

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0315.1
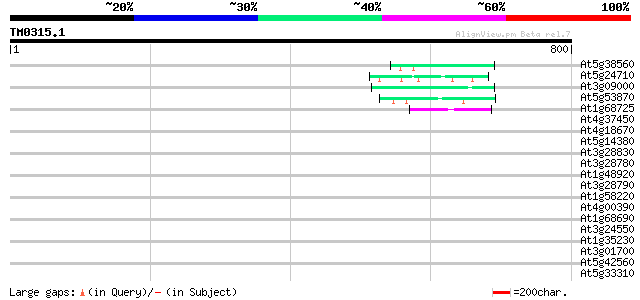
(800 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g38560 putative protein 46 9e-05
At5g24710 unknown protein 45 2e-04
At3g09000 unknown protein 45 2e-04
At5g53870 predicted GPI-anchored protein 44 4e-04
At1g68725 putative arabinogalactan protein AGP19 43 6e-04
At4g37450 arabinogalactan protein AGP18 42 0.001
At4g18670 extensin-like protein 42 0.001
At5g14380 agp6 42 0.001
At3g28830 hypothetical protein 41 0.002
At3g28780 histone-H4-like protein 41 0.002
At1g48920 unknown protein 40 0.004
At3g28790 unknown protein 40 0.005
At1g58220 MYB-family transcription factor-like protein 40 0.005
At4g00390 unknown protein 40 0.007
At1g68690 protein kinase, putative 40 0.007
At3g24550 protein kinase, putative 39 0.009
At1g35230 arabinogalactan-protein AGP5 39 0.009
At3g01700 arabinogalactan-protein AGP11 39 0.012
At5g42560 unknown protein 39 0.015
At5g33310 putative protein 38 0.026
>At5g38560 putative protein
Length = 681
Score = 45.8 bits (107), Expect = 9e-05
Identities = 42/161 (26%), Positives = 62/161 (38%), Gaps = 12/161 (7%)
Query: 543 LTKASSDAGTSNPG---AQPTTAAAPKGKNVAEAS-------VAAATEPTAVPASSPASA 592
L+ SS++ T+ P QPTT +AP + V+++ P V + P+S+
Sbjct: 10 LSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSSS 69
Query: 593 GATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSP--PS 650
+ S A+S A+ TP +T PP +A PSP P+
Sbjct: 70 PPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAPT 129
Query: 651 THDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 691
T + PSP GE P T S P P +S
Sbjct: 130 TTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTS 170
Score = 40.4 bits (93), Expect = 0.004
Identities = 35/143 (24%), Positives = 55/143 (37%), Gaps = 9/143 (6%)
Query: 547 SSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGA 606
SS +S+P P +P +VA++ P V AS P S AT A ++
Sbjct: 62 SSPPPSSSPPPSPPVITSPP------PTVASSPPPPVVIASPPPSTPATTPPAPPQTVSP 115
Query: 607 TAA-SAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSP-PSTHDAGSMPSPPHQ- 663
A + ++ P ETP P +PP P PST + SPP
Sbjct: 116 PPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPP 175
Query: 664 GEKSCPGAATTSEAAQIEQAPAP 686
+ P ++ ++ + + P P
Sbjct: 176 ATSASPPSSNPTDPSTLAPPPTP 198
>At5g24710 unknown protein
Length = 1003
Score = 45.1 bits (105), Expect = 2e-04
Identities = 52/199 (26%), Positives = 74/199 (37%), Gaps = 33/199 (16%)
Query: 514 PKFNTEGDAKKRG-------GAENQAESTKAPKRRRLTKASSDAGTSNPGA--------- 557
PK + DA ++ G + S APK+ LT+ ++ + P A
Sbjct: 785 PKLTSLSDASRKPPIEILPPGMSSIFASITAPKKPLLTQKTAQPEVAKPLALEEPTKPLA 844
Query: 558 --QPTTAAAPKGKNVAEASVAAATEP---TAVPASSPASAGATAAVAAESSIGATAASAG 612
P ++ AP+ ++ E + AAA P TA A SPA A A A S A
Sbjct: 845 IEAPPSSEAPQTESAPETA-AAAESPAPETAAVAESPAPGTAAVAEAPASETAAAPVDGP 903
Query: 613 VNATKAAASSDTPIGDEE---KENETPKSPPRQNAPPSPPSTHDAGSMP-----SPPHQG 664
V T S P+ EE +E P S P S +T + P +PP
Sbjct: 904 VTET---VSEPPPVEKEETSLEEKSDPSSTPNTETATSTENTSQTTTTPESVTTAPPEPI 960
Query: 665 EKSCPGAATTSEAAQIEQA 683
+ P TT+ E A
Sbjct: 961 TTAPPETVTTTAVKPTENA 979
>At3g09000 unknown protein
Length = 541
Score = 44.7 bits (104), Expect = 2e-04
Identities = 36/176 (20%), Positives = 62/176 (34%), Gaps = 5/176 (2%)
Query: 517 NTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVA 576
N D + Q S+ RR + + S TS P + P +
Sbjct: 121 NCREDIVSGNNNKPQTSSSSVAGLRRPSSSGSSRSTSRPATPTRRSTTPTTSTSRPVTTR 180
Query: 577 AATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTP-IGDEEKENET 635
A+ ++ P S A A + + T +S + S+ P +K
Sbjct: 181 ASNSRSSTPTSRATLTAARATTSTAAPRTTTTSSGSARSATPTRSNPRPSSASSKKPVSR 240
Query: 636 PKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 691
P +P R+ + P+ PS + P +G P + S+A +P+P + SS
Sbjct: 241 PATPTRRPSTPTGPSIVSS----KAPSRGTSPSPTVNSLSKAPSRGTSPSPTLNSS 292
Score = 37.0 bits (84), Expect = 0.044
Identities = 41/180 (22%), Positives = 66/180 (35%), Gaps = 22/180 (12%)
Query: 534 STKAPKRRRLTKASSDAGTSNP---GAQPTTA-AAPKGKNVAEASVAAAT------EPTA 583
ST P R + + S TS A+ TT+ AAP+ + S +AT P++
Sbjct: 172 STSRPVTTRASNSRSSTPTSRATLTAARATTSTAAPRTTTTSSGSARSATPTRSNPRPSS 231
Query: 584 VPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQN 643
+ P S AT + G + S+ + + S + T SP +
Sbjct: 232 ASSKKPVSRPATPTRRPSTPTGPSIVSSKAPSRGTSPSPTVNSLSKAPSRGTSPSPTLNS 291
Query: 644 APPSPPSTHDAGSMPSPPH----------QGEKSCPGAATT--SEAAQIEQAPAPEVGSS 691
+ P P S+ +PP+ + PG A+ S + IE+ P G S
Sbjct: 292 SRPWKPPEMPGFSLEAPPNLRTTLADRPVSASRGRPGVASAPGSRSGSIERGGGPTSGGS 351
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 43.9 bits (102), Expect = 4e-04
Identities = 48/190 (25%), Positives = 70/190 (36%), Gaps = 29/190 (15%)
Query: 528 AENQAESTKAPKRRRLTKA----SSDAGTSNPGAQPTTAAA-------PKGKNVAEASVA 576
A A S P R ++ A SS + P P+ A + P K+ + S +
Sbjct: 161 APASAPSKSQPPRSSVSPAQPPKSSSPISHTPALSPSHATSHSPATPSPSPKSPSPVSHS 220
Query: 577 AATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETP 636
+ P P+ SPA + + A S A A S + A A S +P TP
Sbjct: 221 PSHSPAHTPSHSPAHTPSHSPAHAPSHSPAHAPSH----SPAHAPSHSPAHSPSHSPATP 276
Query: 637 KSPPRQNAP--------------PSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQ 682
KSP ++P PSP S+ +P Q P + T+ A
Sbjct: 277 KSPSPSSSPAQSPATPSPMTPQSPSPVSSPSPDQSAAPSDQSTPLAPSPSETTPTADNIT 336
Query: 683 APAPEVGSSS 692
APAP ++S
Sbjct: 337 APAPSPRTNS 346
Score = 33.9 bits (76), Expect = 0.37
Identities = 35/134 (26%), Positives = 49/134 (36%), Gaps = 3/134 (2%)
Query: 86 RSNSPLHATHLPKTIMTSNLPHASNTRHPSSFSPSPIKTLLPHCHPFPRPLFTSLIAYQT 145
+S+SP+ +H P + H+ T PS SPSP+ H P P +
Sbjct: 183 KSSSPI--SHTPALSPSHATSHSPATPSPSPKSPSPVSHSPSH-SPAHTPSHSPAHTPSH 239
Query: 146 PSAHFRSGAVAYHPFRYPLFTSYSGESHCPYLYIAHILIFSFLSLHCLLKMASNPTSPNH 205
AH S + A+ P P +H P A S S +P +P
Sbjct: 240 SPAHAPSHSPAHAPSHSPAHAPSHSPAHSPSHSPATPKSPSPSSSPAQSPATPSPMTPQS 299
Query: 206 SSSSSESDPDSTAA 219
S S PD +AA
Sbjct: 300 PSPVSSPSPDQSAA 313
Score = 31.2 bits (69), Expect = 2.4
Identities = 31/119 (26%), Positives = 48/119 (40%), Gaps = 7/119 (5%)
Query: 574 SVAAATEPTAVPAS-SPASAGATAAVAAESSIGATAASAGVNATKAAASSDTP-IGDEEK 631
S + PT P S SP S A A+ ++S ++ S ++ S TP +
Sbjct: 140 SPVPSVSPTQPPKSHSPVSPVAPASAPSKSQPPRSSVSPAQPPKSSSPISHTPALSPSHA 199
Query: 632 ENETPKSP---PRQNAPPSPPSTHDAGSMP--SPPHQGEKSCPGAATTSEAAQIEQAPA 685
+ +P +P P+ +P S +H P SP H S A + S A +PA
Sbjct: 200 TSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHSPAHTPSHSPAHAPSHSPAHAPSHSPA 258
Score = 29.3 bits (64), Expect = 9.1
Identities = 25/108 (23%), Positives = 42/108 (38%), Gaps = 5/108 (4%)
Query: 566 KGKNVAEASVAAATEPTAVPASSPA-SAGATAAVAAESSIGATAASAGVNATKAAASSDT 624
KG+ + +A +P+A PA SP S T + S + A ++ + ++ SS +
Sbjct: 119 KGQKLIVVVLAVRNQPSA-PAHSPVPSVSPTQPPKSHSPVSPVAPASAPSKSQPPRSSVS 177
Query: 625 PIGDEEKEN---ETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCP 669
P + + TP P SP + + PSP P
Sbjct: 178 PAQPPKSSSPISHTPALSPSHATSHSPATPSPSPKSPSPVSHSPSHSP 225
>At1g68725 putative arabinogalactan protein AGP19
Length = 248
Score = 43.1 bits (100), Expect = 6e-04
Identities = 32/119 (26%), Positives = 52/119 (42%), Gaps = 10/119 (8%)
Query: 570 VAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDE 629
+A A +++ + PA+SP ++ TA ++ TAA T +++ P
Sbjct: 12 LASALISSFSVNAQGPAASPVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPP---- 67
Query: 630 EKENETPKSPPRQNAPPSPPSTHDAG--SMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 686
+P +PP P SPP+ A S +PP Q +S P +A T + PAP
Sbjct: 68 ----ASPVTPPPAVTPTSPPAPKVAPVISPATPPPQPPQSPPASAPTVSPPPVSPPPAP 122
Score = 37.4 bits (85), Expect = 0.033
Identities = 40/145 (27%), Positives = 54/145 (36%), Gaps = 21/145 (14%)
Query: 546 ASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIG 605
+++ A A PTTAA P S A +P A P + P + T+ A +
Sbjct: 34 STTTAPPPTTAAPPTTAAPPPTTTTPPVS---AAQPPASPVTPPPAVTPTSPPAPK---- 86
Query: 606 ATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGE 665
A AT +P + P SPP AP SPP T P+ P
Sbjct: 87 --VAPVISPATPPPQPPQSPPASAPTVSPPPVSPP--PAPTSPPPT------PASPPPAP 136
Query: 666 KSCPGAATTSEAAQIE----QAPAP 686
S P A + A + QAP+P
Sbjct: 137 ASPPPAPASPPPAPVSPPPVQAPSP 161
Score = 35.8 bits (81), Expect = 0.097
Identities = 38/143 (26%), Positives = 45/143 (30%), Gaps = 12/143 (8%)
Query: 546 ASSDAGTSNPGAQPTTAA--APKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESS 603
+S P A P T+ AP A + AA T P P SA A
Sbjct: 18 SSFSVNAQGPAASPVTSTTTAPPPTTAAPPTTAAPPPTTTTP---PVSAAQPPASPVTPP 74
Query: 604 IGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQ 663
T S S TP + P+SPP SPP + SPP
Sbjct: 75 PAVTPTSPPAPKVAPVISPATP------PPQPPQSPPASAPTVSPPPVSPPPAPTSPP-P 127
Query: 664 GEKSCPGAATTSEAAQIEQAPAP 686
S P A + A PAP
Sbjct: 128 TPASPPPAPASPPPAPASPPPAP 150
Score = 32.7 bits (73), Expect = 0.82
Identities = 29/117 (24%), Positives = 42/117 (35%), Gaps = 5/117 (4%)
Query: 553 SNPGAQPTTAAAPKGKNVAEASV--AAATEPTAVPASSPASAGATAAVAAESSIGATAAS 610
S P + PT + P A S A+ P A + PA A A + + A +
Sbjct: 103 SPPASAPTVSPPPVSPPPAPTSPPPTPASPPPAPASPPPAPASPPPAPVSPPPVQAPSPI 162
Query: 611 AGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPP---STHDAGSMPSPPHQG 664
+ A A + ++ + P P +PPSPP D PSP G
Sbjct: 163 SLPPAPAPAPTKHKRKHKHKRHHHAPAPAPIPPSPPSPPVLTDPQDTAPAPSPNTNG 219
>At4g37450 arabinogalactan protein AGP18
Length = 209
Score = 42.4 bits (98), Expect = 0.001
Identities = 36/135 (26%), Positives = 58/135 (42%), Gaps = 11/135 (8%)
Query: 553 SNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAG 612
S+P PTT +AP + A T PT PA +P +A A++ V + S + S+
Sbjct: 26 SSPTKSPTTPSAP---TTSPTKSPAVTSPTTAPAKTP-TASASSPVESPKSPAPVSESSP 81
Query: 613 VNATKAAASSDTPIGDEEKENETPKSP-PRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGA 671
+S P +P P P ++PP+P + +P+P K
Sbjct: 82 PPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAA-PVADVPAPAPSKHKK---- 136
Query: 672 ATTSEAAQIEQAPAP 686
TT ++ + + APAP
Sbjct: 137 -TTKKSKKHQAAPAP 150
Score = 36.2 bits (82), Expect = 0.075
Identities = 37/148 (25%), Positives = 50/148 (33%), Gaps = 9/148 (6%)
Query: 535 TKAPKRRRLTKASSD---AGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVP-ASSPA 590
TK+P T A + A S+P P + A + V ++ P P SSP
Sbjct: 43 TKSPAVTSPTTAPAKTPTASASSPVESPKSPAPVSESSPPPTPVPESSPPVPAPMVSSPV 102
Query: 591 SAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPS 650
S+ A A+S AA A + +K P P PP+PP+
Sbjct: 103 SSPPVPAPVADSPPAPVAAPVADVPAPAPSKHKKTTKKSKKHQAAPAPAPELLGPPAPPT 162
Query: 651 THDAGSMPSPPHQGEKSCPGAATTSEAA 678
P P P A S AA
Sbjct: 163 -----ESPGPNSDAFSPGPSADDQSGAA 185
Score = 33.1 bits (74), Expect = 0.63
Identities = 28/115 (24%), Positives = 43/115 (37%), Gaps = 18/115 (15%)
Query: 581 PTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPP 640
P + P SP + A +S + +A A+ASS E+PKSP
Sbjct: 24 PISSPTKSPTTPSAPTTSPTKSPAVTSPTTAPAKTPTASASSPV---------ESPKSPA 74
Query: 641 --RQNAPPSPPSTHDAGSMPSPPHQGEKSCP-------GAATTSEAAQIEQAPAP 686
+++PP P + +P+P S P + AA + PAP
Sbjct: 75 PVSESSPPPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAAPVADVPAP 129
>At4g18670 extensin-like protein
Length = 839
Score = 42.4 bits (98), Expect = 0.001
Identities = 35/115 (30%), Positives = 47/115 (40%), Gaps = 9/115 (7%)
Query: 547 SSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGA 606
S + T +PG+ PT+ P +S T PT P SP S+ T
Sbjct: 457 SPPSTTPSPGSPPTSPTTPTPGGSPPSS---PTTPT--PGGSPPSSPTTPTPGGSPPSSP 511
Query: 607 TAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPP 661
T S G + + S PI + TP SP +PPSP S + +PSPP
Sbjct: 512 TTPSPGGSPPSPSISPSPPITVPSPPS-TPTSPG---SPPSPSSPTPSSPIPSPP 562
Score = 41.6 bits (96), Expect = 0.002
Identities = 35/133 (26%), Positives = 52/133 (38%), Gaps = 12/133 (9%)
Query: 559 PTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKA 618
P T +P G + + V + T P S P S + S T G + +
Sbjct: 439 PPTTPSPGGSPPSPSIVPSPPSTTPSPGSPPTSPTTPTPGGSPPSSPTTPTPGG--SPPS 496
Query: 619 AASSDTPIGDEEKENETPK---SPPRQNAPPSPPSTHDAGSMPSPPH--QGEKSCPGAAT 673
+ ++ TP G TP SPP + PSPP T +PSPP S P ++
Sbjct: 497 SPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSPPIT-----VPSPPSTPTSPGSPPSPSS 551
Query: 674 TSEAAQIEQAPAP 686
+ ++ I P P
Sbjct: 552 PTPSSPIPSPPTP 564
Score = 35.4 bits (80), Expect = 0.13
Identities = 32/140 (22%), Positives = 58/140 (40%), Gaps = 8/140 (5%)
Query: 526 GGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVP 585
GG+ + +T P + ++ +P + PTT + P G + + + P
Sbjct: 478 GGSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPS-PGGSPPSPSISPSPPITVPSP 536
Query: 586 ASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQN-A 644
S+P S G+ + ++ + S+ + + ++ TPI + SPP +
Sbjct: 537 PSTPTSPGSPPSPSSPTP------SSPIPSPPTPSTPPTPISPGQNSPPIIPSPPFTGPS 590
Query: 645 PPSPPSTHDAGSMPSPPHQG 664
PPS PS +PSPP G
Sbjct: 591 PPSSPSPPLPPVIPSPPIVG 610
Score = 30.4 bits (67), Expect = 4.1
Identities = 31/135 (22%), Positives = 48/135 (34%), Gaps = 7/135 (5%)
Query: 526 GGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVP 585
GG+ + +T P + SS S G+ P+ + +P + + T P + P
Sbjct: 491 GGSPPSSPTTPTPGG---SPPSSPTTPSPGGSPPSPSISPSPPITVPSPPSTPTSPGSPP 547
Query: 586 ASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAP 645
+ S + + S T S G N+ S G + +P PP
Sbjct: 548 SPSSPTPSSPIPSPPTPSTPPTPISPGQNSPPIIPSPPFT-GPSPPSSPSPPLPP---VI 603
Query: 646 PSPPSTHDAGSMPSP 660
PSPP S P P
Sbjct: 604 PSPPIVGPTPSSPPP 618
>At5g14380 agp6
Length = 150
Score = 42.0 bits (97), Expect = 0.001
Identities = 38/136 (27%), Positives = 56/136 (40%), Gaps = 22/136 (16%)
Query: 544 TKASSDAGTSNPGAQPTTAAAPKGKNVA--EASVAAATEPTAVPASSPASAGATAAVAAE 601
T ++DA +++P P+ AAP A +A A P A P SS AS+ ++ AAE
Sbjct: 17 TAFAADAPSASPKKSPSPTAAPTKAPTATTKAPSAPTKAPAAAPKSSSASSPKASSPAAE 76
Query: 602 SSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSP-PSTHDAGSMPSP 660
+ SA + A A P ++PP+P P + A PS
Sbjct: 77 GPVPEDDYSASSPSDSAEA-------------------PTVSSPPAPTPDSTSAADGPSD 117
Query: 661 PHQGEKSCPGAATTSE 676
E GA TT++
Sbjct: 118 GPTAESPKSGAVTTAK 133
Score = 32.7 bits (73), Expect = 0.82
Identities = 22/101 (21%), Positives = 41/101 (39%), Gaps = 3/101 (2%)
Query: 595 TAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSP---PRQNAPPSPPST 651
T +A + A +AS + + AA + P + + K+P P+ ++ SP ++
Sbjct: 12 TLTIATAFAADAPSASPKKSPSPTAAPTKAPTATTKAPSAPTKAPAAAPKSSSASSPKAS 71
Query: 652 HDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 692
A P P S P + + AP P+ S++
Sbjct: 72 SPAAEGPVPEDDYSASSPSDSAEAPTVSSPPAPTPDSTSAA 112
>At3g28830 hypothetical protein
Length = 536
Score = 41.2 bits (95), Expect = 0.002
Identities = 32/138 (23%), Positives = 67/138 (48%), Gaps = 7/138 (5%)
Query: 546 ASSDAGTSNPGAQPTTAA-APKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSI 604
ASS++ ++ G+ ++ + + K K E+S + ++ +V S S+G +AA ++ S
Sbjct: 197 ASSESSSTKSGSVSSSGSVSTKSK---ESSSSGSSASGSVATKSKESSGGSAATKSKESS 253
Query: 605 GATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQG 664
G +AA+ ++ +A++ G + +PK+ P + S A + S QG
Sbjct: 254 GGSAATKSKESSGGSATTGKTSGSP---SGSPKASPSGSVSGKSSSKGSASAQGSASAQG 310
Query: 665 EKSCPGAATTSEAAQIEQ 682
S G+A+ +A ++
Sbjct: 311 SASAQGSASAQGSASAQR 328
Score = 36.2 bits (82), Expect = 0.075
Identities = 34/178 (19%), Positives = 66/178 (36%), Gaps = 20/178 (11%)
Query: 515 KFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEAS 574
K ++ + G +++ + +K SS + + + +A GK S
Sbjct: 220 KESSSSGSSASGSVATKSKESSGGSAATKSKESSGGSAATKSKESSGGSATTGKTSGSPS 279
Query: 575 VAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDE--EKE 632
+ P+ + +S G+ +A + S+ G+ +A +A +A++ G K
Sbjct: 280 GSPKASPSGSVSGKSSSKGSASAQGSASAQGSASAQGSASAQGSASAQRRESGAMAMSKS 339
Query: 633 NETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGS 690
ET S RQ+ S S+ + TT+ Q+E + EV S
Sbjct: 340 RETKTSSQRQSKSSSESSS------------------SSTTTTTVKQVESETSKEVMS 379
Score = 33.5 bits (75), Expect = 0.48
Identities = 51/245 (20%), Positives = 94/245 (37%), Gaps = 41/245 (16%)
Query: 498 SAELLKARKRRMARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRL------TKASSDAG 551
SA K + +A + + + G A +++ STK+ +K SS +G
Sbjct: 167 SASEQKGKSINIASYGLDVDVNDSSIVGGAASSESSSTKSGSVSSSGSVSTKSKESSSSG 226
Query: 552 TSNPGAQPTTAAAPKGKNVA----------------EASVAAAT------EPTAVPASSP 589
+S G+ T + G + A E+S +AT P+ P +SP
Sbjct: 227 SSASGSVATKSKESSGGSAATKSKESSGGSAATKSKESSGGSATTGKTSGSPSGSPKASP 286
Query: 590 --------ASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIG--DEEKENETPKSP 639
+S G+ +A + S+ G+ +A +A +A++ G K ET S
Sbjct: 287 SGSVSGKSSSKGSASAQGSASAQGSASAQGSASAQGSASAQRRESGAMAMSKSRETKTSS 346
Query: 640 PRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPN 699
RQ+ S S+ + + E S S Q+E+ A + ++ L +
Sbjct: 347 QRQSKSSSESSSSSTTTTTVKQVESETS---KEVMSFIMQLEKKYAAKAELKVFFESLKS 403
Query: 700 AIEPS 704
+++ S
Sbjct: 404 SMQAS 408
>At3g28780 histone-H4-like protein
Length = 614
Score = 41.2 bits (95), Expect = 0.002
Identities = 34/132 (25%), Positives = 55/132 (40%), Gaps = 3/132 (2%)
Query: 547 SSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGA 606
S+ G ++ + A+A G EA+ + E AS+ A++G +A ES G
Sbjct: 281 SASGGAASGAGAASGASAKTGGESGEAASGGSAETGGESASAGAASGGSAETGGESGSGG 340
Query: 607 TAASAGVNATKAAASSDTPI--GDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQG 664
AAS G +A+ A S +P G E E+ + +GS+ S
Sbjct: 341 -AASGGESASGGATSGGSPETGGSAETGGESASGGAASGGESASGGAASSGSVESGGEST 399
Query: 665 EKSCPGAATTSE 676
+ G+A TS+
Sbjct: 400 GATSGGSAETSD 411
Score = 39.7 bits (91), Expect = 0.007
Identities = 47/183 (25%), Positives = 70/183 (37%), Gaps = 18/183 (9%)
Query: 517 NTEGDAKKRGGA----ENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAE 572
+T GD+ G+ + + A T S G S+ G ++A A
Sbjct: 165 DTTGDSGSSAGSPSYPSDDGSGSTAGGPSGSTTDGSAGGESSMGGDSSSAGAAG----ES 220
Query: 573 ASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKE 632
S A A A A S +AGA + AA + G A+AG A+ AA++DT G E
Sbjct: 221 GSAATADSGDAAGADSGGAAGADSGGAASADSGG--AAAGETASGGAAAADTSGGSAETG 278
Query: 633 NETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATT---SEAAQIEQAPAPEVG 689
E+ A + ++ G GE + G+A T S +A + E G
Sbjct: 279 GESASGGAASGAGAASGASAKTGG-----ESGEAASGGSAETGGESASAGAASGGSAETG 333
Query: 690 SSS 692
S
Sbjct: 334 GES 336
Score = 37.0 bits (84), Expect = 0.044
Identities = 43/174 (24%), Positives = 62/174 (34%), Gaps = 23/174 (13%)
Query: 520 GDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAAT 579
G+A G AE ES A ++ G++ G + + A G A +
Sbjct: 305 GEAASGGSAETGGESASA--------GAASGGSAETGGESGSGGAASGGESASGGATSGG 356
Query: 580 EPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSP 639
P G +A ES+ G AAS G +A+ AASS G E E+ +
Sbjct: 357 SP---------ETGGSAETGGESASGG-AASGGESASGGAASS----GSVESGGESTGAT 402
Query: 640 PRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATT-SEAAQIEQAPAPEVGSSS 692
+A S S + G + G+A T E+ A GS S
Sbjct: 403 SGGSAETSDESASGGAASGGESASGGAASGGSAETGGESTSSGVASGGSTGSES 456
Score = 32.7 bits (73), Expect = 0.82
Identities = 46/204 (22%), Positives = 75/204 (36%), Gaps = 31/204 (15%)
Query: 517 NTEGDAKKRGGAENQAESTKAPKRRRLTKASSDA-------GTSNPGAQPTTAAAPKGKN 569
+ G A G E+ EST A + A G S G + +A G
Sbjct: 381 SASGGAASSGSVESGGESTGATSGGSAETSDESASGGAASGGESASGGAASGGSAETGGE 440
Query: 570 VAEASVAAATEPTAVPASS-PASAGATAAVAAESSIGATAASAGVN---------ATKAA 619
+ VA+ + AS+ AS G+T A ++ G+T A +G + + A
Sbjct: 441 STSSGVASGGSTGSESASAGAASGGSTEANGGAAAGGSTEAGSGTSTETSSMGGGSAAAG 500
Query: 620 ASSDTPIGDEEKENETPKSPPRQNAPPSPPS----THDAGSMPSPPHQGEKSCPGAATTS 675
S++ G T +S +A S T G P+ G S G + ++
Sbjct: 501 GVSESSSGGSTAAGGTSESASGGSATAGGASGGTYTDSTGGSPT----GSPSAGGPSGSA 556
Query: 676 EAAQIE------QAPAPEVGSSSY 693
+ +E Q+ + GS+SY
Sbjct: 557 SESSMEGGTFGGQSMGGQAGSASY 580
Score = 32.3 bits (72), Expect = 1.1
Identities = 43/186 (23%), Positives = 65/186 (34%), Gaps = 13/186 (6%)
Query: 518 TEGDAKKRGG-AENQAESTKAPKRRRLTKASSDAGTSNP---GAQPTTAAAPKGKNVAEA 573
T G + + GG AE ES AS A +S G + T A + ++
Sbjct: 353 TSGGSPETGGSAETGGESASGGAASGGESASGGAASSGSVESGGESTGATSGGSAETSDE 412
Query: 574 SVAAATEPTAVPASSPASAGATAAVAAESSIGATAA--SAGVNATKAAASSDTPIGDEEK 631
S + AS A++G +A ES+ A+ S G + A A+S G E
Sbjct: 413 SASGGAASGGESASGGAASGGSAETGGESTSSGVASGGSTGSESASAGAASG---GSTEA 469
Query: 632 ENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAAT----TSEAAQIEQAPAPE 687
+ + T G + +S G +T TSE+A A A
Sbjct: 470 NGGAAAGGSTEAGSGTSTETSSMGGGSAAAGGVSESSSGGSTAAGGTSESASGGSATAGG 529
Query: 688 VGSSSY 693
+Y
Sbjct: 530 ASGGTY 535
>At1g48920 unknown protein
Length = 557
Score = 40.4 bits (93), Expect = 0.004
Identities = 40/188 (21%), Positives = 69/188 (36%), Gaps = 17/188 (9%)
Query: 505 RKRRMARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAA 564
+++ + + K + D+ E +A+ A K + SSD +S+ P A A
Sbjct: 50 KEKAVKKVPKKVESSDDSDSESEEEEKAKKVPAKKAASSSDESSDDSSSDDEPAPKKAVA 109
Query: 565 PKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDT 624
VA+ S +SS + VA A A + V A K ++S D
Sbjct: 110 ATNGTVAKKSKD--------DSSSSDDDSSDEEVAVTKKPAAAAKNGSVKAKKESSSED- 160
Query: 625 PIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGE-----KSCPGAATTSEAAQ 679
D E+E K P + A P+ + + + E K+ P AA + ++
Sbjct: 161 ---DSSSEDEPAKKPAAKIAKPAAKDSSSSDDDSDEDSEDEKPATKKAAPAAAKAASSSD 217
Query: 680 IEQAPAPE 687
+ E
Sbjct: 218 SSDEDSDE 225
Score = 34.3 bits (77), Expect = 0.28
Identities = 44/226 (19%), Positives = 81/226 (35%), Gaps = 26/226 (11%)
Query: 484 LSRTPKYLEDMNFTSAELLKARKRRMARFSPKFNTEGDAKKRGGAENQAESTKA------ 537
+ + PK +E + + +E + K + ++ ++ ++++ KA
Sbjct: 54 VKKVPKKVESSDDSDSESEEEEKAKKVPAKKAASSSDESSDDSSSDDEPAPKKAVAATNG 113
Query: 538 --PKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVA-----EASVAAATEPTAVPASSPA 590
K+ + +SSD +S+ T A KN + E+S + PA PA
Sbjct: 114 TVAKKSKDDSSSSDDDSSDEEVAVTKKPAAAAKNGSVKAKKESSSEDDSSSEDEPAKKPA 173
Query: 591 SAGATAAVAAESSIG-----------ATAASAGVNATKAAASSDTPIGDEEKENETPKSP 639
+ A A SS A A KAA+SSD+ DE+ + E+
Sbjct: 174 AKIAKPAAKDSSSSDDDSDEDSEDEKPATKKAAPAAAKAASSSDS--SDEDSDEESEDEK 231
Query: 640 PRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPA 685
P Q + S + S + E +++ +E A
Sbjct: 232 PAQKKADTKASKKSSSDESSESEEDESEDEEETPKKKSSDVEMVDA 277
Score = 30.8 bits (68), Expect = 3.1
Identities = 45/212 (21%), Positives = 82/212 (38%), Gaps = 21/212 (9%)
Query: 443 EKPNGITDPSEDSLSANDKAFLNLLAPLPILDCTTVLEGADLSRTPKYLEDMNFTSAELL 502
+K +D S D S++D+ AP + T A S+ D + + E+
Sbjct: 83 KKAASSSDESSDDSSSDDEP-----APKKAVAATNGTV-AKKSKDDSSSSDDDSSDEEVA 136
Query: 503 KARKRRMARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSN-------- 554
+K A + + ++ + ++ E K P + A+ D+ +S+
Sbjct: 137 VTKKPAAAAKNGSVKAKKESSSEDDSSSEDEPAKKPAAKIAKPAAKDSSSSDDDSDEDSE 196
Query: 555 ---PGAQPTTAAAPKGKNVAEASVAAATEPTA--VPASSPASAGATAAVAA-ESSIGATA 608
P + AA K + +++S + E + PA A A+ ++ ESS
Sbjct: 197 DEKPATKKAAPAAAKAASSSDSSDEDSDEESEDEKPAQKKADTKASKKSSSDESSESEED 256
Query: 609 ASAGVNATKAAASSDTPIGDEEKEN-ETPKSP 639
S T SSD + D EK + + PK+P
Sbjct: 257 ESEDEEETPKKKSSDVEMVDAEKSSAKQPKTP 288
>At3g28790 unknown protein
Length = 608
Score = 40.0 bits (92), Expect = 0.005
Identities = 45/189 (23%), Positives = 75/189 (38%), Gaps = 36/189 (19%)
Query: 517 NTEGDAKKRGGAENQA--ESTKAPKRRRLTKASS-----DAGTSNPGAQPT--------- 560
+T G + G++ +A +S+ + K + ++ SS D S+ GA P+
Sbjct: 226 STAGSIETNTGSKTEAGSKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPST 285
Query: 561 -TAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAES----------SIGATAA 609
T + P ++ +T + PA S +AG T+ +ES S +AA
Sbjct: 286 PTPSTPTPSTPTPSTPTPSTPTPSTPAPSTPAAGKTSEKGSESASMKKESNSKSESESAA 345
Query: 610 SAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCP 669
S V+ TK + GD K+ S +P P+ PS G+ S
Sbjct: 346 SGSVSKTKETNKGSS--GDTYKDTTGTSSGSPSGSPSGSPT-------PSTSTDGKASSK 396
Query: 670 GAATTSEAA 678
G+A+ S A
Sbjct: 397 GSASASAGA 405
Score = 38.1 bits (87), Expect = 0.020
Identities = 34/133 (25%), Positives = 63/133 (46%), Gaps = 15/133 (11%)
Query: 513 SPKFNTEGDAKKRGGAENQAESTKA---PKRRRLTKASSDAGTSNPGAQPTTAAAPKGKN 569
S K + KK +++++ES + K + K SS G + T++ +P G
Sbjct: 322 SEKGSESASMKKESNSKSESESAASGSVSKTKETNKGSS--GDTYKDTTGTSSGSPSGSP 379
Query: 570 VAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNA-------TKAAASS 622
+ + +T+ A SS SA A+A +A +S GA+A++ A +K+++SS
Sbjct: 380 SGSPTPSTSTDGKA---SSKGSASASAGASASASAGASASAEESAASQKKESNSKSSSSS 436
Query: 623 DTPIGDEEKENET 635
+ +E E +T
Sbjct: 437 SSTTSVKEVETQT 449
Score = 36.6 bits (83), Expect = 0.057
Identities = 34/149 (22%), Positives = 54/149 (35%), Gaps = 11/149 (7%)
Query: 544 TKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESS 603
+ +S + +SN +Q T++ + G S+ T S +S+ T V+ SS
Sbjct: 202 SSSSDNESSSNTKSQGTSSKS--GSESTAGSIETNTGSKTEAGSKSSSSAKTKEVSGGSS 259
Query: 604 IGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQ 663
+ G ++ + + S TP TP +P PS P+ P
Sbjct: 260 GNTYKDTTGSSSGASPSGSPTP---------TPSTPTPSTPTPSTPTPSTPTPSTPTPST 310
Query: 664 GEKSCPGAATTSEAAQIEQAPAPEVGSSS 692
S P A TSE + E S S
Sbjct: 311 PAPSTPAAGKTSEKGSESASMKKESNSKS 339
Score = 33.5 bits (75), Expect = 0.48
Identities = 26/120 (21%), Positives = 52/120 (42%), Gaps = 4/120 (3%)
Query: 528 AENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPAS 587
+E +ES K S A + + T G + + ++ P+ P+
Sbjct: 322 SEKGSESASMKKESNSKSESESAASGSVSKTKETNKGSSGDTYKDTTGTSSGSPSGSPSG 381
Query: 588 SPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPS 647
SP + +T A SS G+ +ASAG A+ +A++ + +E ++ +S + ++ S
Sbjct: 382 SPTPSTSTDGKA--SSKGSASASAG--ASASASAGASASAEESAASQKKESNSKSSSSSS 437
Score = 30.8 bits (68), Expect = 3.1
Identities = 32/191 (16%), Positives = 65/191 (33%), Gaps = 4/191 (2%)
Query: 504 ARKRRMARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAA 563
A KR + + + D G+ + ++ + + + S +S G++ T +
Sbjct: 171 AEKRGKSIDESSYGLDVDVNASIGSSSGSDGSSSSDNESSSNTKSQGTSSKSGSESTAGS 230
Query: 564 APKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSD 623
+ + ++ S S+G T SS GA+ + + S
Sbjct: 231 IETNTGSKTEAGSKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPST 290
Query: 624 TPIGDEEKENETPKSPPRQNAPPSPPS---THDAGSMPSPPHQGEKSCPGAATTSEAAQI 680
TP +P PS P+ T + GS S + E + + ++ + +
Sbjct: 291 PTPSTPTPSTPTPSTPTPSTPAPSTPAAGKTSEKGS-ESASMKKESNSKSESESAASGSV 349
Query: 681 EQAPAPEVGSS 691
+ GSS
Sbjct: 350 SKTKETNKGSS 360
>At1g58220 MYB-family transcription factor-like protein
Length = 834
Score = 40.0 bits (92), Expect = 0.005
Identities = 43/151 (28%), Positives = 59/151 (38%), Gaps = 10/151 (6%)
Query: 543 LTKASSDAGTSNPGAQPTTAAAPK---GKNVAEASVAAATEP--TAVPASSPASAGATAA 597
+ A+S A + T PK GKN A V TEP TA S P +G ++A
Sbjct: 353 MVTANSVAAAACMSGLATAVTVPKIEPGKNAVSALVPK-TEPVKTASTVSMPRPSGISSA 411
Query: 598 VAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPP--STHDAG 655
+ E A AAS ++ +A P+ P+ +AP P +T
Sbjct: 412 LNTEPVKTAVAASLPRSSGIISAPKVEPVKTAASAASLPRPSGMISAPKVEPVKTTASVA 471
Query: 656 SMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 686
S+P P G S P A AA +P P
Sbjct: 472 SLPRP--SGIISAPKAEPVKTAASAASSPRP 500
Score = 37.0 bits (84), Expect = 0.044
Identities = 46/181 (25%), Positives = 63/181 (34%), Gaps = 14/181 (7%)
Query: 514 PKFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEA 573
P T G++ R A S A + PG +A PK + V A
Sbjct: 338 PVKKTTGNSTSRADLMVTANSVAAAACMSGLATAVTVPKIEPGKNAVSALVPKTEPVKTA 397
Query: 574 SV-----------AAATEP--TAVPASSPASAGATAAVAAES-SIGATAASAGVNATKAA 619
S A TEP TAV AS P S+G +A E A+AAS + +
Sbjct: 398 STVSMPRPSGISSALNTEPVKTAVAASLPRSSGIISAPKVEPVKTAASAASLPRPSGMIS 457
Query: 620 ASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQ 679
A P+ P+ +AP + P A + SP G S P + A
Sbjct: 458 APKVEPVKTTASVASLPRPSGIISAPKAEPVKTAASAASSPRPSGMISAPKVESVKTTAS 517
Query: 680 I 680
+
Sbjct: 518 M 518
Score = 33.1 bits (74), Expect = 0.63
Identities = 42/183 (22%), Positives = 69/183 (36%), Gaps = 20/183 (10%)
Query: 522 AKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTA---AAPKGKNVAEASVAAA 578
A K + A + +P+ + A + P + +APK + V A+ AA+
Sbjct: 482 APKAEPVKTAASAASSPRPSGMISAPKVESVKTTASMPRPSGIISAPKAELVKSAASAAS 541
Query: 579 TEPTAVPASSP-----ASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIG------ 627
T+ SSP SA + A+ SS+ + + V AAA++ +G
Sbjct: 542 LPCTSGIISSPKAELVKSAASAASFPRPSSMLSAPKADPVKIVPAAATNTKSVGPLNLRH 601
Query: 628 --DEEKENETPKSP---PRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQ 682
+ + P SP P AP S ST + S+P P + P + Q
Sbjct: 602 AVNGSPNHTIPSSPFTKPLHMAPLSKGSTIQSNSVP-PSFASSRLVPTQRAPAATVVTPQ 660
Query: 683 APA 685
P+
Sbjct: 661 KPS 663
Score = 31.2 bits (69), Expect = 2.4
Identities = 35/139 (25%), Positives = 49/139 (35%), Gaps = 7/139 (5%)
Query: 553 SNPGAQPTTAAA-----PKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGAT 607
S P +P AA P+ + A + TA AS P +G +A AE T
Sbjct: 433 SAPKVEPVKTAASAASLPRPSGMISAPKVEPVKTTASVASLPRPSGIISAPKAEPV--KT 490
Query: 608 AASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKS 667
AASA + + S + + P+ +AP + A + P G S
Sbjct: 491 AASAASSPRPSGMISAPKVESVKTTASMPRPSGIISAPKAELVKSAASAASLPCTSGIIS 550
Query: 668 CPGAATTSEAAQIEQAPAP 686
P A AA P P
Sbjct: 551 SPKAELVKSAASAASFPRP 569
Score = 29.3 bits (64), Expect = 9.1
Identities = 37/167 (22%), Positives = 65/167 (38%), Gaps = 9/167 (5%)
Query: 513 SPKFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAE 572
+PK A + APK + +S A P +APK + V
Sbjct: 434 APKVEPVKTAASAASLPRPSGMISAPKVEPVKTTASVASLPRPSG---IISAPKAEPVKT 490
Query: 573 ASVAAATEPTAVPASSP--ASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEE 630
A+ AA++ + S+P S TA++ S I + + V + +AAS G
Sbjct: 491 AASAASSPRPSGMISAPKVESVKTTASMPRPSGIISAPKAELVKSAASAASLPCTSG--- 547
Query: 631 KENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEA 677
+PK+ ++A + + + +P K P AAT +++
Sbjct: 548 -IISSPKAELVKSAASAASFPRPSSMLSAPKADPVKIVPAAATNTKS 593
>At4g00390 unknown protein
Length = 364
Score = 39.7 bits (91), Expect = 0.007
Identities = 27/108 (25%), Positives = 52/108 (48%), Gaps = 2/108 (1%)
Query: 568 KNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIG 627
K++ + AA + TAV A++PA + + +A A S +AA+ + +++ SD+
Sbjct: 35 KSLPATTAAAPAKSTAVSAATPAKSTSVSAAAPSKSTAVSAAADSDSGSESETDSDSEST 94
Query: 628 DEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTS 675
D K ++ PS S+ ++P+ +++ AATTS
Sbjct: 95 DPPKSGSGKTIASKKKDDPS--SSTATLALPAVKSGAKRAASEAATTS 140
>At1g68690 protein kinase, putative
Length = 708
Score = 39.7 bits (91), Expect = 0.007
Identities = 37/171 (21%), Positives = 64/171 (36%), Gaps = 24/171 (14%)
Query: 552 TSNPGAQPTTAAAPKGKNVAEASV----AAATEPTAVPASSPASAGATAAVAAESS---I 604
+++P QP + P + A V ++ P +VP P+ + + S I
Sbjct: 93 STSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILVRSPPPSVRPI 152
Query: 605 GATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQN--------APPSPPSTHDAGS 656
+ T++ P E+ ++P SPP + +PPSPPS + S
Sbjct: 153 QSPPPPPSDRPTQSPPPPSPPSPPSERPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQS 212
Query: 657 MPSPPHQGEKSCPGAATTSEAAQIEQAP--APEVGSSSYYNMLPNAIEPSE 705
P PP + P + S P PE+ ++P + PS+
Sbjct: 213 PPPPPEDTKPQPPRRSPNSPPPTFSSPPRSPPEI-------LVPGSNNPSQ 256
>At3g24550 protein kinase, putative
Length = 652
Score = 39.3 bits (90), Expect = 0.009
Identities = 29/120 (24%), Positives = 44/120 (36%), Gaps = 14/120 (11%)
Query: 579 TEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKS 638
T P+ P S P ++ T A SS T + + + S+ P + P S
Sbjct: 9 TTPSPSPPSPPTNSTTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSLPPPS 68
Query: 639 PPRQNAPPSPPSTHDAGSMPSPP--------------HQGEKSCPGAATTSEAAQIEQAP 684
PP PP P + A PSPP +QG + P +T + + +P
Sbjct: 69 PPGSLTPPLPQPSPSAPITPSPPSPTTPSNPRSPPSPNQGPPNTPSGSTPRTPSNTKPSP 128
Score = 32.7 bits (73), Expect = 0.82
Identities = 27/124 (21%), Positives = 42/124 (33%), Gaps = 8/124 (6%)
Query: 554 NPGAQPTTAAAPKGKNVAEAS---VAAATEPTAVPASSPASAGATAAVAAESS-----IG 605
+PG P+ + N + A++ PT P+S P S + SS +
Sbjct: 6 SPGTTPSPSPPSPPTNSTTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSLP 65
Query: 606 ATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGE 665
+ + S PI TP +P +P P +GS P P +
Sbjct: 66 PPSPPGSLTPPLPQPSPSAPITPSPPSPTTPSNPRSPPSPNQGPPNTPSGSTPRTPSNTK 125
Query: 666 KSCP 669
S P
Sbjct: 126 PSPP 129
Score = 32.0 bits (71), Expect = 1.4
Identities = 23/92 (25%), Positives = 34/92 (36%), Gaps = 3/92 (3%)
Query: 616 TKAAASSDTPIGDEEKENETPKSPPRQ---NAPPSPPSTHDAGSMPSPPHQGEKSCPGAA 672
T + ++ P TP SPP N+ PPS+ S+P P G + P
Sbjct: 20 TNSTTTTPPPAASSPPPTTTPSSPPPSPSTNSTSPPPSSPLPPSLPPPSPPGSLTPPLPQ 79
Query: 673 TTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPS 704
+ A P+P S+ PN P+
Sbjct: 80 PSPSAPITPSPPSPTTPSNPRSPPSPNQGPPN 111
>At1g35230 arabinogalactan-protein AGP5
Length = 133
Score = 39.3 bits (90), Expect = 0.009
Identities = 27/108 (25%), Positives = 42/108 (38%), Gaps = 4/108 (3%)
Query: 573 ASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKE 632
AS A P P SP A T + + ++ A A S N +A ++ P+ +
Sbjct: 16 ASSVVAQAPGPAPTISPLPATPTPSQSPRAT--APAPSPSANPPPSAPTTAPPVS--QPP 71
Query: 633 NETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQI 680
E+P +PP +P P T+ P P AA S + +
Sbjct: 72 TESPPAPPTSTSPSGAPGTNVPSGEAGPAQSPLSGSPNAAAVSRVSLV 119
>At3g01700 arabinogalactan-protein AGP11
Length = 136
Score = 38.9 bits (89), Expect = 0.012
Identities = 31/110 (28%), Positives = 47/110 (42%), Gaps = 9/110 (8%)
Query: 570 VAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDE 629
+A +V AA P+A P +SP + A AA S ++A A+ A TP D
Sbjct: 13 LAVGTVFAADAPSAAPTASPTKSPTKAPAAAPKS-----SAAAPKASSPVAEEPTPEDDY 67
Query: 630 EKENETPKS-PPRQNAPPSPPSTHDAGSMPSPPHQG---EKSCPGAATTS 675
+ + + P ++PP+P D S P G +S ATT+
Sbjct: 68 SAASPSDSAEAPTVSSPPAPTPEADGPSSDGPSSDGPAAAESPKSGATTN 117
Score = 37.4 bits (85), Expect = 0.033
Identities = 39/115 (33%), Positives = 46/115 (39%), Gaps = 13/115 (11%)
Query: 544 TKASSDAGTSNPGAQPT------TAAAPKGKNVA-EASVAAATEPTAVPASSPASAGATA 596
T ++DA ++ P A PT AAAPK A +AS A EPT S AS +A
Sbjct: 17 TVFAADAPSAAPTASPTKSPTKAPAAAPKSSAAAPKASSPVAEEPTPEDDYSAASPSDSA 76
Query: 597 AVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPST 651
SS A A SSD P D E+PKS N S T
Sbjct: 77 EAPTVSSPPAPTPEAD------GPSSDGPSSDGPAAAESPKSGATTNVKLSIAGT 125
>At5g42560 unknown protein
Length = 296
Score = 38.5 bits (88), Expect = 0.015
Identities = 34/123 (27%), Positives = 55/123 (44%), Gaps = 19/123 (15%)
Query: 523 KKRGGAENQAEST--------KAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEAS 574
+KRGG NQA + P+ L+ +SS + + N G +PT + V+ S
Sbjct: 154 QKRGGRANQAPAKPKKAPVPQSEPEEVSLSSSSSSSSSENEGNEPT-------RKVSGPS 206
Query: 575 VAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPI-GDEEKEN 633
T T+VPA+ P +AG T A+ S+ + + + T+ + G+ E N
Sbjct: 207 RPRPT-VTSVPAADPKNAGTTQ--IAQKSVASPIVNPPQSTTQVEPMQIEEVEGEAESGN 263
Query: 634 ETP 636
E P
Sbjct: 264 ENP 266
>At5g33310 putative protein
Length = 1137
Score = 37.7 bits (86), Expect = 0.026
Identities = 40/158 (25%), Positives = 59/158 (37%), Gaps = 14/158 (8%)
Query: 292 PGDRPWLHKPDGDVKRSHFFFAYEYMFSELGIRLPFSPFVQTVLRDINAAPRQLHPNAWA 351
P + P H+P FF YE F G+ P + L + A QL PN
Sbjct: 91 PTESPEDHRPG-------FFCVYEIYFKGCGLTFPLPEALVRYLAALEIALPQLTPNLLR 143
Query: 352 FIRCFEILSAAVGIAPSPTSFFYLYDVDPKSIKNKGWISLKARAGRKCLHPHKSNAKSSF 411
I C ++A G L V S K G+ S A + L + N ++
Sbjct: 144 TILCIITIAAEAGFVIRVPELNELLSVRSSS-KKTGYFSAYPNANQN-LISNLLNKDENW 201
Query: 412 ARKYFRVAVHPAYPEAFTLRDGTALFPLYWTEKPNGIT 449
+F V PA ++ + + + P W+ KP IT
Sbjct: 202 HHPWFLVKKTPA-----SIGNLSDMLPSKWSAKPPFIT 234
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,057,327
Number of Sequences: 26719
Number of extensions: 875391
Number of successful extensions: 5521
Number of sequences better than 10.0: 233
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 180
Number of HSP's that attempted gapping in prelim test: 4203
Number of HSP's gapped (non-prelim): 862
length of query: 800
length of database: 11,318,596
effective HSP length: 107
effective length of query: 693
effective length of database: 8,459,663
effective search space: 5862546459
effective search space used: 5862546459
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0315.1


