDMKMRITSYWKIV-- 589
Query: 538 TLRNTIPKAVVFCQVRQAKQSLLNHFYTQ------IGKKEGKQLSEMLDEDPALMERRQQ 591
LR + ++ Q+ + +SL+N + + + ++G + +ML+E P + +R++
Sbjct: 590 -LRRIVDNLALYLQL--SVKSLVNTRFQKEIVAEMVDPRDGGGVEKMLEESPLVASKREK 646
Query: 592 CAKRLELYKAARDEIDSV 609
++L K ++D + ++
Sbjct: 647 LQNSIKLLKESKDAVAAI 664
>At1g60540
Length = 648
Score = 168 bits (425), Expect = 9e-42
Identities = 167/614 (27%), Positives = 280/614 (45%), Gaps = 82/614 (13%)
Query: 37 LPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQE----YAEF 92
LPT+ VVG QSSGKSSVLES+ G LPRG GI TR PLV++L + + E Y +
Sbjct: 66 LPTIVVVGDQSSGKSSVLESLAGIS-LPRGQGICTRVPLVMRLQRSRSPEPEIWLEYGDK 124
Query: 93 LHMPRRKFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKV 152
+ +P TD + Q I TD + G K +S P+ L + V ++T+VDLPG+T+V
Sbjct: 125 I-VP----TDEEHIAQTICAATDVIAGSGKGVSDTPLTLHVKKAGVPDITMVDLPGITRV 179
Query: 153 AVEGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGV 212
V GQPE+I ++I MV Y+E SIIL + A D T ++I+++++VD TGERT V
Sbjct: 180 PVNGQPENIYEQISRMVMKYIEPQESIILNVLSATVDFTTCESIRMSKQVDKTGERTLAV 239
Query: 213 LTKLDLMDKG----TNALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYF 268
+TK D+ +G A DV G Y V + NR + + AR +E F
Sbjct: 240 VTKADMAPEGLLQKVTADDVSIGLGY------VCVRNRIGEETYEE-----ARMQEELLF 288
Query: 269 ANSPDYGHLANK-MGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPI 327
P + ++ +G LA+ L+Q +I +P I IN +E E + L PI
Sbjct: 289 RTHPMLSMINDEIVGIPVLAQKLTQIQGMMISRCLPEIERKINVKMEISVLEFNKL--PI 346
Query: 328 AL-DAGAQLYTILELC-RAFERIFK--EHLDGGRPGGDRIYHV----------FDNQLPA 373
+ G L +++++ A E + + H D D+ H F + L A
Sbjct: 347 VMASTGEALMSLMDIIGSAKESLLRILVHGDFSEYPNDQKMHCTARLAEMLNRFSDNLQA 406
Query: 374 ALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSYFRG-PAEASVDAV 432
+ + + L + KV+ E P + ++ + + P E +
Sbjct: 407 QPQAVTEE---FLMDEIKVLEERKCIGLPNFIPRSSFLAILSQHVDGIQDKPVEFIKEIW 463
Query: 433 NFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTLRLVDMESSYLTV 492
+++ E+V S+ T FP Q+++ A + R +E SK T+ +
Sbjct: 464 DYI--EVVLSSVITTYS-DNFPQIQSSIKRAGRNLMSRTKERSKTTSQQ----------- 509
Query: 493 DFFRRLPQEVEKAGGPATPNVDRYAEGHFRR-----------IASNVSSYIGLVADTLRN 541
+F + + K A P H R+ + ++SY +V LR
Sbjct: 510 NFINSVLHDENKPENFALPGFGDVKISHLRKYHAHLLQQAFDMKMRITSYWTIV---LRR 566
Query: 542 TIPKAVVFCQVRQAKQSLLNHFYTQ------IGKKEGKQLSEMLDEDPALMERRQQCAKR 595
+ ++ Q + ++L+N + + + + G + ML+E P++ +R++
Sbjct: 567 VVDNLALYLQF--SVKNLVNSQFQKEIVAEMVDPRAGGGIDRMLEESPSVASKREKLKNS 624
Query: 596 LELYKAARDEIDSV 609
L+L K ++D + ++
Sbjct: 625 LKLLKESKDVVAAI 638
>At1g53140 dynamin-like protein
Length = 260
Score = 94.4 bits (233), Expect = 2e-19
Identities = 65/205 (31%), Positives = 105/205 (50%), Gaps = 19/205 (9%)
Query: 37 LPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQL-HKL----------ETG 85
+P + +GGQS GKSS+LE+++G F R + TRRPL+LQ+ H L +
Sbjct: 62 IPEIVAIGGQSDGKSSLLEALLGFRFNVREVEMGTRRPLILQMVHDLSALEPRCRFQDED 121
Query: 86 SQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGKTKQ-ISPIPIHLSIYSPNVVNLTLV 144
S+EY + V I+ T+ + KTK +SP PI + + NLT++
Sbjct: 122 SEEYGS-------PIVSATAVADVIRSRTEALLKKTKTAVSPKPIVMRAEYAHCPNLTII 174
Query: 145 DLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDP 204
D PG A +G+PE+ EI +MV++ P+ I+L + ++ + +S + RE+D
Sbjct: 175 DTPGFVLKAKKGEPETTPDEILSMVKSLASPPHRILLFLQQSSVEWCSSLWLDAVREIDS 234
Query: 205 TGERTFGVLTKLDLMDKGTNALDVL 229
+ RT V++K D K N L +
Sbjct: 235 SFRRTIVVVSKFDNRLKVQNFLTTI 259
>At1g60530 unknown protein
Length = 175
Score = 59.7 bits (143), Expect = 5e-09
Identities = 41/85 (48%), Positives = 52/85 (60%), Gaps = 4/85 (4%)
Query: 37 LPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMP 96
LPT+ VVG QSSGKSSVLES+ G + LPRG GI TR PLV++L + + S E +L
Sbjct: 62 LPTIVVVGDQSSGKSSVLESLAGIN-LPRGQGICTRVPLVMRLQR--SSSPEPEIWLEYS 118
Query: 97 RRKF-TDFSLVRQEIQDETDRVTGK 120
+ TD V + I TD + GK
Sbjct: 119 DKVVPTDEEHVAEAICAATDVIAGK 143
>At3g19730 dynamin-like protein
Length = 157
Score = 41.2 bits (95), Expect = 0.002
Identities = 19/42 (45%), Positives = 28/42 (66%)
Query: 38 PTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQL 79
P V VVG Q+ GKS+++E+++G F G G TRRP+ L +
Sbjct: 49 PAVLVVGQQTDGKSALVEALMGFQFNHVGGGTKTRRPITLHM 90
>At3g19720 hypothetical protein
Length = 649
Score = 38.5 bits (88), Expect = 0.011
Identities = 49/215 (22%), Positives = 87/215 (39%), Gaps = 27/215 (12%)
Query: 140 NLTLVDLPGLTKVAVEGQPESI---VQEIETMVRTYVEKPNSIILAISPANQDIATSDAI 196
NLT++D PGL A + ++ + +E +VR ++ IIL + ++ D + +
Sbjct: 27 NLTIIDTPGLIAPAPGLKNRALQVQARAVEALVRAKMQHKEFIILCLEDSS-DWSIATTR 85
Query: 197 KLAREVDPTGERTFGVLTKLD--------------LMDKGTNALD-VLEGRSYRLQHPWV 241
++ +VDP RT V TKLD + +ALD L G S
Sbjct: 86 RIVMQVDPELSRTIVVSTKLDTKIPQFSCSSDVEVFLSPPASALDSSLLGDSPFFTSVPS 145
Query: 242 GIVNRSQADINKNVD---MIIARRKEREYFANSPDYGHLANK-----MGSEYLAKLLSQH 293
G V Q + K+ D ++ R+ + + G L K +G L L +
Sbjct: 146 GRVGYGQDSVYKSNDEFKQAVSLREMEDIASLEKKLGRLLTKQEKSRIGISKLRLFLEEL 205
Query: 294 LESVIRARIPSITSLINKSLEELESEMDHLGRPIA 328
L + +P I L+ K ++D + + ++
Sbjct: 206 LWKRYKESVPLIIPLLGKEYRSTVRKLDTVSKELS 240
>At2g13610 putative ABC transporter
Length = 649
Score = 32.0 bits (71), Expect = 1.0
Identities = 18/42 (42%), Positives = 24/42 (56%), Gaps = 3/42 (7%)
Query: 34 WEALPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPL 75
WE L A+VG +GKSS+LE + R GS V +RP+
Sbjct: 73 WEIL---AIVGPSGAGKSSLLEILAARLIPQTGSVYVNKRPV 111
>At5g28410 putative protein
Length = 106
Score = 31.6 bits (70), Expect = 1.4
Identities = 17/36 (47%), Positives = 25/36 (69%), Gaps = 7/36 (19%)
Query: 201 EVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRL 236
EVDP G+ +TK+DL D+GTNA++ GRS ++
Sbjct: 34 EVDPKGD-----MTKIDLKDQGTNAVET--GRSSKV 62
>At1g47250 20S proteasome subunit PAF2
Length = 277
Score = 31.6 bits (70), Expect = 1.4
Identities = 56/258 (21%), Positives = 99/258 (37%), Gaps = 42/258 (16%)
Query: 284 EYLAKLLSQHLESV-IRARIPSITSLINKSLEELESEM-------DHLGRPIA-LDAGAQ 334
EY + + Q ++ +R+R + + +NK+ EL S DH+G IA L A +
Sbjct: 23 EYAMEAVKQGSAAIGLRSRSHVVLACVNKAQSELSSHQRKIFKVDDHIGVAIAGLTADGR 82
Query: 335 LYTILELCRAFERIFKEHLDGGRPGGDRIYHVFDNQLPAALR--KLPFDRHLSLQNVRKV 392
+ + + F + P G + H+ D R K P+ V +
Sbjct: 83 VLSRYMRSESINHSFT--YESPLPVGRLVVHLADKAQVCTQRSWKRPY-------GVGLL 133
Query: 393 VSEADGYQPHLI--APEQGYRRL--------IDSALSYFRGPAEASVDAVNFVLKELVRK 442
V D HL P Y +A +Y E+ ++ ++L++
Sbjct: 134 VGGLDESGAHLYYNCPSGNYFEYQAFAIGSRSQAAKTYLERKFESFQESSK---EDLIKD 190
Query: 443 SIAETKELKRFPTFQAALATAANEALERFREESKKTTLRLVDMESSYLTVDFFRRLPQEV 502
+I +E + T +++L T + ++ +D ES +D F ++P+E
Sbjct: 191 AIMAIRETLQGETLKSSLCTVSVLGVDE--------PFHFLDQESIQKVIDTFEKVPEEE 242
Query: 503 EKAG-GPATPNVDRYAEG 519
E AG G A P A G
Sbjct: 243 EDAGEGEAEPEAAPGAAG 260
>At5g01500 unknown protein
Length = 415
Score = 30.4 bits (67), Expect = 3.0
Identities = 23/75 (30%), Positives = 36/75 (47%), Gaps = 14/75 (18%)
Query: 404 IAPEQGYRRLIDSALSYFR-----------GPAEASVD---AVNFVLKELVRKSIAETKE 449
+A E GYR + AL+ R GP+ S+ A+NF + +LV+KS+ E +
Sbjct: 243 LAVEPGYRTMSQVALNMLREEGVASFYNGLGPSLLSIAPYIAINFCVFDLVKKSLPEKYQ 302
Query: 450 LKRFPTFQAALATAA 464
K + A+ AA
Sbjct: 303 QKTQSSLLTAVVAAA 317
>At3g28860 P-glycoprotein, putative
Length = 1252
Score = 30.0 bits (66), Expect = 3.9
Identities = 17/42 (40%), Positives = 27/42 (63%), Gaps = 1/42 (2%)
Query: 29 SFSSLWEALPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIV 70
+F+ + + TVAVVGG SGKS+V+ S++ R + P I+
Sbjct: 385 NFNIFFPSGKTVAVVGGSGSGKSTVV-SLIERFYDPNSGQIL 425
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,955,031
Number of Sequences: 26719
Number of extensions: 548145
Number of successful extensions: 1758
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1698
Number of HSP's gapped (non-prelim): 39
length of query: 613
length of database: 11,318,596
effective HSP length: 105
effective length of query: 508
effective length of database: 8,513,101
effective search space: 4324655308
effective search space used: 4324655308
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0316.4


