

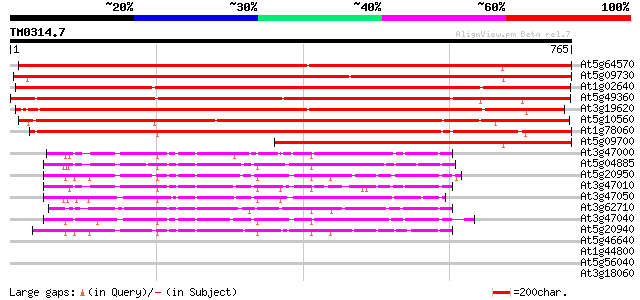
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.7
(765 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g64570 beta-xylosidase 1127 0.0
At5g09730 beta-xylosidase - like protein 1073 0.0
At1g02640 beta-xylosidase, putative 858 0.0
At5g49360 xylosidase 849 0.0
At3g19620 beta-xylosidase, putative 843 0.0
At5g10560 beta-xylosidase - like protein 719 0.0
At1g78060 putative protein 712 0.0
At5g09700 beta-glucosidase - like protein 535 e-152
At3g47000 beta-D-glucan exohydrolase - like protein 140 2e-33
At5g04885 unknown protein 140 3e-33
At5g20950 beta-D-glucan exohydrolase-like protein 139 8e-33
At3g47010 beta-D-glucan exohydrolase - like protein 130 2e-30
At3g47050 beta-D-glucan exohydrolase - like protein 129 6e-30
At3g62710 beta-D-glucan exohydrolase-like protein 126 5e-29
At3g47040 beta-D-glucan exohydrolase - like protein 125 7e-29
At5g20940 beta-glucosidase - like protein 125 9e-29
At5g46640 unknown protein 33 0.46
At1g44800 nodulin like protein 32 1.7
At5g56040 receptor protein kinase-like protein 31 3.0
At3g18060 WD40-repeat protein 30 5.1
>At5g64570 beta-xylosidase
Length = 784
Score = 1127 bits (2914), Expect = 0.0
Identities = 546/760 (71%), Positives = 634/760 (82%), Gaps = 7/760 (0%)
Query: 12 LVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQEKIGN 71
L F +F Q+SP FACDV N S A +GFC+ L + RVADLV RLTLQEKIG
Sbjct: 26 LCFFLYFLNFSNAQSSPVFACDVAANPSLAAYGFCNTVLKIEYRVADLVARLTLQEKIGF 85
Query: 72 LGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFNSTLF 131
L A V RLGIP YEWWSEALHGVS IGPGT FSS VPGATSFP ILTAASFN +LF
Sbjct: 86 LVSKANGVTRLGIPTYEWWSEALHGVSYIGPGTHFSSQVPGATSFPQVILTAASFNVSLF 145
Query: 132 QAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKYAAGYVK 191
QAIGKVVSTEARAMYNVGLAGLTYWSPN+NIFRDPRWGRGQETPGEDPLL+SKYA+GYVK
Sbjct: 146 QAIGKVVSTEARAMYNVGLAGLTYWSPNVNIFRDPRWGRGQETPGEDPLLASKYASGYVK 205
Query: 192 GLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKSCVI 251
GLQ+TD GDSN+LKVAACCKHYTAYDVDNWKGV+RY+FNAVV+QQD+DDT+QPPFKSCV+
Sbjct: 206 GLQETDGGDSNRLKVAACCKHYTAYDVDNWKGVERYSFNAVVTQQDMDDTYQPPFKSCVV 265
Query: 252 DGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTP 311
DGNVASVMCSYNQVNGKPTCADPDLL GVIRG+WKLNGYIVSDCDSV+VL+K+QHYTKTP
Sbjct: 266 DGNVASVMCSYNQVNGKPTCADPDLLSGVIRGEWKLNGYIVSDCDSVDVLYKNQHYTKTP 325
Query: 312 EEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPS 371
EAAA SILAGLDL+CGS+LG++TE AVK GL++EA+I+ AISNNF TLMRLGFFDGNP
Sbjct: 326 AEAAAISILAGLDLNCGSFLGQHTEEAVKSGLVNEAAIDKAISNNFLTLMRLGFFDGNPK 385
Query: 372 TLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATR 431
YGGLGP DVCTS NQELA +AARQGIVLLKN+ G LPLS KSIK+LAVIGPNAN T+
Sbjct: 386 NQIYGGLGPTDVCTSANQELAADAARQGIVLLKNT-GCLPLSPKSIKTLAVIGPNANVTK 444
Query: 432 VMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAASSDATVIVVG 491
MIGNYEG PCKY +PLQGL V T+Y+PGC +V CA A + ATK+AA++D +V+V+G
Sbjct: 445 TMIGNYEGTPCKYTTPLQGLAGTVSTTYLPGCSNVACAVADVAGATKLAATADVSVLVIG 504
Query: 492 ASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSI 551
A +IEAES DRV++ LPGQQQ LV +VA A++GPV+LVIMSGGG D++FAK + KI I
Sbjct: 505 ADQSIEAESRDRVDLHLPGQQQELVIQVAKAAKGPVLLVIMSGGGFDITFAKNDPKIAGI 564
Query: 552 LWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRT 611
LWVGYPGEAGG AIAD+IFG YNPSG+LPMTWYP+SYVE VPMT MNMR D ++GYPGRT
Sbjct: 565 LWVGYPGEAGGIAIADIIFGRYNPSGKLPMTWYPQSYVEKVPMTIMNMRPDKASGYPGRT 624
Query: 612 YRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDVADEHCQN- 670
YRFY GETV++FGDG+ Y+ H +VKAP LV + L E+HVCRSSEC+SLD HC+N
Sbjct: 625 YRFYTGETVYAFGDGLSYTKFSHTLVKAPSLVSLGLEENHVCRSSECQSLDAIGPHCENA 684
Query: 671 -----MAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVR 725
AF++H++V+N G HTV LF TPPA+H +P+KHL+G EK+ L + EA VR
Sbjct: 685 VSGGGSAFEVHIKVRNGGDREGIHTVFLFTTPPAIHGSPRKHLVGFEKIRLGKREEAVVR 744
Query: 726 FMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
F V++CKDLSVVDE+G R++ LG+HLLHVG+LK+ LS+RI
Sbjct: 745 FKVEICKDLSVVDEIGKRKIGLGKHLLHVGDLKHSLSIRI 784
>At5g09730 beta-xylosidase - like protein
Length = 773
Score = 1073 bits (2776), Expect = 0.0
Identities = 524/771 (67%), Positives = 612/771 (78%), Gaps = 12/771 (1%)
Query: 6 QNRVSVLVCFSFFCATVC------GQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADL 59
+NR V F C VC Q+SP FACDVT N S AG FC+ L + RV DL
Sbjct: 4 RNRALFSVSTLFLCFIVCISEQSNNQSSPVFACDVTGNPSLAGLRFCNAGLSIKARVTDL 63
Query: 60 VKRLTLQEKIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMP 119
V RLTL+EKIG L A+ V RLGIP Y+WWSEALHGVSN+G G+RF+ VPGATSFP
Sbjct: 64 VGRLTLEEKIGFLTSKAIGVSRLGIPSYKWWSEALHGVSNVGGGSRFTGQVPGATSFPQV 123
Query: 120 ILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDP 179
ILTAASFN +LFQAIGKVVSTEARAMYNVG AGLT+WSPN+NIFRDPRWGRGQETPGEDP
Sbjct: 124 ILTAASFNVSLFQAIGKVVSTEARAMYNVGSAGLTFWSPNVNIFRDPRWGRGQETPGEDP 183
Query: 180 LLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLD 239
LSSKYA YVKGLQ+TD GD N+LKVAACCKHYTAYD+DNW+ V R TFNAVV+QQDL
Sbjct: 184 TLSSKYAVAYVKGLQETDGGDPNRLKVAACCKHYTAYDIDNWRNVNRLTFNAVVNQQDLA 243
Query: 240 DTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVE 299
DTFQPPFKSCV+DG+VASVMCSYNQVNGKPTCADPDLL GVIRGQW+LNGYIVSDCDSV+
Sbjct: 244 DTFQPPFKSCVVDGHVASVMCSYNQVNGKPTCADPDLLSGVIRGQWQLNGYIVSDCDSVD 303
Query: 300 VLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFAT 359
VLF+ QHY KTPEEA AKS+LAGLDL+C + G++ GAVK GL++E +I+ AISNNFAT
Sbjct: 304 VLFRKQHYAKTPEEAVAKSLLAGLDLNCDHFNGQHAMGAVKAGLVNETAIDKAISNNFAT 363
Query: 360 LMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKS 419
LMRLGFFDG+P YGGLGPKDVCT++NQELAR+ ARQGIVLLKNS GSLPLS +IK+
Sbjct: 364 LMRLGFFDGDPKKQLYGGLGPKDVCTADNQELARDGARQGIVLLKNSAGSLPLSPSAIKT 423
Query: 420 LAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKI 479
LAVIGPNANAT MIGNY G+PCKY +PLQGL V ++Y GC +V C A + A +
Sbjct: 424 LAVIGPNANATETMIGNYHGVPCKYTTPLQGLAETVSSTYQLGC-NVACVDADIGSAVDL 482
Query: 480 AASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDV 539
AAS+DA V+VVGA +IE E DRV++ LPG+QQ LV+ VA A+RGPV+LVIMSGGG D+
Sbjct: 483 AASADAVVLVVGADQSIEREGHDRVDLYLPGKQQELVTRVAMAARGPVVLVIMSGGGFDI 542
Query: 540 SFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNM 599
+FAK + KITSI+WVGYPGEAGG AIADVIFG +NPSG LPMTWYP+SYVE VPM+NMNM
Sbjct: 543 TFAKNDKKITSIMWVGYPGEAGGLAIADVIFGRHNPSGNLPMTWYPQSYVEKVPMSNMNM 602
Query: 600 RADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECK 659
R D S GYPGR+YRFY GETV++F D + Y+ +H+++KAP+LV + L E+H CRSSEC+
Sbjct: 603 RPDKSKGYPGRSYRFYTGETVYAFADALTYTKFDHQLIKAPRLVSLSLDENHPCRSSECQ 662
Query: 660 SLDVADEHCQNMA-----FDIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKV 714
SLD HC+N F++HL VKN G + SHTV LF T P VH +P K LLG EK+
Sbjct: 663 SLDAIGPHCENAVEGGSDFEVHLNVKNTGDRAGSHTVFLFTTSPQVHGSPIKQLLGFEKI 722
Query: 715 HLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
L EA VRF V+VCKDLSVVDE G R++ LG HLLHVG+LK+ L++ +
Sbjct: 723 RLGKSEEAVVRFNVNVCKDLSVVDETGKRKIALGHHLLHVGSLKHSLNISV 773
>At1g02640 beta-xylosidase, putative
Length = 768
Score = 858 bits (2217), Expect = 0.0
Identities = 420/757 (55%), Positives = 548/757 (71%), Gaps = 7/757 (0%)
Query: 9 VSVLVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQEK 68
++V++ F ++VC + FACD TK+A+ A FC S+P+ +RV DL+ RLTL EK
Sbjct: 9 LAVILFFLISSSSVCVHSRETFACD-TKDAATATLRFCQLSVPIPERVRDLIGRLTLAEK 67
Query: 69 IGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFNS 128
+ LG++A + RLGI YEWWSEALHGVSN+GPGT+F V P ATSFP I T ASFN+
Sbjct: 68 VSLLGNTAAAIPRLGIKGYEWWSEALHGVSNVGPGTKFGGVYPAATSFPQVITTVASFNA 127
Query: 129 TLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKYAAG 188
+L+++IG+VVS EARAMYN G+ GLTYWSPN+NI RDPRWGRGQETPGEDP+++ KYAA
Sbjct: 128 SLWESIGRVVSNEARAMYNGGVGGLTYWSPNVNILRDPRWGRGQETPGEDPVVAGKYAAS 187
Query: 189 YVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKS 248
YV+GLQ D ++LKVAACCKH+TAYD+DNW GV R+ FNA VS+QD++DTF PF+
Sbjct: 188 YVRGLQ---GNDRSRLKVAACCKHFTAYDLDNWNGVDRFHFNAKVSKQDIEDTFDVPFRM 244
Query: 249 CVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYT 308
CV +GNVAS+MCSYNQVNG PTCADP+LLK IR QW LNGYIVSDCDSV VL+ QHYT
Sbjct: 245 CVKEGNVASIMCSYNQVNGVPTCADPNLLKKTIRNQWGLNGYIVSDCDSVGVLYDTQHYT 304
Query: 309 KTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDG 368
TPEEAAA SI AGLDLDCG +LG +T AVK+ LL E+ ++NA+ N MRLG FDG
Sbjct: 305 GTPEEAAADSIKAGLDLDCGPFLGAHTIDAVKKNLLRESDVDNALINTLTVQMRLGMFDG 364
Query: 369 NPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNAN 428
+ + PYG LGP VCT ++ LA EAA+QGIVLLKN SLPLS++ +++AVIGPN++
Sbjct: 365 DIAAQPYGHLGPAHVCTPVHKGLALEAAQQGIVLLKNHGSSLPLSSQRHRTVAVIGPNSD 424
Query: 429 ATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQL-DDATKIAASSDATV 487
AT MIGNY G+ C Y SP+QG+T T + GC DVHC +L D A + A +DATV
Sbjct: 425 ATVTMIGNYAGVACGYTSPVQGITGYARTIHQKGCVDVHCMDDRLFDAAVEAARGADATV 484
Query: 488 IVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDK 547
+V+G +IEAE DR +++LPG+QQ LVS VA A++GPVILV+MSGG +D+SFA+ + K
Sbjct: 485 LVMGLDQSIEAEFKDRNSLLLPGKQQELVSRVAKAAKGPVILVLMSGGPIDISFAEKDRK 544
Query: 548 ITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGY 607
I +I+W GYPG+ GG AIAD++FG NP G+LPMTWYP+ Y+ N+PMT M+MR S
Sbjct: 545 IPAIVWAGYPGQEGGTAIADILFGSANPGGKLPMTWYPQDYLTNLPMTEMSMRPVHSKRI 604
Query: 608 PGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDVADEH 667
PGRTYRFY G V+ FG G+ Y+ H I AP++ +P+A + KS+ V
Sbjct: 605 PGRTYRFYDGPVVYPFGHGLSYTRFTHNIADAPKV--IPIAVRGRNGTVSGKSIRVTHAR 662
Query: 668 CQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFM 727
C ++ +H+ V N+G +HT+L+F PP AP+K L+ E+VH+A + +V+
Sbjct: 663 CDRLSLGVHVEVTNVGSRDGTHTMLVFSAPPGGEWAPKKQLVAFERVHVAVGEKKRVQVN 722
Query: 728 VDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
+ VCK LSVVD GNRR+P+G H +H+G+ + +S++
Sbjct: 723 IHVCKYLSVVDRAGNRRIPIGDHGIHIGDESHTVSLQ 759
>At5g49360 xylosidase
Length = 774
Score = 849 bits (2193), Expect = 0.0
Identities = 435/772 (56%), Positives = 539/772 (69%), Gaps = 16/772 (2%)
Query: 2 ACLLQNRVSVLVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVK 61
A L+ N+V V++ F P FACD N FC ++P+ RV DL+
Sbjct: 7 ALLIGNKVVVILVFLLCLVHSSESLRPLFACDPA-NGLTRTLRFCRANVPIHVRVQDLLG 65
Query: 62 RLTLQEKIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPIL 121
RLTLQEKI NL ++A V RLGI YEWWSEALHG+S++GPG +F PGATSFP I
Sbjct: 66 RLTLQEKIRNLVNNAAAVPRLGIGGYEWWSEALHGISDVGPGAKFGGAFPGATSFPQVIT 125
Query: 122 TAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLL 181
TAASFN +L++ IG+VVS EARAMYN G+AGLTYWSPN+NI RDPRWGRGQETPGEDP++
Sbjct: 126 TAASFNQSLWEEIGRVVSDEARAMYNGGVAGLTYWSPNVNILRDPRWGRGQETPGEDPIV 185
Query: 182 SSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDT 241
++KYAA YV+GLQ T G N+LKVAACCKHYTAYD+DNW GV R+ FNA V+QQDL+DT
Sbjct: 186 AAKYAASYVRGLQGTAAG--NRLKVAACCKHYTAYDLDNWNGVDRFHFNAKVTQQDLEDT 243
Query: 242 FQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVL 301
+ PFKSCV +G VASVMCSYNQVNGKPTCAD +LLK IRGQW+LNGYIVSDCDSV+V
Sbjct: 244 YNVPFKSCVYEGKVASVMCSYNQVNGKPTCADENLLKNTIRGQWRLNGYIVSDCDSVDVF 303
Query: 302 FKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLM 361
F QHYT TPEEAAA+SI AGLDLDCG +L +TEGAVK+GLL E IN A++N M
Sbjct: 304 FNQQHYTSTPEEAAARSIKAGLDLDCGPFLAIFTEGAVKKGLLTENDINLALANTLTVQM 363
Query: 362 RLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLA 421
RLG FDGN PY LGP+DVCT ++ LA EAA QGIVLLKNS SLPLS + +++A
Sbjct: 364 RLGMFDGNLG--PYANLGPRDVCTPAHKHLALEAAHQGIVLLKNSARSLPLSPRRHRTVA 421
Query: 422 VIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAA 481
VIGPN++ T MIGNY G C Y SPLQG++ T + GC V C Q A + AA
Sbjct: 422 VIGPNSDVTETMIGNYAGKACAYTSPLQGISRYARTLHQAGCAGVACKGNQGFGAAEAAA 481
Query: 482 -SSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVS 540
+DATV+V+G +IEAE+ DR ++LPG QQ LV+ VA ASRGPVILV+MSGG +DV+
Sbjct: 482 READATVLVMGLDQSIEAETRDRTGLLLPGYQQDLVTRVAQASRGPVILVLMSGGPIDVT 541
Query: 541 FAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMR 600
FAK + ++ +I+W GYPG+AGGAAIA++IFG NP G+LPMTWYP+ YV VPMT M MR
Sbjct: 542 FAKNDPRVAAIIWAGYPGQAGGAAIANIIFGAANPGGKLPMTWYPQDYVAKVPMTVMAMR 601
Query: 601 ADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAP----QLVFVPLAEDHVCRSS 656
A S YPGRTYRFYKG VF FG G+ Y+ H + K+P + L + +S
Sbjct: 602 A--SGNYPGRTYRFYKGPVVFPFGFGLSYTTFTHSLAKSPLAQLSVSLSNLNSANTILNS 659
Query: 657 ECKSLDVADEHCQNM-AFDIHLRVKNMGKMSSSHTVLLFFTPP---AVHNAPQKHLLGIE 712
S+ V+ +C + +H+ V N G+ +HTV +F PP K L+ E
Sbjct: 660 SSHSIKVSHTNCNSFPKMPLHVEVSNTGEFDGTHTVFVFAEPPINGIKGLGVNKQLIAFE 719
Query: 713 KVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
KVH+ ++ V+ VD CK L VVDE G RR+P+G+H LH+G+LK+ + V+
Sbjct: 720 KVHVMAGAKQTVQVDVDACKHLGVVDEYGKRRIPMGEHKLHIGDLKHTILVQ 771
>At3g19620 beta-xylosidase, putative
Length = 781
Score = 843 bits (2178), Expect = 0.0
Identities = 417/759 (54%), Positives = 550/759 (71%), Gaps = 17/759 (2%)
Query: 8 RVSVLVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQE 67
R+S+L+ +++C ++ FACD++ A+ A +GFC+ SL R DLV RL+L+E
Sbjct: 8 RLSLLIIA--LVSSLC-ESQKNFACDISAPAT-AKYGFCNVSLSYEARAKDLVSRLSLKE 63
Query: 68 KIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFN 127
K+ L + A V RLG+P YEWWSEALHGVS++GPG F+ VPGATSFP ILTAASFN
Sbjct: 64 KVQQLVNKATGVPRLGVPPYEWWSEALHGVSDVGPGVHFNGTVPGATSFPATILTAASFN 123
Query: 128 STLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKYAA 187
++L+ +G+VVSTEARAM+NVGLAGLTYWSPN+N+FRDPRWGRGQETPGEDPL+ SKYA
Sbjct: 124 TSLWLKMGEVVSTEARAMHNVGLAGLTYWSPNVNVFRDPRWGRGQETPGEDPLVVSKYAV 183
Query: 188 GYVKGLQQTDD-GDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPF 246
YVKGLQ D G S +LKV++CCKHYTAYD+DNWKG+ R+ F+A V++QDL+DT+Q PF
Sbjct: 184 NYVKGLQDVHDAGKSRRLKVSSCCKHYTAYDLDNWKGIDRFHFDAKVTKQDLEDTYQTPF 243
Query: 247 KSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQH 306
KSCV +G+V+SVMCSYN+VNG PTCADP+LL+GVIRGQW+L+GYIVSDCDS++V F D H
Sbjct: 244 KSCVEEGDVSSVMCSYNRVNGIPTCADPNLLRGVIRGQWRLDGYIVSDCDSIQVYFNDIH 303
Query: 307 YTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFF 366
YTKT E+A A ++ AGL+++CG +LGKYTE AVK L+ + ++ A+ N+ LMRLGFF
Sbjct: 304 YTKTREDAVALALKAGLNMNCGDFLGKYTENAVKLKKLNGSDVDEALIYNYIVLMRLGFF 363
Query: 367 DGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPN 426
DG+P +LP+G LGP DVC+ ++Q LA EAA+QGIVLL+N G LPL ++K LAVIGPN
Sbjct: 364 DGDPKSLPFGNLGPSDVCSKDHQMLALEAAKQGIVLLENR-GDLPLPKTTVKKLAVIGPN 422
Query: 427 ANATRVMIGNYEGIPCKYISPLQGLTALVPTS--YVPGCPDVHCASAQL-DDATKIAASS 483
ANAT+VMI NY G+PCKY SP+QGL VP Y PGC DV C L A K + +
Sbjct: 423 ANATKVMISNYAGVPCKYTSPIQGLQKYVPEKIVYEPGCKDVKCGDQTLISAAVKAVSEA 482
Query: 484 DATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAK 543
D TV+VVG +EAE LDRVN+ LPG Q+ LV +VANA++ V+LVIMS G +D+SFAK
Sbjct: 483 DVTVLVVGLDQTVEAEGLDRVNLTLPGYQEKLVRDVANAAKKTVVLVIMSAGPIDISFAK 542
Query: 544 ANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADP 603
I ++LWVGYPGEAGG AIA VIFG YNPSGRLP TWYP+ + + V MT+MNMR +
Sbjct: 543 NLSTIRAVLWVGYPGEAGGDAIAQVIFGDYNPSGRLPETWYPQEFADKVAMTDMNMRPNS 602
Query: 604 STGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDV 663
++G+PGR+YRFY G+ ++ FG G+ YS+ ++ AP ++ + + + ++ S+D+
Sbjct: 603 TSGFPGRSYRFYTGKPIYKFGYGLSYSSFSTFVLSAPSIIHI--KTNPIMNLNKTTSVDI 660
Query: 664 ADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHNA------PQKHLLGIEKVHLA 717
+ +C ++ I + VKN G S SH VL+F+ PP + P L+G E+V +
Sbjct: 661 STVNCHDLKIRIVIGVKNHGLRSGSHVVLVFWKPPKCSKSLVGGGVPLTQLVGFERVEVG 720
Query: 718 GKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGN 756
+ DVCK LS+VD G R++ G H L +G+
Sbjct: 721 RSMTEKFTVDFDVCKALSLVDTHGKRKLVTGHHKLVIGS 759
>At5g10560 beta-xylosidase - like protein
Length = 792
Score = 719 bits (1857), Expect = 0.0
Identities = 368/791 (46%), Positives = 508/791 (63%), Gaps = 46/791 (5%)
Query: 12 LVCFSFFCATVCG-----QTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQ 66
L+ FF + + + P F C K F+ + FC+ SL + R LV L L
Sbjct: 7 LISLLFFTSAIAETFKNLDSHPQFPC---KPPHFSSYPFCNVSLSIKQRAISLVSLLMLP 63
Query: 67 EKIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASF 126
EKIG L ++A V RLGIP YEWWSE+LHG+++ GPG F+ + ATSFP I++AASF
Sbjct: 64 EKIGQLSNTAASVPRLGIPPYEWWSESLHGLADNGPGVSFNGSISAATSFPQVIVSAASF 123
Query: 127 NSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKYA 186
N TL+ IG V+ E RAMYN G AGLT+W+PNIN+FRDPRWGRGQETPGEDP + S+Y
Sbjct: 124 NRTLWYEIGSAVAVEGRAMYNGGQAGLTFWAPNINVFRDPRWGRGQETPGEDPKVVSEYG 183
Query: 187 AGYVKGLQQT-------------------DDGDSNKLKVAACCKHYTAYDVDNWKGVQRY 227
+V+G Q+ DD KL ++ACCKH+TAYD++ W RY
Sbjct: 184 VEFVRGFQEKKKRKVLKRRFSDDVDDDRHDDDADGKLMLSACCKHFTAYDLEKWGNFTRY 243
Query: 228 TFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKL 287
FNAVV++QD++DT+QPPF++C+ DG + +MCSYN VNG P CA DLL+ R +W
Sbjct: 244 DFNAVVTEQDMEDTYQPPFETCIRDGKASCLMCSYNAVNGVPACAQGDLLQKA-RVEWGF 302
Query: 288 NGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEA 347
GYI SDCD+V +F Q YTK+PEEA A +I AG+D++CG+Y+ ++T+ A++QG + E
Sbjct: 303 EGYITSDCDAVATIFAYQGYTKSPEEAVADAIKAGVDINCGTYMLRHTQSAIEQGKVSEE 362
Query: 348 SINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSP 407
++ A+ N FA +RLG FDG+P YG LG D+C+S++++LA EA RQGIVLLKN
Sbjct: 363 LVDRALLNLFAVQLRLGLFDGDPRRGQYGKLGSNDICSSDHRKLALEATRQGIVLLKNDH 422
Query: 408 GSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALV-PTSYVPGCPDV 466
LPL+ + SLA++GP AN M G Y G PC+ + L V TSY GC DV
Sbjct: 423 KLLPLNKNHVSSLAIVGPMANNISNMGGTYTGKPCQRKTLFTELLEYVKKTSYASGCSDV 482
Query: 467 HCAS-AQLDDATKIAASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRG 525
C S +A IA +D ++V G L+ E E DRV++ LPG+Q+ LVS VA S+
Sbjct: 483 SCDSDTGFGEAVAIAKGADFVIVVAGLDLSQETEDKDRVSLSLPGKQKDLVSHVAAVSKK 542
Query: 526 PVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYP 585
PVILV+ GG +DV+FAK + +I SI+W+GYPGE GG A+A++IFG +NP GRLP TWYP
Sbjct: 543 PVILVLTGGGPVDVTFAKNDPRIGSIIWIGYPGETGGQALAEIIFGDFNPGGRLPTTWYP 602
Query: 586 ESYVENVPMTNMNMRADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFV 645
ES+ + V M++M+MRA+ S GYPGRTYRFY G V+SFG G+ Y+ E+KI+ AP + +
Sbjct: 603 ESFTD-VAMSDMHMRANSSRGYPGRTYRFYTGPQVYSFGTGLSYTKFEYKILSAP--IRL 659
Query: 646 PLAEDHVCRSSECKSL------------DVADEHCQNMAFDIHLRVKNMGKMSSSHTVLL 693
L+E +SS K L DV C+++ F++ + V N G++ SH V+L
Sbjct: 660 SLSELLPQQSSHKKQLQHGEELRYLQLDDVIVNSCESLRFNVRVHVSNTGEIDGSHVVML 719
Query: 694 FF-TPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLL 752
F PP + P+K L+G ++VH+ + F++D CK LSV +++G R +PLG H+L
Sbjct: 720 FSKMPPVLSGVPEKQLIGYDRVHVRSNEMMETVFVIDPCKQLSVANDVGKRVIPLGSHVL 779
Query: 753 HVGNLKYPLSV 763
+G+L++ LSV
Sbjct: 780 FLGDLQHSLSV 790
>At1g78060 putative protein
Length = 767
Score = 712 bits (1837), Expect = 0.0
Identities = 357/754 (47%), Positives = 505/754 (66%), Gaps = 24/754 (3%)
Query: 28 PPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQEKIGNLGDSAVDVGRLGIPRY 87
PP +CD + N + + FC LP+ R DLV RLT+ EKI L ++A + RLG+P Y
Sbjct: 22 PPHSCDPS-NPTTKLYQFCRTDLPIGKRARDLVSRLTIDEKISQLVNTAPGIPRLGVPAY 80
Query: 88 EWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFNSTLFQAIGKVVSTEARAMYN 147
EWWSEALHGV+ GPG RF+ V ATSFP ILTAASF+S + I +V+ EAR +YN
Sbjct: 81 EWWSEALHGVAYAGPGIRFNGTVKAATSFPQVILTAASFDSYEWFRIAQVIGKEARGVYN 140
Query: 148 VGLA-GLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKYAAGYVKGLQ-QTDDGD---SN 202
G A G+T+W+PNINIFRDPRWGRGQETPGEDP+++ YA YV+GLQ + DG SN
Sbjct: 141 AGQANGMTFWAPNINIFRDPRWGRGQETPGEDPMMTGTYAVAYVRGLQGDSFDGRKTLSN 200
Query: 203 KLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSY 262
L+ +ACCKH+TAYD+D WKG+ RY FNA VS DL +T+QPPFK C+ +G + +MC+Y
Sbjct: 201 HLQASACCKHFTAYDLDRWKGITRYVFNAQVSLADLAETYQPPFKKCIEEGRASGIMCAY 260
Query: 263 NQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAG 322
N+VNG P+CADP+LL RGQW GYI SDCD+V +++ Q Y K+PE+A A + AG
Sbjct: 261 NRVNGIPSCADPNLLTRTARGQWAFRGYITSDCDAVSIIYDAQGYAKSPEDAVADVLKAG 320
Query: 323 LDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKD 382
+D++CGSYL K+T+ A++Q + E I+ A+ N F+ +RLG F+G+P+ LPYG + P +
Sbjct: 321 MDVNCGSYLQKHTKSALQQKKVSETDIDRALLNLFSVRIRLGLFNGDPTKLPYGNISPNE 380
Query: 383 VCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPC 442
VC+ +Q LA +AAR GIVLLKN+ LP S +S+ SLAVIGPNA+ + ++GNY G PC
Sbjct: 381 VCSPAHQALALDAARNGIVLLKNNLKLLPFSKRSVSSLAVIGPNAHVVKTLLGNYAGPPC 440
Query: 443 KYISPLQGLTALVPTS-YVPGCPDVHCASAQLDDATKIAASSDATVIVVGASLAIEAESL 501
K ++PL L + V + Y GC V C++A +D A IA ++D V+++G E E
Sbjct: 441 KTVTPLDALRSYVKNAVYHQGCDSVACSNAAIDQAVAIAKNADHVVLIMGLDQTQEKEDF 500
Query: 502 DRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAG 561
DRV++ LPG+QQ L++ VANA++ PV+LV++ GG +D+SFA N+KI SI+W GYPGEAG
Sbjct: 501 DRVDLSLPGKQQELITSVANAAKKPVVLVLICGGPVDISFAANNNKIGSIIWAGYPGEAG 560
Query: 562 GAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYKGETVF 621
G AI+++IFG +NP GRLP+TWYP+S+V N+ MT+M MR+ +TGYPGRTY+FYKG V+
Sbjct: 561 GIAISEIIFGDHNPGGRLPVTWYPQSFV-NIQMTDMRMRS--ATGYPGRTYKFYKGPKVY 617
Query: 622 SFGDGIGYSNVEHKI-VKAPQLVFVPLAEDHVCRSSECKSL--DVADEHCQNMAFDIHLR 678
FG G+ YS ++ A +++ ++ S +L ++ E C + +
Sbjct: 618 EFGHGLSYSAYSYRFKTLAETNLYLNQSKAQTNSDSVRYTLVSEMGKEGCDVAKTKVTVE 677
Query: 679 VKNMGKMSSSHTVLLFFTPPAVH-------NAPQKHLLGIEKVHLAGKSEAQVRFMVDVC 731
V+N G+M+ H VL+F A H +K L+G + + L+ +A++ F + +C
Sbjct: 678 VENQGEMAGKHPVLMF----ARHERGGEDGKRAEKQLVGFKSIVLSNGEKAEMEFEIGLC 733
Query: 732 KDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
+ LS +E G + G++ L VG+ + PL V +
Sbjct: 734 EHLSRANEFGVMVLEEGKYFLTVGDSELPLIVNV 767
>At5g09700 beta-glucosidase - like protein
Length = 411
Score = 535 bits (1379), Expect = e-152
Identities = 260/411 (63%), Positives = 316/411 (76%), Gaps = 6/411 (1%)
Query: 361 MRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSL 420
MRLGFFDGNP PYGGLGPKDVCT EN+ELA E ARQGIVLLKNS GSLPLS +IK+L
Sbjct: 1 MRLGFFDGNPKNQPYGGLGPKDVCTVENRELAVETARQGIVLLKNSAGSLPLSPSAIKTL 60
Query: 421 AVIGPNANATRVMIGNYEGIPCKYISPLQGLTALV-PTSYVPGCPDVHCASAQLDDATKI 479
AVIGPNAN T+ MIGNYEG+ CKY +PLQGL V T Y GC +V C A LD A +
Sbjct: 61 AVIGPNANVTKTMIGNYEGVACKYTTPLQGLERTVLTTKYHRGCFNVTCTEADLDSAKTL 120
Query: 480 AASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDV 539
AAS+DATV+V+GA IE E+LDR+++ LPG+QQ LV++VA A+RGPV+LVIMSGGG D+
Sbjct: 121 AASADATVLVMGADQTIEKETLDRIDLNLPGKQQELVTQVAKAARGPVVLVIMSGGGFDI 180
Query: 540 SFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNM 599
+FAK ++KITSI+WVGYPGEAGG AIADVIFG +NPSG+LPMTWYP+SYVE VPMTNMNM
Sbjct: 181 TFAKNDEKITSIMWVGYPGEAGGIAIADVIFGRHNPSGKLPMTWYPQSYVEKVPMTNMNM 240
Query: 600 RADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECK 659
R D S GY GRTYRFY GETV++FGDG+ Y+N H+++KAP+ V + L E CRS EC+
Sbjct: 241 RPDKSNGYLGRTYRFYIGETVYAFGDGLSYTNFSHQLIKAPKFVSLNLDESQSCRSPECQ 300
Query: 660 SLDVADEHCQNMA-----FDIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKV 714
SLD HC+ F++ L+V+N+G + TV LF TPP VH +P+K LLG EK+
Sbjct: 301 SLDAIGPHCEKAVGERSDFEVQLKVRNVGDREGTETVFLFTTPPEVHGSPRKQLLGFEKI 360
Query: 715 HLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
L K E VRF VDVCKDL VVDE+G R++ LG HLLHVG+LK+ ++ +
Sbjct: 361 RLGKKEETVVRFKVDVCKDLGVVDEIGKRKLALGHHLLHVGSLKHSFNISV 411
>At3g47000 beta-D-glucan exohydrolase - like protein
Length = 608
Score = 140 bits (354), Expect = 2e-33
Identities = 162/616 (26%), Positives = 270/616 (43%), Gaps = 99/616 (16%)
Query: 51 PVADRVADLVKRLTLQEKIGNLGD-----------------SAVDVG------------- 80
PV RV DL+ R+TL EKIG + S ++ G
Sbjct: 16 PVEARVKDLLSRMTLPEKIGQMTQIERRVASPSAFTDFFIGSVLNAGGSVPFEDAKSSDW 75
Query: 81 --------------RLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASF 126
RLGIP + ++A+HG +N V GAT FP I A+
Sbjct: 76 ADMIDGFQRSALASRLGIPII-YGTDAVHGNNN----------VYGATVFPHNIGLGATR 124
Query: 127 NSTLFQAIGKVVSTEARAMYNVGLAGLTY-WSPNINIFRDPRWGRGQETPGEDPLLSSKY 185
++ L + IG + E RA +G+ + +SP + + RDPRWGR E+ GEDP L +
Sbjct: 125 DADLVRRIGAATALEVRA------SGVHWAFSPCVAVLRDPRWGRCYESYGEDPELVCEM 178
Query: 186 AAGYVKGLQQTDDGD--------SNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQD 237
+ V GLQ + + + V AC KH+ D KG+ N + S ++
Sbjct: 179 TS-LVSGLQGVPPEEHPNGYPFVAGRNNVVACVKHFVG-DGGTDKGINE--GNTIASYEE 234
Query: 238 LDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDS 297
L+ PP+ C+ G V++VM SY+ NG AD LL +++ + G++VSD +
Sbjct: 235 LEKIHIPPYLKCLAQG-VSTVMASYSSWNGTRLHADRFLLTEILKEKLGFKGFLVSDWEG 293
Query: 298 VEVLFKDQ-----HYTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNA 352
++ L + Q + KT A ++ + ++ T+ V+ G + A IN+A
Sbjct: 294 LDRLSEPQGSNYRYCIKTAVNAGIDMVMVPFKYE--QFIQDMTD-LVESGEIPMARINDA 350
Query: 353 ISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGS--- 409
+ G F G+P L L P C E++ELA+EA R+ +VLLK+ +
Sbjct: 351 VERILRVKFVAGLF-GHP--LTDRSLLPTVGC-KEHRELAQEAVRKSLVLLKSGKNADKP 406
Query: 410 -LPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHC 468
LPL ++ K + V G +A+ G + + T L+ +
Sbjct: 407 FLPLD-RNAKRILVTGTHADDLGYQCGGWTKTWFGLSGRITIGTTLLDAIKEAVGDETEV 465
Query: 469 ASAQLDDATKIAASSDATVIVVGASLAIEAESL-DRVNIVLPGQQQLLVSEVANASRGPV 527
+ +A+S + +V AE++ D + +P +V+ VA P
Sbjct: 466 IYEKTPSKETLASSEGFSYAIVAVGEPPYAETMGDNSELRIPFNGTDIVTAVAEII--PT 523
Query: 528 ILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPES 587
+++++SG + V +K +++ PG G +ADV+FG Y+ G+LP++W+
Sbjct: 524 LVILISGRPV-VLEPTVLEKTEALVAAWLPG-TEGQGVADVVFGDYDFKGKLPVSWF--K 579
Query: 588 YVENVPMTNMNMRADP 603
+VE++P+ DP
Sbjct: 580 HVEHLPLDAHANSYDP 595
>At5g04885 unknown protein
Length = 665
Score = 140 bits (352), Expect = 3e-33
Identities = 172/627 (27%), Positives = 266/627 (41%), Gaps = 107/627 (17%)
Query: 47 DKSLPVADRVADLVKRLTLQEKIGNL------------------------GDSA------ 76
D V+DRVADL R+TL+EKIG + G SA
Sbjct: 32 DPKQTVSDRVADLFGRMTLEEKIGQMVQIDRSVATVNIMRDYFIGSVLSGGGSAPLPEAS 91
Query: 77 ----VD----------VGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILT 122
VD V RLGIP + +A+HG +N V AT FP +
Sbjct: 92 AQNWVDMINEYQKGALVSRLGIPMI-YGIDAVHGHNN----------VYNATIFPHNVGL 140
Query: 123 AASFNSTLFQAIGKVVSTEARAMYNVGLAGLTY-WSPNINIFRDPRWGRGQETPGEDPLL 181
A+ + L + IG + E RA G+ Y ++P I + RDPRWGR E+ ED +
Sbjct: 141 GATRDPDLVKRIGAATAVEVRA------TGIPYTFAPCIAVCRDPRWGRCYESYSEDHKV 194
Query: 182 SSKYAAGYVKGLQQTDDGD--------SNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVV 233
+ GLQ + + KVAAC KHY D +GV N V
Sbjct: 195 VEDMT-DVILGLQGEPPSNYKHGVPFVGGRDKVAACAKHYVG-DGGTTRGVNEN--NTVT 250
Query: 234 SQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVS 293
L P + V G V++VM SY+ NG+ A+ +L+ G ++G K G+++S
Sbjct: 251 DLHGLLSVHMPAYADAVYKG-VSTVMVSYSSWNGEKMHANTELITGYLKGTLKFKGFVIS 309
Query: 294 DCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE------GAVKQGLLDEA 347
D V+ + H T AA I AG+D+ + +TE VK +
Sbjct: 310 DWQGVDKISTPPHTHYTASVRAA--IQAGIDMVMVPF--NFTEFVNDLTTLVKNNSIPVT 365
Query: 348 SINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSP 407
I++A+ +G F+ + + ++ + +++LAREA R+ +VLLKN
Sbjct: 366 RIDDAVRRILLVKFTMGLFENPLADYSFS----SELGSQAHRDLAREAVRKSLVLLKNGN 421
Query: 408 GS---LPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCP 464
+ LPL K+ K L V G +A+ G + I + S + S V
Sbjct: 422 KTNPMLPLPRKTSKIL-VAGTHADNLGYQCGGWT-ITWQGFSGNKNTRGTTLLSAVKSAV 479
Query: 465 D--VHCASAQLDDATKIAASSDA-TVIVVG-ASLAIEAESLDRVNIVLPGQQQLLVSEVA 520
D + DA I +++ A +I VG A A D++ ++ PG ++S
Sbjct: 480 DQSTEVVFRENPDAEFIKSNNFAYAIIAVGEPPYAETAGDSDKLTMLDPG--PAIISSTC 537
Query: 521 NASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLP 580
A + ++V++SG + + A+ W+ PG G I D +FG + SG+LP
Sbjct: 538 QAVK--CVVVVISGRPLVMEPYVASIDALVAAWL--PG-TEGQGITDALFGDHGFSGKLP 592
Query: 581 MTWYPESYVENVPMTNMNMRADPSTGY 607
+TW+ + E +PM+ + DP Y
Sbjct: 593 VTWFRNT--EQLPMSYGDTHYDPLFAY 617
>At5g20950 beta-D-glucan exohydrolase-like protein
Length = 624
Score = 139 bits (349), Expect = 8e-33
Identities = 169/634 (26%), Positives = 266/634 (41%), Gaps = 102/634 (16%)
Query: 47 DKSLPVADRVADLVKRLTLQEKIGNLGD----------------SAVDVGRLGIPRY--- 87
D P+ R+ DL+ R+TLQEKIG + +V G +P
Sbjct: 28 DPKQPLGARIRDLMNRMTLQEKIGQMVQIERSVATPEVMKKYFIGSVLSGGGSVPSEKAT 87
Query: 88 -EWWSEALHGVSNIGPGTRFS-------------SVVPGATSFPMPILTAASFNSTLFQA 133
E W ++ + TR + V GAT FP + + + L +
Sbjct: 88 PETWVNMVNEIQKASLSTRLGIPMIYGIDAVHGHNNVYGATIFPHNVGLGVTRDPNLVKR 147
Query: 134 IGKVVSTEARAMYNVGLAGLTY-WSPNINIFRDPRWGRGQETPGEDPLLSSKYAAGYVKG 192
IG + E RA G+ Y ++P I + RDPRWGR E+ ED + + + G
Sbjct: 148 IGAATALEVRA------TGIPYAFAPCIAVCRDPRWGRCYESYSEDYRIVQQMTE-IIPG 200
Query: 193 LQQTDDGD-----------SNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDT 241
LQ GD K KVAAC KH+ D +G+ N V+ + L
Sbjct: 201 LQ----GDLPTKRKGVPFVGGKTKVAACAKHFVG-DGGTVRGIDEN--NTVIDSKGLFGI 253
Query: 242 FQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVL 301
P + + V G VA++M SY+ NG A+ +L+ G ++ + K G+++SD ++ +
Sbjct: 254 HMPGYYNAVNKG-VATIMVSYSAWNGLRMHANKELVTGFLKNKLKFRGFVISDWQGIDRI 312
Query: 302 FKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE------GAVKQGLLDEASINNAISN 355
H + A I AG+D+ Y YTE +++ L+ + I++A+
Sbjct: 313 TTPPHLNYSYSVYA--GISAGIDMIMVPY--NYTEFIDEISSQIQKKLIPISRIDDALKR 368
Query: 356 NFATLMRLGFFDGNPSTLPYGG-LGPKDVCTSENQELAREAARQGIVLLKNSPGS----L 410
+G F+ + L + LG K E++ELAREA R+ +VLLKN L
Sbjct: 369 ILRVKFTMGLFEEPLADLSFANQLGSK-----EHRELAREAVRKSLVLLKNGKTGAKPLL 423
Query: 411 PLSAKSIKSLAVIGPNANATRVMIG----NYEGIPCKYISPLQGLTALVPTSYVPGCPDV 466
PL KS K L V G +A+ G ++G+ + + A V + P V
Sbjct: 424 PLPKKSGKIL-VAGAHADNLGYQCGGWTITWQGLNGNDHTVGTTILAAVKNTVAPTTQVV 482
Query: 467 HCASAQLDDATKIAASS-DATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRG 525
+ +Q DA + + D ++VVG E D N+ + ++ V G
Sbjct: 483 Y---SQNPDANFVKSGKFDYAIVVVGEPPYAEMFG-DTTNLTISDPGPSIIGNVC----G 534
Query: 526 PVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYP 585
V V++ G V I +++ PG G +AD +FG Y +G+L TW+
Sbjct: 535 SVKCVVVVVSGRPVVIQPYVSTIDALVAAWLPG-TEGQGVADALFGDYGFTGKLARTWFK 593
Query: 586 ESYVENVPMTNMNMRADPSTGYP---GRTYRFYK 616
V+ +PM + DP YP G T + YK
Sbjct: 594 S--VKQLPMNVGDRHYDPL--YPFGFGLTTKPYK 623
>At3g47010 beta-D-glucan exohydrolase - like protein
Length = 609
Score = 130 bits (328), Expect = 2e-30
Identities = 163/636 (25%), Positives = 272/636 (42%), Gaps = 131/636 (20%)
Query: 47 DKSLPVADRVADLVKRLTLQEKIGNLGDSAVDVG-------------------------- 80
++ PV RV DL+ R+TL EKIG + V
Sbjct: 13 NRDAPVEARVKDLLSRMTLPEKIGQMTQIERSVASPQVITNSFIGSVQSGAGSWPLEDAK 72
Query: 81 ------------------RLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILT 122
RLGIP + ++A+HG +N V GAT FP I
Sbjct: 73 SSDWADMIDGFQRSALASRLGIPII-YGTDAVHGNNN----------VYGATVFPHNIGL 121
Query: 123 AASFNSTLFQAIGKVVSTEARAMYNVGLAGLTY-WSPNINIFRDPRWGRGQETPGEDPLL 181
A+ ++ L + IG + E RA +G+ + ++P + + DPRWGR E+ E +
Sbjct: 122 GATRDADLVKRIGAATALEIRA------SGVHWTFAPCVAVLGDPRWGRCYESYSEAAKI 175
Query: 182 SSKYAAGYVKGLQQTDDGD--------SNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVV 233
+ + + GLQ + + + V AC KH+ D KG+ N +
Sbjct: 176 VCEMSL-LISGLQGEPPEEHPYGYPFLAGRNNVIACAKHFVG-DGGTEKGLSEG--NTIT 231
Query: 234 SQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVS 293
S +DL+ P+ +C+ G V++VM S++ NG +D LL V++ + G++VS
Sbjct: 232 SYEDLEKIHVAPYLNCIAQG-VSTVMASFSSWNGSRLHSDYFLLTEVLKQKLGFKGFLVS 290
Query: 294 DCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE------GAVKQGLLDEA 347
D D +E + + + I AG+D+ + KY + V+ G + A
Sbjct: 291 DWDGLETISEPE--GSNYRNCVKLGINAGIDMVMVPF--KYEQFIQDMTDLVESGEIPMA 346
Query: 348 SINNAISNNFATLMRLGFFD---GNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLK 404
+N+A+ G F+ + S L G +G K E++E+AREA R+ +VLLK
Sbjct: 347 RVNDAVERILRVKFVAGLFEHPLADRSLL--GTVGCK-----EHREVAREAVRKSLVLLK 399
Query: 405 NSPGS----LPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYV 460
N + LPL ++ K + V+G +AN +GN G K S G +
Sbjct: 400 NGKNADTPFLPLD-RNAKRILVVGMHAND----LGNQCGGWTKIKSGQSGRITI------ 448
Query: 461 PGCPDVHCASAQLDDATKI----------AASSDA---TVIVVGASLAIEAESLDRVNIV 507
G + A + D T++ ASSD ++ VG E + D +
Sbjct: 449 -GTTLLDSIKAAVGDKTEVIFEKTPTKETLASSDGFSYAIVAVGEPPYAEMKG-DNSELT 506
Query: 508 LPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIAD 567
+P +++ V A + P ++++ SG M V +K +++ +PG G ++D
Sbjct: 507 IPFNGNNIITAV--AEKIPTLVILFSGRPM-VLEPTVLEKTEALVAAWFPG-TEGQGMSD 562
Query: 568 VIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADP 603
VIFG Y+ G+LP++W+ V+ +P+ DP
Sbjct: 563 VIFGDYDFKGKLPVSWFKR--VDQLPLNAEANSYDP 596
>At3g47050 beta-D-glucan exohydrolase - like protein
Length = 612
Score = 129 bits (324), Expect = 6e-30
Identities = 152/603 (25%), Positives = 256/603 (42%), Gaps = 85/603 (14%)
Query: 47 DKSLPVADRVADLVKRLTLQEKIGNL-------GDSAV----DVGRLGIPRYEW------ 89
++ PV RV DL+ R+TL EKIG + AV +G + W
Sbjct: 12 NREAPVEARVKDLLSRMTLAEKIGQMTLIERSVASEAVIRDFSIGSVLNRAGGWPFEDAK 71
Query: 90 ---WSEALHGVSNIGPGTRF-------------SSVVPGATSFPMPILTAASFNSTLFQA 133
W++ + G +R ++ V GAT FP I A+ ++ L +
Sbjct: 72 SSNWADMIDGFQRSALESRLGIPIIYGIDAVHGNNDVYGATIFPHNIGLGATRDADLVKR 131
Query: 134 IGKVVSTEARAMYNVGLAGLTYWS--PNINIFRDPRWGRGQETPGEDPLLSSKYAAGYVK 191
IG + E RA +W+ P + + +DPRWGR E+ GE + S+ + V
Sbjct: 132 IGAATALEVRACG-------AHWAFAPCVAVVKDPRWGRCYESYGEVAQIVSEMTS-LVS 183
Query: 192 GLQQTDDGD--------SNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQ 243
GLQ D + + V AC KH+ N K + N ++ +DL+
Sbjct: 184 GLQGEPSKDHTNGYPFLAGRKNVVACAKHFVGDGGTN-KAINEG--NTILRYEDLERKHI 240
Query: 244 PPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFK 303
P+K C+ G V++VM SY+ NG + LL +++ + GY+VSD + ++ L
Sbjct: 241 APYKKCISQG-VSTVMASYSSWNGDKLHSHYFLLTEILKQKLGFKGYVVSDWEGLDRLSD 299
Query: 304 DQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE------GAVKQGLLDEASINNAISNNF 357
I AG+D+ + KY + V+ G + A +N+A+
Sbjct: 300 PPG--SNYRNCVKIGINAGIDMVMVPF--KYEQFRNDLIDLVESGEVSMARVNDAVERIL 355
Query: 358 ATLMRLGFFD---GNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGS--LPL 412
G F+ + S LP G E++ELAREA R+ +VLLKN LPL
Sbjct: 356 RVKFVAGLFEFPLTDRSLLPTVGC-------KEHRELAREAVRKSLVLLKNGRYGEFLPL 408
Query: 413 SAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQ 472
+ + + L V+G +A+ G + + T L+ + +
Sbjct: 409 NCNAERIL-VVGTHADDLGYQCGGWTKTMYGQSGRITDGTTLLDAIKAAVGDETEVIYEK 467
Query: 473 LDDATKIAASSDATVIVVGASLAIEAESL-DRVNIVLPGQQQLLVSEVANASRGPVILVI 531
+A+ + +V + AE++ D +V+P +++ VA + P ++++
Sbjct: 468 SPSEETLASGYRFSYAIVAVGESPYAETMGDNSELVIPFNGSEIITTVAE--KIPTLVIL 525
Query: 532 MSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVEN 591
SG M + + +K +++ PG G IADVIFG Y+ G+LP TW+ V+
Sbjct: 526 FSGRPMFLE-PQVLEKAEALVAAWLPGTEG-QGIADVIFGDYDFRGKLPATWFKR--VDQ 581
Query: 592 VPM 594
+P+
Sbjct: 582 LPL 584
>At3g62710 beta-D-glucan exohydrolase-like protein
Length = 650
Score = 126 bits (316), Expect = 5e-29
Identities = 149/582 (25%), Positives = 244/582 (41%), Gaps = 75/582 (12%)
Query: 54 DRVADLVKRLTLQEKIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGA 113
D D+ KR+ + L S RLGIP + +A+HG + A
Sbjct: 102 DTGKDIAKRIFQTNAMKKLSLST----RLGIPLL-YAVDAVHG----------HNTFIDA 146
Query: 114 TSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQE 173
T FP + A+ + L + IG + + E RA G+A ++P + + RDPRWGR E
Sbjct: 147 TIFPHNVGLGATRDPQLVKKIGAITAQEVRA---TGVAQA--FAPCVAVCRDPRWGRCYE 201
Query: 174 TPGEDPLLSSKYAAGYVKGLQQTDDGDSN-KLKVAACCKHYTAYDVDNWKGVQRYTFNAV 232
+ EDP + + + GLQ ++ K+ VA C KH+ D G+ N V
Sbjct: 202 SYSEDPAVVNMMTESIIDGLQGNAPYLADPKINVAGCAKHFVG-DGGTINGINEN--NTV 258
Query: 233 VSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIV 292
L PPF+ V G +AS+M SY+ +NG A+ ++ ++ K G+++
Sbjct: 259 ADNATLFGIHMPPFEIAVKKG-IASIMASYSSLNGVKMHANRAMITDYLKNTLKFQGFVI 317
Query: 293 SDCDSVEVL--FKDQHYTKTPEEAAAKSILAGLDL-----DCGSYLGKYTEGAVKQGLLD 345
SD ++ + + +YT + E SI AG+D+ YL K T V G +
Sbjct: 318 SDWLGIDKITPIEKSNYTYSIE----ASINAGIDMVMVPWAYPEYLEKLT-NLVNGGYIP 372
Query: 346 EASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKN 405
+ I++A+ +G F+ ++L L + + ++E+ REA R+ +VLLKN
Sbjct: 373 MSRIDDAVRRILRVKFSIGLFE---NSLADEKLPTTEFGSEAHREVGREAVRKSMVLLKN 429
Query: 406 SPGS----LPLSAKSIKSLAVIGPNANATRVMIGNY-------------------EGIPC 442
+PL K +K + V G +AN G + G+P
Sbjct: 430 GKTDADKIVPL-PKKVKKIVVAGRHANDMGWQCGGFSLTWQGFNGTGEDMPTNTKHGLPT 488
Query: 443 KYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAASSDATVIVVGASLAIEA-ESL 501
I L A+ V +V D K+ A + T++VVG + E
Sbjct: 489 GKIKGTTILEAI--QKAVDPTTEVVYVEEPNQDTAKLHADAAYTIVVVGETPYAETFGDS 546
Query: 502 DRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAG 561
+ I PG L S + +++++ G + D + ++ PG
Sbjct: 547 PTLGITKPGPDTL--SHTCGSGMKCLVILVT---GRPLVIEPYIDMLDALAVAWLPG-TE 600
Query: 562 GAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADP 603
G +ADV+FG + +G LP TW +V +PM + DP
Sbjct: 601 GQGVADVLFGDHPFTGTLPRTWM--KHVTQLPMNVGDKNYDP 640
>At3g47040 beta-D-glucan exohydrolase - like protein
Length = 636
Score = 125 bits (315), Expect = 7e-29
Identities = 162/670 (24%), Positives = 278/670 (41%), Gaps = 128/670 (19%)
Query: 47 DKSLPVADRVADLVKRLTLQEKIGNLGD-------------------------------- 74
+K PV RV DL+ R+TL EKIG +
Sbjct: 12 NKDAPVEARVKDLLSRMTLPEKIGQMTQIERVVTTPPVITDNFIGSVLNGGGSWPFEDAK 71
Query: 75 ------------SAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFS-SVVPGATSFPM--- 118
+A RLGIP + +A+HG +N+ T F ++ GATS M
Sbjct: 72 TSDWADMIDGYQNAALASRLGIPII-YGIDAVHGNNNVYGATIFPHNIGLGATSLVMLLH 130
Query: 119 -----------PILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWS--PNINIFRD 165
++ ++ L + +G + E RA +W+ P + RD
Sbjct: 131 IDLEPKSLGRNKVVVKCDRDADLIRRVGAATALEVRACG-------AHWAFAPCVAALRD 183
Query: 166 PRWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGD--------SNKLKVAACCKHYTAYD 217
PRWGR E+ EDP + + ++ V GLQ + + + V AC KH+ D
Sbjct: 184 PRWGRSYESYSEDPDIICELSS-LVSGLQGEPPKEHPNGYPFLAGRNNVVACAKHFVG-D 241
Query: 218 VDNWKGVQRYTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLL 277
KG+ N +VS ++L+ P+ +C+ G V++VM SY+ NG +D LL
Sbjct: 242 GGTDKGINE--GNTIVSYEELEKIHLAPYLNCLAQG-VSTVMASYSSWNGSKLHSDYFLL 298
Query: 278 KGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE- 336
+++ + G+++SD +++E L + + S+ AG+D+ + KY +
Sbjct: 299 TELLKQKLGFKGFVISDWEALERL--SEPFGSNYRNCVKISVNAGVDMVMVPF--KYEQF 354
Query: 337 -----GAVKQGLLDEASINNAISNNFATLMRLGFFDGNPST--LPYGGLGPKDVCTSENQ 389
V+ G + + I++A+ G F+ +P T G +G K E++
Sbjct: 355 IKDLTDLVESGEVTMSRIDDAVERILRVKFVAGLFE-HPLTDRSLLGTVGCK-----EHR 408
Query: 390 ELAREAARQGIVLLKNSPGS----LPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYI 445
ELARE+ R+ +VLLKN S LPL +++K + V G +A+ G +
Sbjct: 409 ELARESVRKSLVLLKNGTNSEKPFLPLD-RNVKRILVTGTHADDLGYQCGGWTKAWFGLS 467
Query: 446 SPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAASSDATVIVVGASLAIEAESL-DRV 504
+ T L+ + +A+ + +V AE+L D
Sbjct: 468 GRITIGTTLLDAIKEAVGDKTEVIYEKTPSEETLASLQRFSYAIVAVGETPYAETLGDNS 527
Query: 505 NIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAA 564
+ +P +V+ A A + P ++V+ SG + V +K +++ PG G
Sbjct: 528 ELTIPLNGNDIVT--ALAEKIPTLVVLFSGRPL-VLEPLVLEKAEALVAAWLPG-TEGQG 583
Query: 565 IADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYKGETVFSFG 624
+ DVIFG Y+ G+LP++W+ V+ +P+T DP +F G
Sbjct: 584 MTDVIFGDYDFEGKLPVSWFKR--VDQLPLTADANSYDP----------------LFPLG 625
Query: 625 DGIGYSNVEH 634
G+ Y++ E+
Sbjct: 626 FGLNYNSSEN 635
>At5g20940 beta-glucosidase - like protein
Length = 626
Score = 125 bits (314), Expect = 9e-29
Identities = 162/630 (25%), Positives = 267/630 (41%), Gaps = 92/630 (14%)
Query: 32 CDVTKN-ASFAGFGFCDKSLPVADRVADLVKRLTLQEKIGNLGD---------------- 74
C V N A + D P+ R+ +L+ +TL+EKIG +
Sbjct: 18 CTVAANKVPLANAKYKDPKEPLGVRIKNLMSHMTLEEKIGQMVQVERVNATTEVMQKYFV 77
Query: 75 SAVDVGRLGIPRY----EWWSEALHGVSNIGPGTRFS-------------SVVPGATSFP 117
+V G +P+ E W ++ V TR + V AT FP
Sbjct: 78 GSVFSGGGSVPKPYIGPEAWVNMVNEVQKKALSTRLGIPIIYGIDAVHGHNTVYNATIFP 137
Query: 118 MPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTY-WSPNINIFRDPRWGRGQETPG 176
+ + + L + IG+ + E RA G+ Y ++P I + RDPRWGR E+
Sbjct: 138 HNVGLGVTRDPGLVKRIGEATALEVRA------TGIQYVFAPCIAVCRDPRWGRCYESYS 191
Query: 177 EDPLLSSKYAAGYVKGLQQTDDGD-----------SNKLKVAACCKHYTAYDVDNWKGVQ 225
ED + + + GLQ GD + K KVAAC KH+ D +G+
Sbjct: 192 EDHKIVQQMTE-IIPGLQ----GDLPTGQKGVPFVAGKTKVAACAKHFVG-DGGTLRGMN 245
Query: 226 RYTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQW 285
N V++ L P + V G VA+VM SY+ +NG A+ L+ G ++ +
Sbjct: 246 AN--NTVINSNGLLGIHMPAYHDAVNKG-VATVMVSYSSINGLKMHANKKLITGFLKNKL 302
Query: 286 KLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCG-SYLGKYTE---GAVKQ 341
K G ++SD V+ + + AA + AGLD+ G S L K + VK+
Sbjct: 303 KFRGIVISDYLGVDQINTPLGANYSHSVYAATT--AGLDMFMGSSNLTKLIDELTSQVKR 360
Query: 342 GLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIV 401
+ + I++A+ +G F+ NP + L K + + E++ELAREA R+ +V
Sbjct: 361 KFIPMSRIDDAVKRILRVKFTMGLFE-NP--IADHSLA-KKLGSKEHRELAREAVRKSLV 416
Query: 402 LLKNSPGS----LPLSAKSIKSLAVIGPNANATRVMIG----NYEGIPCKYISPLQGLTA 453
LLKN + LPL K+ K L V G +A+ G ++G+ ++ + A
Sbjct: 417 LLKNGENADKPLLPLPKKANKIL-VAGTHADNLGYQCGGWTITWQGLNGNNLTIGTTILA 475
Query: 454 LVPTSYVPGCPDVHCASAQLDDATKIAASSDATVIVVGASLAIEAESLDRVNIVLPGQQQ 513
V + P ++ + D A D ++ VG E D N+ +
Sbjct: 476 AVKKTVDPKTQVIY--NQNPDTNFVKAGDFDYAIVAVGEKPYAEGFG-DSTNLTISEPGP 532
Query: 514 LLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFY 573
+ V + + ++V++SG + + + I +++ PG G +ADV+FG Y
Sbjct: 533 STIGNVCASVK--CVVVVVSGRPVVMQIS----NIDALVAAWLPG-TEGQGVADVLFGDY 585
Query: 574 NPSGRLPMTWYPESYVENVPMTNMNMRADP 603
+G+L TW+ V+ +PM + DP
Sbjct: 586 GFTGKLARTWF--KTVDQLPMNVGDPHYDP 613
>At5g46640 unknown protein
Length = 386
Score = 33.5 bits (75), Expect = 0.46
Identities = 48/153 (31%), Positives = 66/153 (42%), Gaps = 22/153 (14%)
Query: 359 TLMRLGFFDGNPSTLPYGGLGPKDVCTSENQEL--------AREAARQGIVLLKNSPGSL 410
TL LGF DG+PS+ P G D +NQ+L R+ G + L +P S
Sbjct: 72 TLESLGFGDGSPSSQPM-RFGIDD----QNQQLQVKKKRGRPRKYTPDGSIALGLAPTSP 126
Query: 411 PLSAKSIK----SLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDV 466
LSA S + G N N+ + G P S + L AL TS V P V
Sbjct: 127 LLSAASNSYGEGGVGDSGGNGNSVDPPVKRNRGRPPG--SSKKQLDALGGTSGVGFTPHV 184
Query: 467 HCASAQLDDATKIAASSD---ATVIVVGASLAI 496
+ D A+K+ A SD T+ ++ AS A+
Sbjct: 185 IEVNTGEDIASKVMAFSDQGSRTICILSASGAV 217
>At1g44800 nodulin like protein
Length = 370
Score = 31.6 bits (70), Expect = 1.7
Identities = 35/122 (28%), Positives = 52/122 (41%), Gaps = 15/122 (12%)
Query: 437 YEGIPCKYISP-LQGLTA-----LVPTSYVPGCPDVHCASAQLDDATKIAASS--DATVI 488
Y G+ C I+ +Q + + TS+ P C + L A KI S A I
Sbjct: 255 YSGVVCSGIAYYIQSIVIKQRGPVFTTSFSPMCMIITAFLGALVLAEKIHLGSIIGAVFI 314
Query: 489 VVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKI 548
V+G + +S D VN P +++ VA + P+ V+ G DVS A N +
Sbjct: 315 VLGLYSVVWGKSKDEVN---PLDEKI----VAKSQELPITNVVKQTNGHDVSGAPTNGVV 367
Query: 549 TS 550
TS
Sbjct: 368 TS 369
>At5g56040 receptor protein kinase-like protein
Length = 1090
Score = 30.8 bits (68), Expect = 3.0
Identities = 23/93 (24%), Positives = 41/93 (43%), Gaps = 7/93 (7%)
Query: 656 SECKSLDVADEHCQNMAFDIH---LRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIE 712
S+C+ L D N++ I ++N+ K+ L F PP + N + L +
Sbjct: 406 SQCQELQAIDLSYNNLSGSIPNGIFEIRNLTKLLLLSNYLSGFIPPDIGNCTNLYRLRLN 465
Query: 713 KVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRV 745
LAG A++ + K+L+ +D NR +
Sbjct: 466 GNRLAGNIPAEIGNL----KNLNFIDISENRLI 494
>At3g18060 WD40-repeat protein
Length = 609
Score = 30.0 bits (66), Expect = 5.1
Identities = 15/43 (34%), Positives = 25/43 (57%), Gaps = 2/43 (4%)
Query: 403 LKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYI 445
L SP K++ SLAV+ NA ++ G+Y+G+ CK++
Sbjct: 313 LDKSPFQFSGHMKNVSSLAVL--KGNADYILSGSYDGLICKWM 353
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,549,591
Number of Sequences: 26719
Number of extensions: 775355
Number of successful extensions: 1573
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1478
Number of HSP's gapped (non-prelim): 30
length of query: 765
length of database: 11,318,596
effective HSP length: 107
effective length of query: 658
effective length of database: 8,459,663
effective search space: 5566458254
effective search space used: 5566458254
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0314.7


