

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
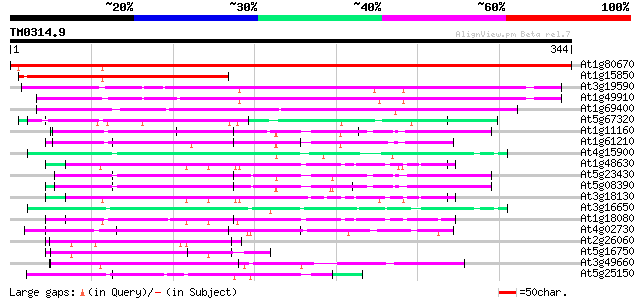
Query= TM0314.9
(344 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g80670 mRNA export protein, putative 613 e-176
At1g15850 hypothetical protein 207 5e-54
At3g19590 mitotic checkpoint protein, putative 205 3e-53
At1g49910 mitotic checkpoint protein, putative 194 6e-50
At1g69400 hypothetical protein 117 1e-26
At5g67320 unknown protein 70 2e-12
At1g11160 hypothetical protein 64 1e-10
At1g61210 64 2e-10
At4g15900 PRL1 protein 62 3e-10
At1g48630 unknown protein 62 5e-10
At5g23430 unknown protein 62 6e-10
At5g08390 katanin p80 subunit - like protein 61 8e-10
At3g18130 protein kinase C-receptor/G-protein, putative 61 1e-09
At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2 60 1e-09
At1g18080 putative guanine nucleotide-binding protein beta sub... 60 1e-09
At4g02730 putative WD-repeat protein 59 3e-09
At2g26060 unknown protein 57 2e-08
At5g16750 WD40-repeat protein 56 3e-08
At3g49660 putative WD-40 repeat - protein 56 3e-08
At5g25150 transcription initiation factor IID-associated factor-... 55 6e-08
>At1g80670 mRNA export protein, putative
Length = 349
Score = 613 bits (1582), Expect = e-176
Identities = 281/349 (80%), Positives = 319/349 (90%), Gaps = 5/349 (1%)
Query: 1 MSTF--LSNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN--- 55
M+TF + N NPNKS+EV P+DS+SSL FSP+A++LVATSWDNQVRCWE++R+
Sbjct: 1 MATFGAPATANSNPNKSYEVTPSPADSISSLSFSPRADILVATSWDNQVRCWEISRSGAS 60
Query: 56 VATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDI 115
+A+APKASISHDQPVLCSAWKDDGTTVFSGGCDKQ KMWPLLSGGQP+TVAMH+ P+ +
Sbjct: 61 LASAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQAKMWPLLSGGQPVTVAMHEGPIAAM 120
Query: 116 AWIPEMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVY 175
AWIP M+LL TGSWD+T+KYWDTRQ NPVHTQQLP++CY +SVKHPLMVVGTADRN++V+
Sbjct: 121 AWIPGMNLLATGSWDKTLKYWDTRQQNPVHTQQLPDKCYTLSVKHPLMVVGTADRNLIVF 180
Query: 176 NLQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCH 235
NLQNPQ EFKRI SPLKYQTRC+ AFPDQQGFLVGSIEGRVGVHHLDDSQQ KNFTFKCH
Sbjct: 181 NLQNPQTEFKRIQSPLKYQTRCVTAFPDQQGFLVGSIEGRVGVHHLDDSQQSKNFTFKCH 240
Query: 236 REGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDG 295
R+GN+IYSVNSLNFHPVH TFATAGSDGAFNFWDKDSKQRLKAM RC+QPIPCS+FN+DG
Sbjct: 241 RDGNDIYSVNSLNFHPVHGTFATAGSDGAFNFWDKDSKQRLKAMSRCNQPIPCSSFNHDG 300
Query: 296 SIFAYSVCYDWSKGAENHNPTTAKTNIYLHLPQESEVKGKPRIGATGRK 344
SI+AY+ CYDWSKGAENHNP TAK++I+LHLPQESEVK KPR+GATGRK
Sbjct: 301 SIYAYAACYDWSKGAENHNPATAKSSIFLHLPQESEVKAKPRVGATGRK 349
>At1g15850 hypothetical protein
Length = 140
Score = 207 bits (528), Expect = 5e-54
Identities = 96/132 (72%), Positives = 112/132 (84%), Gaps = 4/132 (3%)
Query: 6 SNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN---VATAPKA 62
S+TN NPN S+E+ P +DS+SSL FSPKA++LVATSWD QVRCWE+ R+ +A+ PK
Sbjct: 10 SSTN-NPNNSYEITPPATDSISSLSFSPKADILVATSWDCQVRCWEITRSDGSIASEPKV 68
Query: 63 SISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMS 122
S+SHDQPVLCSAWKDDGTTVF+GGCDKQ KMWPLLSG QP TVAMHDAP IAWIP M+
Sbjct: 69 SMSHDQPVLCSAWKDDGTTVFTGGCDKQAKMWPLLSGAQPSTVAMHDAPFNQIAWIPGMN 128
Query: 123 LLVTGSWDRTIK 134
LLVTGSWD+T+K
Sbjct: 129 LLVTGSWDKTLK 140
>At3g19590 mitotic checkpoint protein, putative
Length = 340
Score = 205 bits (522), Expect = 3e-53
Identities = 119/342 (34%), Positives = 189/342 (54%), Gaps = 20/342 (5%)
Query: 8 TNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHD 67
T P+ E++ PPSD +S+L FS ++ L+ +SWD +VR ++V+ N + K H
Sbjct: 2 TTVTPSAGRELSNPPSDGISNLRFSNNSDHLLVSSWDKRVRLYDVSTN---SLKGEFLHG 58
Query: 68 QPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTG 127
VL + DD + FS G D +V+ + + G+ + HD V+ + + ++TG
Sbjct: 59 GAVLDCCFHDDFSG-FSVGADYKVRRI-VFNVGKEDILGTHDKAVRCVEYSYAAGQVITG 116
Query: 128 SWDRTIKYWDTR-----QPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQV 182
SWD+T+K WD R + V T PER Y+MS+ +VV TA R++ +Y+L+N
Sbjct: 117 SWDKTVKCWDPRGASGPERTQVGTYLQPERVYSMSLVGHRLVVATAGRHVNIYDLRNMSQ 176
Query: 183 EFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLD--DSQQGKNFTFKCHREGNE 240
+R S LKYQTRC+ +P+ G+ + S+EGRV + D ++ Q K + FKCHR+
Sbjct: 177 PEQRRESSLKYQTRCVRCYPNGTGYALSSVEGRVAMEFFDLSEAAQAKKYAFKCHRKSEA 236
Query: 241 ----IYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGS 296
+Y VNS+ FHP++ TFAT G DG N WD ++K+RL + I +F+ DG
Sbjct: 237 GRDIVYPVNSIAFHPIYGTFATGGCDGFVNIWDGNNKKRLYQYSKYPTSISALSFSRDGQ 296
Query: 297 IFAYSVCYDWSKGAENHNPTTAKTNIYLHLPQESEVKGKPRI 338
+ A + Y + +G ++ P I++ E EVK KP++
Sbjct: 297 LLAVASSYTFEEGEKSQEPEA----IFVRSVNEIEVKPKPKV 334
>At1g49910 mitotic checkpoint protein, putative
Length = 339
Score = 194 bits (493), Expect = 6e-50
Identities = 111/333 (33%), Positives = 183/333 (54%), Gaps = 20/333 (6%)
Query: 17 EVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWK 76
E++ PPSD +S+L FS ++ L+ +SWD VR ++ ++ + H VL +
Sbjct: 10 ELSNPPSDGISNLRFSNNSDHLLVSSWDKSVRLYDANGDLM---RGEFKHGGAVLDCCFH 66
Query: 77 DDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYW 136
DD + FS D +V+ +G + + + H+ PV+ + + ++TGSWD+TIK W
Sbjct: 67 DDSSG-FSVCADTKVRRIDFNAGKEDV-LGTHEKPVRCVEYSYAAGQVITGSWDKTIKCW 124
Query: 137 DTR-----QPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPL 191
D R + + T PER ++S+ +VV TA R++ +Y+L+N +R S L
Sbjct: 125 DPRGASGTERTQIGTYMQPERVNSLSLVGNRLVVATAGRHVNIYDLRNMSQPEQRRESSL 184
Query: 192 KYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQ--QGKNFTFKCHREGNE----IYSVN 245
KYQTRC+ +P+ G+ + S+EGRV + D S+ Q K + FKCHR+ + +Y VN
Sbjct: 185 KYQTRCVRCYPNGTGYALSSVEGRVSMEFFDLSEAAQAKKYAFKCHRKSEDGRDIVYPVN 244
Query: 246 SLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYD 305
++ FHP++ TFA+ G DG N WD ++K+RL + I +F+ DG + A + Y
Sbjct: 245 AIAFHPIYGTFASGGCDGFVNIWDGNNKKRLYQYSKYPTSIAALSFSRDGGLLAVASSYT 304
Query: 306 WSKGAENHNPTTAKTNIYLHLPQESEVKGKPRI 338
+ +G + H P I++ E EVK KP++
Sbjct: 305 FEEGDKPHEPDA----IFVRSVNEIEVKPKPKV 333
>At1g69400 hypothetical protein
Length = 314
Score = 117 bits (292), Expect = 1e-26
Identities = 84/302 (27%), Positives = 148/302 (48%), Gaps = 13/302 (4%)
Query: 17 EVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWK 76
E P D+VS L FSP++N L+ SWD+ +R ++V + + S Q L
Sbjct: 7 EFENPIEDAVSRLRFSPQSNNLLVASWDSYLRLYDVESSSLSLELNS----QAALLDCCF 62
Query: 77 DDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYW 136
++ +T F+ G D ++ + L + G T+ HD I + E +++ +D IK+W
Sbjct: 63 ENESTSFTSGSDGFIRRYDL-NAGTVDTIGRHDDISTSIVYSYEKGEVISTGFDEKIKFW 121
Query: 137 DTRQPNP-VHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQT 195
DTRQ V + ++V +VV D ++ +Y+L+N F+ S ++
Sbjct: 122 DTRQRESLVFSTDAGGAVGCVTVSGNNLVV-CVDASMHIYDLRNLDEAFQSYASQVEVPI 180
Query: 196 RCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGK-NFTFKCH---REGN-EIYSVNSLNFH 250
RC+ + P +G+ VGS++GRV V + S + ++F+CH R G + +N++ F
Sbjct: 181 RCITSVPYSRGYAVGSVDGRVAVDFPNTSCSSEIKYSFRCHPKSRNGRLDGVCINAIEFS 240
Query: 251 PV-HHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYDWSKG 309
P TF T ++G W+ S++RL + R S I AF++ G + A + + +
Sbjct: 241 PCGSGTFVTGDNEGYVISWNAKSRRRLNELPRYSNSIASLAFDHTGELLAIASSHTYQDA 300
Query: 310 AE 311
E
Sbjct: 301 KE 302
>At5g67320 unknown protein
Length = 613
Score = 69.7 bits (169), Expect = 2e-12
Identities = 68/320 (21%), Positives = 121/320 (37%), Gaps = 36/320 (11%)
Query: 6 SNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEV----------ARN 55
S T+ PN + + + V + +SP A+LL + S D R W + RN
Sbjct: 248 SQTSHIPNSDVRILEGHTSEVCACAWSPSASLLASGSGDATARIWSIPEGSFKAVHTGRN 307
Query: 56 VAT-----APKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDA 110
+ A S + V W +GT + +G CD Q ++W L +G T++ H
Sbjct: 308 INALILKHAKGKSNEKSKDVTTLDWNGEGTLLATGSCDGQARIWTL-NGELISTLSKHKG 366
Query: 111 PVKDIAWIPEMSLLVTGSWDRTIKYWDT-----RQPNPVHTQQLPERCYAMSVKHPLMVV 165
P+ + W + L+TGS DRT WD +Q H+ + + +V
Sbjct: 367 PIFSLKWNKKGDYLLTGSVDRTAVVWDVKAEEWKQQFEFHSGPTLDVDWRNNVS-----F 421
Query: 166 GTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQ 225
T+ + ++Y + + + + + + C+ P S + + ++ S
Sbjct: 422 ATSSTDSMIYLCKIGETRPAKTFTGHQGEVNCVKWDPTGSLLASCSDDSTAKIWNIKQS- 480
Query: 226 QGKNFTFKCHREGNEIYSVN------SLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAM 279
F EIY++ N T A+A D WD + + L +
Sbjct: 481 ---TFVHDLREHTKEIYTIRWSPTGPGTNNPNKQLTLASASFDSTVKLWDAELGKMLCSF 537
Query: 280 LRCSQPIPCSAFNNDGSIFA 299
+P+ AF+ +G A
Sbjct: 538 NGHREPVYSLAFSPNGEYIA 557
Score = 46.6 bits (109), Expect = 2e-05
Identities = 61/258 (23%), Positives = 93/258 (35%), Gaps = 25/258 (9%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEV-ARNVATAPKASISHDQPVLCSAWKDDGTT 81
S V++L ++ + LL S D Q R W + ++T K H P+ W G
Sbjct: 324 SKDVTTLDWNGEGTLLATGSCDGQARIWTLNGELISTLSK----HKGPIFSLKWNKKGDY 379
Query: 82 VFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTI---KYWDT 138
+ +G D+ +W + + H P D+ W +S T S D I K +T
Sbjct: 380 LLTGSVDRTAVVWDVKAEEWKQQFEFHSGPTLDVDWRNNVS-FATSSTDSMIYLCKIGET 438
Query: 139 RQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNL-QNPQVEFKRIISPLKYQTRC 197
R Q C L+ + D ++N+ Q+ V R + Y R
Sbjct: 439 RPAKTFTGHQGEVNCVKWDPTGSLLASCSDDSTAKIWNIKQSTFVHDLREHTKEIYTIRW 498
Query: 198 LAAFP------DQQGFLVGSIEGRVGVHHLDDSQQGKNF-TFKCHREGNEIYSVNSLNFH 250
P Q S + V L D++ GK +F HRE V SL F
Sbjct: 499 SPTGPGTNNPNKQLTLASASFDSTV---KLWDAELGKMLCSFNGHRE-----PVYSLAFS 550
Query: 251 PVHHTFATAGSDGAFNFW 268
P A+ D + + W
Sbjct: 551 PNGEYIASGSLDKSIHIW 568
Score = 45.1 bits (105), Expect = 6e-05
Identities = 34/144 (23%), Positives = 59/144 (40%), Gaps = 14/144 (9%)
Query: 12 PNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVL 71
P K+F +Q V+ + + P +LL + S D+ + W + ++ T H + +
Sbjct: 440 PAKTFTGHQ---GEVNCVKWDPTGSLLASCSDDSTAKIWNIKQS--TFVHDLREHTKEIY 494
Query: 72 CSAWKDDGT---------TVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMS 122
W G T+ S D VK+W G + H PV +A+ P
Sbjct: 495 TIRWSPTGPGTNNPNKQLTLASASFDSTVKLWDAELGKMLCSFNGHREPVYSLAFSPNGE 554
Query: 123 LLVTGSWDRTIKYWDTRQPNPVHT 146
+ +GS D++I W ++ V T
Sbjct: 555 YIASGSLDKSIHIWSIKEGKIVKT 578
>At1g11160 hypothetical protein
Length = 974
Score = 63.9 bits (154), Expect = 1e-10
Identities = 35/111 (31%), Positives = 54/111 (48%), Gaps = 2/111 (1%)
Query: 27 SSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGG 86
S++ F P L + S D +R W+ + + H + + + DG V SGG
Sbjct: 53 SAVEFHPFGEFLASGSSDTNLRVWDTRKKGCI--QTYKGHTRGISTIEFSPDGRWVVSGG 110
Query: 87 CDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWD 137
D VK+W L +G H+ P++ + + P LL TGS DRT+K+WD
Sbjct: 111 LDNVVKVWDLTAGKLLHEFKCHEGPIRSLDFHPLEFLLATGSADRTVKFWD 161
Score = 59.3 bits (142), Expect = 3e-09
Identities = 55/201 (27%), Positives = 89/201 (43%), Gaps = 22/201 (10%)
Query: 103 MTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNPVHT-QQLPERCYAMSVKHP 161
M++ H +PV +A+ E L++ G+ IK WD + V C A+ HP
Sbjct: 1 MSLCGHTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEF-HP 59
Query: 162 L---MVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAF---PDQQGFLVGSIEGR 215
+ G++D N+ V++ + K I K TR ++ PD + + G ++
Sbjct: 60 FGEFLASGSSDTNLRVWDTRK-----KGCIQTYKGHTRGISTIEFSPDGRWVVSGGLDNV 114
Query: 216 VGVHHLDDSQQGKNF-TFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQ 274
V V D GK FKCH EG + SL+FHP+ AT +D FWD ++ +
Sbjct: 115 VKVW---DLTAGKLLHEFKCH-EG----PIRSLDFHPLEFLLATGSADRTVKFWDLETFE 166
Query: 275 RLKAMLRCSQPIPCSAFNNDG 295
+ + + AF+ DG
Sbjct: 167 LIGTTRPEATGVRAIAFHPDG 187
Score = 52.8 bits (125), Expect = 3e-07
Identities = 43/192 (22%), Positives = 76/192 (39%), Gaps = 7/192 (3%)
Query: 26 VSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSG 85
V S+ F+ + L++A + ++ W++ + +A H + G + SG
Sbjct: 10 VDSVAFNSEEVLVLAGASSGVIKLWDLEESKMV--RAFTGHRSNCSAVEFHPFGEFLASG 67
Query: 86 GCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNPVH 145
D +++W G T H + I + P+ +V+G D +K WD +H
Sbjct: 68 SSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVWDLTAGKLLH 127
Query: 146 TQQLPERCYAMSVKHP---LMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFP 202
+ E HP L+ G+ADR + ++L+ E P R +A P
Sbjct: 128 EFKCHEGPIRSLDFHPLEFLLATGSADRTVKFWDLET--FELIGTTRPEATGVRAIAFHP 185
Query: 203 DQQGFLVGSIEG 214
D Q G +G
Sbjct: 186 DGQTLFCGLDDG 197
>At1g61210
Length = 282
Score = 63.5 bits (153), Expect = 2e-10
Identities = 34/111 (30%), Positives = 55/111 (48%), Gaps = 2/111 (1%)
Query: 27 SSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGG 86
S++ F P L + S D ++ W++ + + H + + + DG V SGG
Sbjct: 134 SAVEFHPFGEFLASGSSDANLKIWDIRKKGCI--QTYKGHSRGISTIRFTPDGRWVVSGG 191
Query: 87 CDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWD 137
D VK+W L +G H+ P++ + + P LL TGS DRT+K+WD
Sbjct: 192 LDNVVKVWDLTAGKLLHEFKFHEGPIRSLDFHPLEFLLATGSADRTVKFWD 242
Score = 53.5 bits (127), Expect = 2e-07
Identities = 55/230 (23%), Positives = 95/230 (40%), Gaps = 37/230 (16%)
Query: 64 ISHDQPVLC-SAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDA------------ 110
++H V C S K +GG D +V +W + G+P ++ +DA
Sbjct: 32 LAHSANVNCLSIGKKTSRLFITGGDDYKVNLWAI---GKPTSLMKNDAIAFYWQSLCGHT 88
Query: 111 -PVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNPVHT-QQLPERCYAMSVKHPL---MVV 165
V +A+ L++ G+ IK WD + V C A+ HP +
Sbjct: 89 SAVDSVAFDSAEVLVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEF-HPFGEFLAS 147
Query: 166 GTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAF---PDQQGFLVGSIEGRVGVHHLD 222
G++D N+ +++++ K I K +R ++ PD + + G ++ V V L
Sbjct: 148 GSSDANLKIWDIRK-----KGCIQTYKGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDLT 202
Query: 223 DSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDS 272
+ F F EG + SL+FHP+ AT +D FWD ++
Sbjct: 203 AGKLLHEFKF---HEG----PIRSLDFHPLEFLLATGSADRTVKFWDLET 245
Score = 53.1 bits (126), Expect = 2e-07
Identities = 37/159 (23%), Positives = 66/159 (41%), Gaps = 5/159 (3%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
+ +V S+ F L++A + ++ W+V A +A H + G +
Sbjct: 88 TSAVDSVAFDSAEVLVLAGASSGVIKLWDVEE--AKMVRAFTGHRSNCSAVEFHPFGEFL 145
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPN 142
SG D +K+W + G T H + I + P+ +V+G D +K WD
Sbjct: 146 ASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDLTAGK 205
Query: 143 PVHTQQLPERCYAMSVKHP---LMVVGTADRNILVYNLQ 178
+H + E HP L+ G+ADR + ++L+
Sbjct: 206 LLHEFKFHEGPIRSLDFHPLEFLLATGSADRTVKFWDLE 244
Score = 29.6 bits (65), Expect = 2.5
Identities = 30/142 (21%), Positives = 55/142 (38%), Gaps = 15/142 (10%)
Query: 162 LMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRC-------LAAFPDQQGF-LVGSIE 213
L + G D + ++ + P K +Q+ C AF + L G+
Sbjct: 50 LFITGGDDYKVNLWAIGKPTSLMKNDAIAFYWQSLCGHTSAVDSVAFDSAEVLVLAGASS 109
Query: 214 GRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSK 273
G + + +++++ + FT HR + +++ FHP A+ SD WD K
Sbjct: 110 GVIKLWDVEEAKMVRAFTG--HRS-----NCSAVEFHPFGEFLASGSSDANLKIWDIRKK 162
Query: 274 QRLKAMLRCSQPIPCSAFNNDG 295
++ S+ I F DG
Sbjct: 163 GCIQTYKGHSRGISTIRFTPDG 184
>At4g15900 PRL1 protein
Length = 486
Score = 62.4 bits (150), Expect = 3e-10
Identities = 70/304 (23%), Positives = 112/304 (36%), Gaps = 34/304 (11%)
Query: 12 PNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVL 71
P K++ V Q V S+ F P S D ++ W+VA V H + V
Sbjct: 165 PWKNYRVIQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKLTLTG--HIEQVR 222
Query: 72 CSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDR 131
A + T +FS G DKQVK W L + H + V +A P + +L+TG D
Sbjct: 223 GLAVSNRHTYMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLALHPTLDVLLTGGRDS 282
Query: 132 TIKYWDTRQPNPVHTQQLPERCYAMSVKHPL---MVVGTADRNILVYNLQNPQVEFKRII 188
+ WD R + + P +V G+ D I ++L+ + + +
Sbjct: 283 VCRVWDIRTKMQIFALSGHDNTVCSVFTRPTDPQVVTGSHDTTIKFWDLR-----YGKTM 337
Query: 189 SPL---KYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFK----CHREGNEI 241
S L K R + P + F S + K F+ CH ++
Sbjct: 338 STLTHHKKSVRAMTLHPKENAFASASADNT------------KKFSLPKGEFCHNMLSQQ 385
Query: 242 YSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYS 301
++ + T G +G+ FWD S + QP + ++ I Y+
Sbjct: 386 KTIINAMAVNEDGVMVTGGDNGSIWFWDWKSGHSFQQSETIVQP---GSLESEAGI--YA 440
Query: 302 VCYD 305
CYD
Sbjct: 441 ACYD 444
Score = 28.1 bits (61), Expect = 7.3
Identities = 33/124 (26%), Positives = 49/124 (38%), Gaps = 19/124 (15%)
Query: 25 SVSSLCFSPKANLLVATSWDNQVRC----WEVARNVATAPKASISHDQPVLCSAWKDDGT 80
SV ++ PK N + S DN + E N+ + K I+ A +DG
Sbjct: 346 SVRAMTLHPKENAFASASADNTKKFSLPKGEFCHNMLSQQKTIIN------AMAVNEDGV 399
Query: 81 TVFSGGCDKQVKMWPLLSGG---QPMTVAM-----HDAPVKDIAWIPEMSLLVTGSWDRT 132
V +GG + + W SG Q T+ +A + + S LVT D+T
Sbjct: 400 MV-TGGDNGSIWFWDWKSGHSFQQSETIVQPGSLESEAGIYAACYDNTGSRLVTCEADKT 458
Query: 133 IKYW 136
IK W
Sbjct: 459 IKMW 462
>At1g48630 unknown protein
Length = 326
Score = 62.0 bits (149), Expect = 5e-10
Identities = 60/249 (24%), Positives = 107/249 (42%), Gaps = 21/249 (8%)
Query: 35 ANLLVATSWDNQVRCWEVAR---NVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQV 91
++++V +S D + W++ + + A + H V DG SG D ++
Sbjct: 28 SDVIVTSSRDKSIILWKLTKEDKSYGVAQRRMTGHSHFVQDVVLSSDGQFALSGSWDGEL 87
Query: 92 KMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR-------QPNPV 144
++W L +G H V +A+ + +V+ S DRTIK W+T
Sbjct: 88 RLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLWNTLGECKYTISEADG 147
Query: 145 HTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPDQ 204
H + + ++ + P +V + D+ + V+NLQN + + ++ +A PD
Sbjct: 148 HKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQN--CKLRNTLAGHSGYLNTVAVSPD- 204
Query: 205 QGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGA 264
G L S G+ GV L D +GK E I ++SL F P + + A ++ +
Sbjct: 205 -GSLCAS-GGKDGVILLWDLAEGKKL---YSLEAGSI--IHSLCFSP-NRYWLCAATENS 256
Query: 265 FNFWDKDSK 273
WD +SK
Sbjct: 257 IRIWDLESK 265
Score = 54.3 bits (129), Expect = 1e-07
Identities = 60/268 (22%), Positives = 107/268 (39%), Gaps = 31/268 (11%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S V + S ++ SWD ++R W++A +T + + H + VL A+ D +
Sbjct: 63 SHFVQDVVLSSDGQFALSGSWDGELRLWDLATGEST--RRFVGHTKDVLSVAFSTDNRQI 120
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAM---HDAPVKDIAWIPE--MSLLVTGSWDRTIKYWD 137
S D+ +K+W L G T++ H V + + P + +V+ SWD+T+K W+
Sbjct: 121 VSASRDRTIKLWNTL-GECKYTISEADGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWN 179
Query: 138 -----TRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLK 192
R H+ L A+S L G D IL+++L E K++ S
Sbjct: 180 LQNCKLRNTLAGHSGYL--NTVAVSPDGSLCASGGKDGVILLWDL----AEGKKLYSLEA 233
Query: 193 YQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHRE----------GN--E 240
F + +L + E + + L+ ++ E GN +
Sbjct: 234 GSIIHSLCFSPNRYWLCAATENSIRIWDLESKSVVEDLKVDLKAEAEKTDGSTGIGNKTK 293
Query: 241 IYSVNSLNFHPVHHTFATAGSDGAFNFW 268
+ SLN+ +T + +DG W
Sbjct: 294 VIYCTSLNWSADGNTLFSGYTDGVIRVW 321
>At5g23430 unknown protein
Length = 837
Score = 61.6 bits (148), Expect = 6e-10
Identities = 62/241 (25%), Positives = 102/241 (41%), Gaps = 23/241 (9%)
Query: 64 ISHDQPVLC-SAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMS 122
++H V C + + +GG D +V +W + +++ H + + + +
Sbjct: 13 VAHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEV 72
Query: 123 LLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVK-HPL---MVVGTADRNILVYNLQ 178
L+ G+ TIK WD + V T R +SV HP G+ D N+ +++++
Sbjct: 73 LVAAGAASGTIKLWDLEEAKIVRTLT-GHRSNCISVDFHPFGEFFASGSLDTNLKIWDIR 131
Query: 179 NPQVEFKRIISPLKYQTR---CLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFT-FKC 234
K I K TR L PD + + G + V V D GK T FK
Sbjct: 132 K-----KGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVW---DLTAGKLLTEFKS 183
Query: 235 HREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNND 294
H EG + SL+FHP AT +D FWD ++ + + + + + C +FN D
Sbjct: 184 H-EGQ----IQSLDFHPHEFLLATGSADRTVKFWDLETFELIGSGGPETAGVRCLSFNPD 238
Query: 295 G 295
G
Sbjct: 239 G 239
Score = 58.2 bits (139), Expect = 7e-09
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 2/110 (1%)
Query: 28 SLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGC 87
S+ F P + S D ++ W++ + H + V + DG V SGG
Sbjct: 106 SVDFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKG--HTRGVNVLRFTPDGRWVVSGGE 163
Query: 88 DKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWD 137
D VK+W L +G H+ ++ + + P LL TGS DRT+K+WD
Sbjct: 164 DNIVKVWDLTAGKLLTEFKSHEGQIQSLDFHPHEFLLATGSADRTVKFWD 213
Score = 28.9 bits (63), Expect = 4.3
Identities = 27/108 (25%), Positives = 49/108 (45%), Gaps = 14/108 (12%)
Query: 26 VSSLCFSPKANLLVATSWDNQVRCWEV-ARNVATAPKASISHDQPVLCSAWKDDGTTVFS 84
V+ L F+P +V+ DN V+ W++ A + T K SH+ + + + +
Sbjct: 146 VNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTEFK---SHEGQIQSLDFHPHEFLLAT 202
Query: 85 GGCDKQVKMW-----PLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTG 127
G D+ VK W L+ G P T A V+ +++ P+ ++ G
Sbjct: 203 GSADRTVKFWDLETFELIGSGGPET-----AGVRCLSFNPDGKTVLCG 245
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 61.2 bits (147), Expect = 8e-10
Identities = 61/241 (25%), Positives = 100/241 (41%), Gaps = 23/241 (9%)
Query: 64 ISHDQPVLC-SAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMS 122
++H V C + + +GG D +V +W + +++ H + + + +
Sbjct: 106 VAHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEG 165
Query: 123 LLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVK-HPL---MVVGTADRNILVYNLQ 178
L+ G+ TIK WD + V T R +SV HP G+ D N+ +++++
Sbjct: 166 LVAAGAASGTIKLWDLEEAKVVRTLT-GHRSNCVSVNFHPFGEFFASGSLDTNLKIWDIR 224
Query: 179 NPQVEFKRIISPLKYQTR---CLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNF-TFKC 234
K I K TR L PD + + G + V V D GK FK
Sbjct: 225 K-----KGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVW---DLTAGKLLHEFKS 276
Query: 235 HREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNND 294
H EG + SL+FHP AT +D FWD ++ + + + + + C FN D
Sbjct: 277 H-EGK----IQSLDFHPHEFLLATGSADKTVKFWDLETFELIGSGGTETTGVRCLTFNPD 331
Query: 295 G 295
G
Sbjct: 332 G 332
Score = 56.6 bits (135), Expect = 2e-08
Identities = 31/110 (28%), Positives = 51/110 (46%), Gaps = 2/110 (1%)
Query: 28 SLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGC 87
S+ F P + S D ++ W++ + H + V + DG + SGG
Sbjct: 199 SVNFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKG--HTRGVNVLRFTPDGRWIVSGGE 256
Query: 88 DKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWD 137
D VK+W L +G H+ ++ + + P LL TGS D+T+K+WD
Sbjct: 257 DNVVKVWDLTAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWD 306
Score = 51.2 bits (121), Expect = 8e-07
Identities = 45/193 (23%), Positives = 76/193 (39%), Gaps = 11/193 (5%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S + S+ F L+ A + ++ W++ A + H + + G
Sbjct: 152 SSGIDSVTFDASEGLVAAGAASGTIKLWDLEE--AKVVRTLTGHRSNCVSVNFHPFGEFF 209
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPN 142
SG D +K+W + G T H V + + P+ +V+G D +K WD
Sbjct: 210 ASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLTAGK 269
Query: 143 PVHTQQLPERCYAMSVKHP---LMVVGTADRNILVYNLQNPQVEFKRIISPLKYQT--RC 197
+H + E HP L+ G+AD+ + ++L+ F+ I S T RC
Sbjct: 270 LLHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWDLET----FELIGSGGTETTGVRC 325
Query: 198 LAAFPDQQGFLVG 210
L PD + L G
Sbjct: 326 LTFNPDGKSVLCG 338
>At3g18130 protein kinase C-receptor/G-protein, putative
Length = 326
Score = 60.8 bits (146), Expect = 1e-09
Identities = 61/249 (24%), Positives = 107/249 (42%), Gaps = 21/249 (8%)
Query: 35 ANLLVATSWDNQVRCWEVARN---VATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQV 91
++++V S D + W++ ++ A + H V DG SG D ++
Sbjct: 28 SDIIVTASRDKSIILWKLTKDDKSYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDGEL 87
Query: 92 KMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR-------QPNPV 144
++W L +G H V +A+ + +V+ S DRTIK W+T
Sbjct: 88 RLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLWNTLGECKYTISEGDG 147
Query: 145 HTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPDQ 204
H + + ++ + P +V + D+ + V+NLQN ++ ++ Y +A PD
Sbjct: 148 HKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLR-NSLVGHSGY-LNTVAVSPD- 204
Query: 205 QGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGA 264
G L S G+ GV L D +GK E I ++SL F P + + A ++ +
Sbjct: 205 -GSLCAS-GGKDGVILLWDLAEGKKL---YSLEAGSI--IHSLCFSP-NRYWLCAATENS 256
Query: 265 FNFWDKDSK 273
WD +SK
Sbjct: 257 IRIWDLESK 265
Score = 54.3 bits (129), Expect = 1e-07
Identities = 61/268 (22%), Positives = 104/268 (38%), Gaps = 31/268 (11%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S V + S ++ SWD ++R W++A T + + H + VL A+ D +
Sbjct: 63 SHFVEDVVLSSDGQFALSGSWDGELRLWDLATGETT--RRFVGHTKDVLSVAFSTDNRQI 120
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAM---HDAPVKDIAWIPE--MSLLVTGSWDRTIKYWD 137
S D+ +K+W L G T++ H V + + P + +V+ SWD+T+K W+
Sbjct: 121 VSASRDRTIKLWNTL-GECKYTISEGDGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWN 179
Query: 138 -----TRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLK 192
R H+ L A+S L G D IL+++L E K++ S
Sbjct: 180 LQNCKLRNSLVGHSGYL--NTVAVSPDGSLCASGGKDGVILLWDL----AEGKKLYSLEA 233
Query: 193 YQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQ--QGKNFTFKCHREGNE---------- 240
F + +L + E + + L+ + K E NE
Sbjct: 234 GSIIHSLCFSPNRYWLCAATENSIRIWDLESKSVVEDLKVDLKSEAEKNEGGVGTGNQKK 293
Query: 241 IYSVNSLNFHPVHHTFATAGSDGAFNFW 268
+ SLN+ T + +DG W
Sbjct: 294 VIYCTSLNWSADGSTLFSGYTDGVVRVW 321
>At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2
Length = 479
Score = 60.5 bits (145), Expect = 1e-09
Identities = 73/298 (24%), Positives = 110/298 (36%), Gaps = 23/298 (7%)
Query: 12 PNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVL 71
P K++ V Q V S+ F P S D ++ W+VA V H V
Sbjct: 159 PWKNYRVLQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKL--TLTGHIGQVR 216
Query: 72 CSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDR 131
A + T +FS G DKQVK W L + H V +A P + +++TG D
Sbjct: 217 GLAVSNRHTYMFSAGDDKQVKCWDLEQNKVIRSYHGHLHGVYCLALHPTLDVVLTGGRDS 276
Query: 132 TIKYWDTRQPNPVHTQQLPERCYAMSV----KHPLMVVGTADRNILVYNLQNPQVEFKRI 187
+ WD R + LP SV P ++ G+ D I ++L+ +
Sbjct: 277 VCRVWDIRTKMQIFV--LPHDSDVFSVLARPTDPQVITGSHDSTIKFWDLR--YGKSMAT 332
Query: 188 ISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSL 247
I+ K R +A P + F+ S + + L + N R+ +VN
Sbjct: 333 ITNHKKTVRAMALHPKENDFVSASAD-NIKKFSLPKGEFCHNM-LSLQRDIINAVAVNE- 389
Query: 248 NFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYD 305
T G G FWD S + QP + ++ I Y+ CYD
Sbjct: 390 -----DGVMVTGGDKGGLWFWDWKSGHNFQRAETIVQP---GSLESEAGI--YAACYD 437
>At1g18080 putative guanine nucleotide-binding protein beta
subunit
Length = 327
Score = 60.5 bits (145), Expect = 1e-09
Identities = 60/250 (24%), Positives = 104/250 (41%), Gaps = 22/250 (8%)
Query: 35 ANLLVATSWDNQVRCWEVARN---VATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQV 91
A+++V+ S D + W++ ++ A + H V DG SG D ++
Sbjct: 28 ADIIVSASRDKSIILWKLTKDDKAYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDGEL 87
Query: 92 KMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDT--------RQPNP 143
++W L +G H V +A+ + +V+ S DRTIK W+T +
Sbjct: 88 RLWDLAAGVSTRRFVGHTKDVLSVAFSLDNRQIVSASRDRTIKLWNTLGECKYTISEGGE 147
Query: 144 VHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPD 203
H + ++ + P +V + D+ + V+NL N + + ++ +A PD
Sbjct: 148 GHRDWVSCVRFSPNTLQPTIVSASWDKTVKVWNLSN--CKLRSTLAGHTGYVSTVAVSPD 205
Query: 204 QQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDG 263
G L S G+ GV L D +GK E N + +++L F P + A G
Sbjct: 206 --GSLCAS-GGKDGVVLLWDLAEGKKL---YSLEANSV--IHALCFSPNRYWLCAATEHG 257
Query: 264 AFNFWDKDSK 273
WD +SK
Sbjct: 258 -IKIWDLESK 266
Score = 53.1 bits (126), Expect = 2e-07
Identities = 33/121 (27%), Positives = 58/121 (47%), Gaps = 9/121 (7%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S V + S ++ SWD ++R W++A V+T + + H + VL A+ D +
Sbjct: 63 SHFVEDVVLSSDGQFALSGSWDGELRLWDLAAGVST--RRFVGHTKDVLSVAFSLDNRQI 120
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAM----HDAPVKDIAWIPE--MSLLVTGSWDRTIKYW 136
S D+ +K+W L G T++ H V + + P +V+ SWD+T+K W
Sbjct: 121 VSASRDRTIKLWNTL-GECKYTISEGGEGHRDWVSCVRFSPNTLQPTIVSASWDKTVKVW 179
Query: 137 D 137
+
Sbjct: 180 N 180
Score = 32.7 bits (73), Expect = 0.30
Identities = 23/86 (26%), Positives = 41/86 (46%), Gaps = 18/86 (20%)
Query: 26 VSSLCFSPKANLLVATSWDNQVRCWEVARNV----------ATAPKASISHD-----QPV 70
+ +LCFSP L A + ++ ++ W++ A A KA S + +
Sbjct: 238 IHALCFSPNRYWLCAAT-EHGIKIWDLESKSIVEDLKVDLKAEAEKADNSGPAATKRKVI 296
Query: 71 LCSA--WKDDGTTVFSGGCDKQVKMW 94
C++ W DG+T+FSG D +++W
Sbjct: 297 YCTSLNWSADGSTLFSGYTDGVIRVW 322
>At4g02730 putative WD-repeat protein
Length = 333
Score = 59.3 bits (142), Expect = 3e-09
Identities = 57/259 (22%), Positives = 112/259 (43%), Gaps = 19/259 (7%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S +S L +S ++ + S D +R W+ AR+ K H V C + +
Sbjct: 85 SSGISDLAWSSDSHYTCSASDDCTLRIWD-ARSPYECLKVLRGHTNFVFCVNFNPPSNLI 143
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPN 142
SG D+ +++W + +G + H P+ + + + SL+V+ S D + K WD ++
Sbjct: 144 VSGSFDETIRIWEVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEGT 203
Query: 143 PVHT---QQLPERCYA-MSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCL 198
+ T + P +A S ++V T D + + N +F ++ + + C+
Sbjct: 204 CLKTLIDDKSPAVSFAKFSPNGKFILVATLDSTLKLSNYATG--KFLKVYTGHTNKVFCI 261
Query: 199 -AAFPDQQG--FLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHT 255
+AF G + GS + V + L Q +N + + + SV+ HPV +
Sbjct: 262 TSAFSVTNGKYIVSGSEDNCVYLWDL----QARNILQRLEGHTDAVISVSC---HPVQNE 314
Query: 256 FATAGS--DGAFNFWDKDS 272
+++G+ D W +D+
Sbjct: 315 ISSSGNHLDKTIRIWKQDA 333
Score = 47.0 bits (110), Expect = 2e-05
Identities = 30/126 (23%), Positives = 58/126 (45%), Gaps = 3/126 (2%)
Query: 10 QNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQP 69
++P + +V + ++ V + F+P +NL+V+ S+D +R WEV + +H P
Sbjct: 115 RSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRIWEV--KTGKCVRMIKAHSMP 172
Query: 70 VLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIA-WIPEMSLLVTGS 128
+ + DG+ + S D K+W G T+ +P A + P ++ +
Sbjct: 173 ISSVHFNRDGSLIVSASHDGSCKIWDAKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVAT 232
Query: 129 WDRTIK 134
D T+K
Sbjct: 233 LDSTLK 238
Score = 43.1 bits (100), Expect = 2e-04
Identities = 27/119 (22%), Positives = 49/119 (40%), Gaps = 8/119 (6%)
Query: 66 HDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLV 125
H + C + +DG + S DK + +W + H + + D+AW +
Sbjct: 42 HTAAISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYEGHSSGISDLAWSSDSHYTC 101
Query: 126 TGSWDRTIKYWDTRQPNPV------HTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQ 178
+ S D T++ WD R P HT + C + L+V G+ D I ++ ++
Sbjct: 102 SASDDCTLRIWDARSPYECLKVLRGHTNFV--FCVNFNPPSNLIVSGSFDETIRIWEVK 158
Score = 30.0 bits (66), Expect = 1.9
Identities = 25/91 (27%), Positives = 38/91 (41%), Gaps = 2/91 (2%)
Query: 247 LNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYDW 306
+NF+P + + D W+ + + ++ + S PI FN DGS+ S +D
Sbjct: 134 VNFNPPSNLIVSGSFDETIRIWEVKTGKCVRMIKAHSMPISSVHFNRDGSLIV-SASHDG 192
Query: 307 S-KGAENHNPTTAKTNIYLHLPQESEVKGKP 336
S K + T KT I P S K P
Sbjct: 193 SCKIWDAKEGTCLKTLIDDKSPAVSFAKFSP 223
>At2g26060 unknown protein
Length = 352
Score = 56.6 bits (135), Expect = 2e-08
Identities = 33/115 (28%), Positives = 51/115 (43%), Gaps = 3/115 (2%)
Query: 25 SVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFS 84
+V S +SP LL S+D W+ + H+ V +W G+ + +
Sbjct: 76 TVRSCAWSPSGQLLATASFDGTTGIWKNYGSEFECISTLEGHENEVKSVSWNASGSCLAT 135
Query: 85 GGCDKQVKMWPLLSGGQPMTVAM---HDAPVKDIAWIPEMSLLVTGSWDRTIKYW 136
DK V +W +L G + A+ H VK + W P M +L + S+D TIK W
Sbjct: 136 CSRDKSVWIWEVLEGNEYDCAAVLTGHTQDVKMVQWHPTMDVLFSCSYDNTIKVW 190
Score = 43.5 bits (101), Expect = 2e-04
Identities = 32/132 (24%), Positives = 57/132 (42%), Gaps = 12/132 (9%)
Query: 23 SDSVSSLCFSPKAN-------LLVATSWDNQVRCWE---VARNVATAPKASISHDQPVLC 72
+D V S+ ++P ++ +L + S DN VR WE ++R+ +H + V
Sbjct: 20 TDRVWSVAWNPVSSHADGVSPILASCSGDNTVRIWEQSSLSRSWTCKTVLEETHTRTVRS 79
Query: 73 SAWKDDGTTVFSGGCDKQVKMWPLLSGGQPM--TVAMHDAPVKDIAWIPEMSLLVTGSWD 130
AW G + + D +W T+ H+ VK ++W S L T S D
Sbjct: 80 CAWSPSGQLLATASFDGTTGIWKNYGSEFECISTLEGHENEVKSVSWNASGSCLATCSRD 139
Query: 131 RTIKYWDTRQPN 142
+++ W+ + N
Sbjct: 140 KSVWIWEVLEGN 151
>At5g16750 WD40-repeat protein
Length = 876
Score = 56.2 bits (134), Expect = 3e-08
Identities = 35/140 (25%), Positives = 67/140 (47%), Gaps = 10/140 (7%)
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
SD++++L SP LL + Q+R W++ ++ H+ PV+ A G +
Sbjct: 60 SDTLTALALSPDDKLLFSAGHSRQIRVWDL--ETLKCIRSWKGHEGPVMGMACHASGGLL 117
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPE--MSLLVTGSWDRTIKYWDTRQ 140
+ G D++V +W + G H V I + P+ ++L++GS D T++ WD
Sbjct: 118 ATAGADRKVLVWDVDGGFCTHYFRGHKGVVSSILFHPDSNKNILISGSDDATVRVWD--- 174
Query: 141 PNPVHTQQLPERCYAMSVKH 160
++ + ++C A+ KH
Sbjct: 175 ---LNAKNTEKKCLAIMEKH 191
Score = 45.4 bits (106), Expect = 4e-05
Identities = 32/88 (36%), Positives = 46/88 (51%), Gaps = 4/88 (4%)
Query: 26 VSSLCFSPKAN--LLVATSWDNQVRCWEV-ARNVATAPKASIS-HDQPVLCSAWKDDGTT 81
VSS+ F P +N +L++ S D VR W++ A+N A + H V A +DG T
Sbjct: 147 VSSILFHPDSNKNILISGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLT 206
Query: 82 VFSGGCDKQVKMWPLLSGGQPMTVAMHD 109
+FS G DK V +W L TVA ++
Sbjct: 207 LFSAGRDKVVNLWDLHDYSCKATVATYE 234
Score = 41.2 bits (95), Expect = 8e-04
Identities = 28/111 (25%), Positives = 48/111 (43%), Gaps = 2/111 (1%)
Query: 26 VSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSG 85
+ S+ FS ++ S D V+ W ++ + K H VL +++ DGT S
Sbjct: 544 IFSVEFSTVDQCVMTASGDKTVKIWAISDG--SCLKTFEGHTSSVLRASFITDGTQFVSC 601
Query: 86 GCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYW 136
G D +K+W + + T H+ V +A + ++ TG D I W
Sbjct: 602 GADGLLKLWNVNTSECIATYDQHEDKVWALAVGKKTEMIATGGGDAVINLW 652
Score = 33.5 bits (75), Expect = 0.17
Identities = 42/192 (21%), Positives = 71/192 (36%), Gaps = 27/192 (14%)
Query: 123 LLVTGSWDRTIKYWDTRQPNPV-----HTQQLPERCYAMSVKHPLMVVGTADRNILVYNL 177
L+VTGS D+T++ W+ + + H + +A V G+ DR + V++L
Sbjct: 418 LIVTGSKDKTVRLWNATSKSCIGVGTGHNGDILAVAFAKK-SFSFFVSGSGDRTLKVWSL 476
Query: 178 QNPQVEFKRIISPLKYQTRCLAAFPDQQGFLV-----------GSIEGRVGVHHLDDSQQ 226
+ + P+ +TR + A D+ V GS + + L D
Sbjct: 477 DGISEDSE---EPINLKTRSVVAAHDKDINSVAVARNDSLVCTGSEDRTASIWRLPDLVH 533
Query: 227 GKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPI 286
T K H+ + S+ F V TA D W LK + +
Sbjct: 534 --VVTLKGHKR-----RIFSVEFSTVDQCVMTASGDKTVKIWAISDGSCLKTFEGHTSSV 586
Query: 287 PCSAFNNDGSIF 298
++F DG+ F
Sbjct: 587 LRASFITDGTQF 598
Score = 30.4 bits (67), Expect = 1.5
Identities = 25/121 (20%), Positives = 53/121 (43%), Gaps = 11/121 (9%)
Query: 26 VSSLCFSPKA-NLLVATSWDNQVRCW---------EVARNVATAPKASISHDQPVLCSAW 75
+ ++ F+ K+ + V+ S D ++ W E N+ T + +HD+ + A
Sbjct: 449 ILAVAFAKKSFSFFVSGSGDRTLKVWSLDGISEDSEEPINLKTRSVVA-AHDKDINSVAV 507
Query: 76 KDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKY 135
+ + V +G D+ +W L +T+ H + + + ++T S D+T+K
Sbjct: 508 ARNDSLVCTGSEDRTASIWRLPDLVHVVTLKGHKRRIFSVEFSTVDQCVMTASGDKTVKI 567
Query: 136 W 136
W
Sbjct: 568 W 568
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 55.8 bits (133), Expect = 3e-08
Identities = 55/275 (20%), Positives = 111/275 (40%), Gaps = 39/275 (14%)
Query: 25 SVSSLCFSPKANLLVATSWDNQVRCWEVAR---NVATAPKASISHDQPVLCSAWKDDGTT 81
+VSS+ FS LL + S D +R + + +A + H+ + A+ D
Sbjct: 26 AVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSSDARF 85
Query: 82 VFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQP 141
+ S DK +K+W + +G T+ H + + P+ +++V+GS+D T++ WD
Sbjct: 86 IVSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMIVSGSFDETVRIWDVTTG 145
Query: 142 N-----PVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNL------------QNPQVEF 184
P H+ P + L+V + D +++ +NP V F
Sbjct: 146 KCLKVLPAHSD--PVTAVDFNRDGSLIVSSSYDGLCRIWDSGTGHCVKTLIDDENPPVSF 203
Query: 185 KRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSV 244
R P+ + LVG+++ + + ++ ++ K +T N Y +
Sbjct: 204 VRF-------------SPNGKFILVGTLDNTLRLWNISSAKFLKTYT----GHVNAQYCI 246
Query: 245 NSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAM 279
+S + D + W+ +SK+ L+ +
Sbjct: 247 SSAFSVTNGKRIVSGSEDNCVHMWELNSKKLLQKL 281
Score = 50.1 bits (118), Expect = 2e-06
Identities = 31/116 (26%), Positives = 54/116 (45%), Gaps = 1/116 (0%)
Query: 26 VSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWK-DDGTTVFS 84
VS + FSP ++ + DN +R W ++ + Q + SA+ +G + S
Sbjct: 201 VSFVRFSPNGKFILVGTLDNTLRLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKRIVS 260
Query: 85 GGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQ 140
G D V MW L S + H V ++A P +L+ +GS D+T++ W ++
Sbjct: 261 GSEDNCVHMWELNSKKLLQKLEGHTETVMNVACHPTENLIASGSLDKTVRIWTQKK 316
Score = 40.8 bits (94), Expect = 0.001
Identities = 26/122 (21%), Positives = 58/122 (47%), Gaps = 10/122 (8%)
Query: 65 SHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAM-----HDAPVKDIAWIP 119
SH++ V + DG + S DK ++ + + + P+ + H+ + D+A+
Sbjct: 22 SHNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSS 81
Query: 120 EMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSV----KHPLMVVGTADRNILVY 175
+ +V+ S D+T+K WD + + T + YA V + ++V G+ D + ++
Sbjct: 82 DARFIVSASDDKTLKLWDVETGSLIKT-LIGHTNYAFCVNFNPQSNMIVSGSFDETVRIW 140
Query: 176 NL 177
++
Sbjct: 141 DV 142
Score = 36.6 bits (83), Expect = 0.021
Identities = 19/67 (28%), Positives = 29/67 (42%), Gaps = 4/67 (5%)
Query: 247 LNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYS----V 302
+NF+P + + D WD + + LK + S P+ FN DGS+ S +
Sbjct: 119 VNFNPQSNMIVSGSFDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGL 178
Query: 303 CYDWSKG 309
C W G
Sbjct: 179 CRIWDSG 185
>At5g25150 transcription initiation factor IID-associated
factor-like protein
Length = 700
Score = 55.1 bits (131), Expect = 6e-08
Identities = 46/191 (24%), Positives = 79/191 (41%), Gaps = 13/191 (6%)
Query: 11 NPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPV 70
N +S+ + S V S FSP + ++++S D +R W N H+ PV
Sbjct: 406 NGRRSYTLLLGHSGPVYSATFSPPGDFVLSSSADTTIRLWSTKLNANLV--CYKGHNYPV 463
Query: 71 LCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWD 130
+ + G S D+ ++W + QP+ + + D+ W P + + TGS D
Sbjct: 464 WDAQFSPFGHYFASCSHDRTARIWS-MDRIQPLRIMA--GHLSDVDWHPNCNYIATGSSD 520
Query: 131 RTIKYWDTRQPNPVHT---QQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRI 187
+T++ WD + V + AMS M G D I++++L R
Sbjct: 521 KTVRLWDVQTGECVRIFIGHRSMVLSLAMSPDGRYMASGDEDGTIMMWDLST-----ARC 575
Query: 188 ISPLKYQTRCL 198
I+PL C+
Sbjct: 576 ITPLMGHNSCV 586
Score = 42.4 bits (98), Expect = 4e-04
Identities = 34/155 (21%), Positives = 63/155 (39%), Gaps = 6/155 (3%)
Query: 64 ISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSL 123
+ H PV + + G V S D +++W + H+ PV D + P
Sbjct: 415 LGHSGPVYSATFSPPGDFVLSSSADTTIRLWSTKLNANLVCYKGHNYPVWDAQFSPFGHY 474
Query: 124 LVTGSWDRTIKYW--DTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQ 181
+ S DRT + W D QP + L + + + + + G++D+ + ++++Q
Sbjct: 475 FASCSHDRTARIWSMDRIQPLRIMAGHLSDVDWHPNCNY--IATGSSDKTVRLWDVQTG- 531
Query: 182 VEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRV 216
E RI + LA PD + G +G +
Sbjct: 532 -ECVRIFIGHRSMVLSLAMSPDGRYMASGDEDGTI 565
Score = 35.0 bits (79), Expect = 0.060
Identities = 22/92 (23%), Positives = 35/92 (37%), Gaps = 24/92 (26%)
Query: 72 CSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAM------------------------ 107
CS+ DG+ V G D +K+W + GQ + A+
Sbjct: 357 CSSISHDGSLVAGGFSDSSIKVWDMAKIGQAGSGALQAENDSSDQSIGPNGRRSYTLLLG 416
Query: 108 HDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR 139
H PV + P +++ S D TI+ W T+
Sbjct: 417 HSGPVYSATFSPPGDFVLSSSADTTIRLWSTK 448
Score = 28.1 bits (61), Expect = 7.3
Identities = 16/51 (31%), Positives = 20/51 (38%), Gaps = 2/51 (3%)
Query: 229 NFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAM 279
N C++ N Y V F P H FA+ D W D Q L+ M
Sbjct: 450 NANLVCYKGHN--YPVWDAQFSPFGHYFASCSHDRTARIWSMDRIQPLRIM 498
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.132 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,672,420
Number of Sequences: 26719
Number of extensions: 382563
Number of successful extensions: 1783
Number of sequences better than 10.0: 185
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 76
Number of HSP's that attempted gapping in prelim test: 901
Number of HSP's gapped (non-prelim): 663
length of query: 344
length of database: 11,318,596
effective HSP length: 100
effective length of query: 244
effective length of database: 8,646,696
effective search space: 2109793824
effective search space used: 2109793824
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0314.9


