

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.14
(587 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
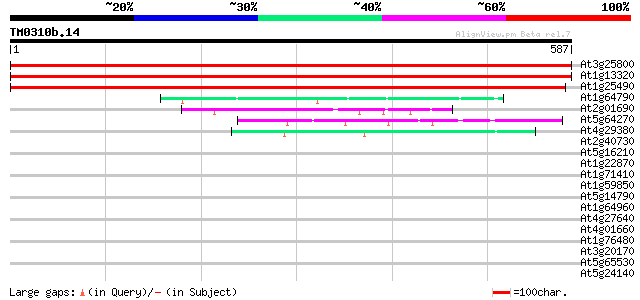
Score E
Sequences producing significant alignments: (bits) Value
At3g25800 protein phosphatase 2A 65 kDa regulatory subunit 1049 0.0
At1g13320 unknown protein 1034 0.0
At1g25490 phosphoprotein phosphatase 2A regulatory subunit A (R... 994 0.0
At1g64790 hypothetical protein 52 9e-07
At2g01690 unknown protein 51 2e-06
At5g64270 nuclear protein-like 50 3e-06
At4g29380 unknown protein 45 9e-05
At2g40730 unknown protein 42 0.001
At5g16210 unknown protein 39 0.011
At1g22870 hypothetical protein 37 0.031
At1g71410 hypothetical protein 36 0.068
At1g59850 hypothetical protein 34 0.20
At5g14790 unknown protein 32 0.75
At1g64960 hypothetical protein 32 0.75
At4g27640 putative protein 32 0.98
At4g01660 putative ABC transporter 31 1.7
At1g76480 unknown protein 31 1.7
At3g20170 unknown protein 30 3.7
At5g65530 unknown protein 30 4.9
At5g24140 squalene monooxygenase 2 (squalene epoxidase 2) (SE 2)... 29 8.3
>At3g25800 protein phosphatase 2A 65 kDa regulatory subunit
Length = 587
Score = 1049 bits (2712), Expect = 0.0
Identities = 526/587 (89%), Positives = 570/587 (96%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+M+D+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD
Sbjct: 1 MSMIDEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIPYVGGVE+A+VLLPPLETL TVEETCVR+K+VESLCR+G+QMRE
Sbjct: 61 DEVLLAMAEELGVFIPYVGGVEYAHVLLPPLETLSTVEETCVREKAVESLCRVGSQMRES 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLV+HFI LVKRLAAGEWFTARVS+CG+FHIAYPSAP+ K ELR++Y QLCQDDMPMVR
Sbjct: 121 DLVDHFISLVKRLAAGEWFTARVSACGVFHIAYPSAPDMLKTELRSLYTQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AATNLGKFAATVE+AHLK+D+MS+F+DLTQDDQDSVRLLAVEGCAALGKLLEPQ+CV
Sbjct: 181 RAAATNLGKFAATVESAHLKTDVMSMFEDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVQ 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RIL+PE+AI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGKDATIE LL
Sbjct: 301 AGKVTKFCRILNPEIAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFDDKLGALCMQWL+DKV+SIRDAAANN+KRLAEEFGP+WAMQHI+PQV
Sbjct: 421 IPLLASQLGVGFFDDKLGALCMQWLQDKVHSIRDAAANNLKRLAEEFGPEWAMQHIVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L+M+N+PHYLYRMTIL+A+SLLAPV+GSEIT SKLLP+V+ ASKDRVPNIKFNVAKVLQS
Sbjct: 481 LEMVNNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPVVMTASKDRVPNIKFNVAKVLQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LIPIV++SVVE T+RP LVELSEDPDVDVRFFA+QALQS D V MSS
Sbjct: 541 LIPIVDQSVVEKTIRPGLVELSEDPDVDVRFFANQALQSIDNVMMSS 587
>At1g13320 unknown protein
Length = 587
Score = 1034 bits (2673), Expect = 0.0
Identities = 518/587 (88%), Positives = 564/587 (95%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+MVD+PLYPIAVLIDELKN+DIQ RLNSI+RLS IARALGEERTRKELIPFLSENNDDD
Sbjct: 1 MSMVDEPLYPIAVLIDELKNDDIQRRLNSIKRLSIIARALGEERTRKELIPFLSENNDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELG FI YVGGVE+A VLLPPLETL TVEETCVR+K+V+SLCRIGAQMRE
Sbjct: 61 DEVLLAMAEELGGFILYVGGVEYAYVLLPPLETLSTVEETCVREKAVDSLCRIGAQMRES 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVEHF PL KRL+AGEWFTARVS+CG+FHIAYPSAP+ K ELR++YGQLCQDDMPMVR
Sbjct: 121 DLVEHFTPLAKRLSAGEWFTARVSACGIFHIAYPSAPDVLKTELRSIYGQLCQDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AATNLGKFAAT+E+AHLK+DIMS+F+DLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAATNLGKFAATIESAHLKTDIMSMFEDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRT+LVPAY RLL DNEAEVRIAA
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYARLLCDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF RIL+PELAI+HILPCVKELS+DSSQHVRSALASVIMGMAPVLGKDATIE LL
Sbjct: 301 AGKVTKFCRILNPELAIQHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
IPLLASQLGVGFFD+KLGALCMQWL+DKV+SIR+AAANN+KRLAEEFGP+WAMQHI+PQV
Sbjct: 421 IPLLASQLGVGFFDEKLGALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
L+MIN+PHYLYRMTIL+A+SLLAPV+GSEIT SKLLP V+ ASKDRVPNIKFNVAK++QS
Sbjct: 481 LEMINNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPAVITASKDRVPNIKFNVAKMMQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
LIPIV+++VVEN +RPCLVELSEDPDVDVR+FA+QALQS D V MSS
Sbjct: 541 LIPIVDQAVVENMIRPCLVELSEDPDVDVRYFANQALQSIDNVMMSS 587
>At1g25490 phosphoprotein phosphatase 2A regulatory subunit A
(RCN1)
Length = 588
Score = 994 bits (2571), Expect = 0.0
Identities = 492/581 (84%), Positives = 551/581 (94%)
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MAMVD+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTRKELIPFLSEN+DDD
Sbjct: 1 MAMVDEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENSDDD 60
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMREQ 120
DEVLLAMAEELGVFIP+VGG+E A+VLLPPLE+LCTVEETCVR+K+VESLC+IG+QM+E
Sbjct: 61 DEVLLAMAEELGVFIPFVGGIEFAHVLLPPLESLCTVEETCVREKAVESLCKIGSQMKEN 120
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
DLVE F+PLVKRLA GEWF ARVS+CG+FH+AY + K ELRA Y QLC+DDMPMVR
Sbjct: 121 DLVESFVPLVKRLAGGEWFAARVSACGIFHVAYQGCTDVLKTELRATYSQLCKDDMPMVR 180
Query: 181 RSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVA 240
R+AA+NLGKFA TVE+ L ++IM++FDDLT+DDQDSVRLLAVEGCAALGKLLEPQ+CVA
Sbjct: 181 RAAASNLGKFATTVESTFLIAEIMTMFDDLTKDDQDSVRLLAVEGCAALGKLLEPQDCVA 240
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
ILPVIVNFSQDKSWRVRYMVANQLYELCEAVGP+ TRT+LVPAYVRLLRDNEAEVRIAA
Sbjct: 241 RILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPDCTRTDLVPAYVRLLRDNEAEVRIAA 300
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
AGKVTKF R+L+PELAI+HILPCVKELS+DSSQHVRSALASVIMGMAP+LGKD+TIE LL
Sbjct: 301 AGKVTKFCRLLNPELAIQHILPCVKELSSDSSQHVRSALASVIMGMAPILGKDSTIEHLL 360
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY
Sbjct: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEY 420
Query: 421 IPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
+PLLASQLG+GFFDDKLGALCMQWL+DKVYSIR+AAANN+KRLAEEFGP+WAMQH++PQV
Sbjct: 421 VPLLASQLGIGFFDDKLGALCMQWLQDKVYSIREAAANNLKRLAEEFGPEWAMQHLVPQV 480
Query: 481 LDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQS 540
LDM+N+PHYL+RM +L+AISL+APV+GSEIT SK LP+VV ASKDRVPNIKFNVAK+LQS
Sbjct: 481 LDMVNNPHYLHRMMVLRAISLMAPVMGSEITCSKFLPVVVEASKDRVPNIKFNVAKLLQS 540
Query: 541 LIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSD 581
LIPIV++SVV+ T+R CLV+LSEDPDVDVR+FA+QAL S D
Sbjct: 541 LIPIVDQSVVDKTIRQCLVDLSEDPDVDVRYFANQALNSID 581
>At1g64790 hypothetical protein
Length = 2428
Score = 52.0 bits (123), Expect = 9e-07
Identities = 86/384 (22%), Positives = 154/384 (39%), Gaps = 32/384 (8%)
Query: 158 EAQKAELRAMYGQLCQDDMPMV---------------RRSAATNLGK--FAATVEAAHLK 200
EA + RAM QL + +V ++S+ LG F A + +
Sbjct: 1238 EAAECAARAMMSQLSAYGVKLVLPSLLKGLEDKAWRTKQSSVQLLGAMAFCAPQQLSQCL 1297
Query: 201 SDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYM 260
++ ++ D V+ +G +++ E ++ ++P ++ D + R+
Sbjct: 1298 PRVVPKLTEVLTDTHPKVQSAGQLALQQVGSVIKNPE-ISSLVPTLLLALTDPNEYTRHA 1356
Query: 261 VANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKF-SRILSPELAIEH 319
+ L P+ LVP R LR+ +E + A+ V S + P+ I +
Sbjct: 1357 LDTLLQTTFVNSVDAPSLALLVPIVHRGLRERSSETKKKASQIVGNMCSLVTEPKDMIPY 1416
Query: 320 I---LPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDV-R 375
I LP VK++ D VRS A + + +G+D L+P LK + +V R
Sbjct: 1417 IGLLLPEVKKVLVDPIPEVRSVAARAVGSLIRGMGED-NFPDLVPWLFETLKSDTSNVER 1475
Query: 376 LNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGF--F 433
L +V +G D +++LP ++ + VR + L LG F +
Sbjct: 1476 YGAAQGLSEVIAALGTDYF-ENILPDLIRHCSHQKASVRDGYLTLFKFLPRSLGAQFQKY 1534
Query: 434 DDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRM 493
+ + L D+ S+RDAA L E ++ ++P V D I + ++ R
Sbjct: 1535 LQLVLPAILDGLADENESVRDAALGAGHVLVEHHATT-SLPLLLPAVEDGIFNDNWRIRQ 1593
Query: 494 TILQAI-SLLAPVLGSEITSSKLL 516
+ ++ + LL V G TS K L
Sbjct: 1594 SSVELLGDLLFKVAG---TSGKAL 1614
Score = 41.2 bits (95), Expect = 0.002
Identities = 111/532 (20%), Positives = 213/532 (39%), Gaps = 85/532 (15%)
Query: 11 IAVLIDELKNEDI----QLRLNSIRRLSTIARALGEERTRKELIPFLSEN--NDDDDEVL 64
I +L+ E+K + ++R + R + ++ R +GE+ +L+P+L E +D +
Sbjct: 1417 IGLLLPEVKKVLVDPIPEVRSVAARAVGSLIRGMGEDNF-PDLVPWLFETLKSDTSNVER 1475
Query: 65 LAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSV---ESLCR-IGAQMREQ 120
A+ L I +G ++ +LP L C+ ++ VRD + + L R +GAQ Q
Sbjct: 1476 YGAAQGLSEVIAALG-TDYFENILPDLIRHCSHQKASVRDGYLTLFKFLPRSLGAQF--Q 1532
Query: 121 DLVEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVR 180
++ +P + A E + R ++ G H+ L +D +R
Sbjct: 1533 KYLQLVLPAILDGLADENESVRDAALGAGHVLVEHHATTSLPLLLPAVEDGIFNDNWRIR 1592
Query: 181 RSAATNLG----KFAATV-----------EAAHLKSDIMSVFDDLTQDDQDSVRLLAVEG 225
+S+ LG K A T E A ++ ++ D L D ++ V LA
Sbjct: 1593 QSSVELLGDLLFKVAGTSGKALLEGGSDDEGASTEAQGRAIIDILGMDKRNEV--LAALY 1650
Query: 226 CAALGKLLEPQECVAH---------------ILPVI----VNFSQDKSWRVRYMVANQLY 266
L ++ H I+P++ ++ S R + L
Sbjct: 1651 MVRTDVSLSVRQAALHVWKTIVANTPKTLKEIMPILMSTLISSLASPSSERRQVAGRSLG 1710
Query: 267 ELCEAVGPEPTRTELVPAYVRLLRDNEAEVR----------IAAAGKVTKFSRILSPELA 316
EL +G E ++P + L+D + + R +A+AG+ S++LS
Sbjct: 1711 ELVRKLG-ERVLPLIIPILSKGLKDPDVDKRQGVCIGLNEVMASAGR----SQLLS---F 1762
Query: 317 IEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRL 376
++ ++P ++ DS+ VR + + G A +++++P L L+D+
Sbjct: 1763 MDQLIPTIRTALCDSALEVRESAGLAFSTLYKSAGLQA-MDEIIPTLLEALEDDEMST-- 1819
Query: 377 NIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDK 436
+ LD + Q+I + + ++LP I L + H + + LA G G F+
Sbjct: 1820 ---TALDGLKQIISV--RTAAVLPHI--LPKLVHLPLSALNAHALGALAEVAGAG-FNTH 1871
Query: 437 LGAL------CMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLD 482
LG + M +V + AA V + +E G + + ++ V D
Sbjct: 1872 LGTILPALLSAMGGENKEVQELAQEAAERVVLVIDEEGVETLLSELLKGVSD 1923
Score = 33.9 bits (76), Expect = 0.26
Identities = 43/203 (21%), Positives = 84/203 (41%), Gaps = 44/203 (21%)
Query: 268 LCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKEL 327
L +G P E++P+Y++L+RD + R
Sbjct: 1977 LARVIGSVPK--EVLPSYIKLVRDAVSTAR------------------------------ 2004
Query: 328 SADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQ 387
D + R VI G+ +++ LLP+FL L ++R L ++ +
Sbjct: 2005 --DKERRKRKGGYVVIPGLC----LPKSLKPLLPVFLQGLISGSAELREQAAIGLGELIE 2058
Query: 388 VIGIDLLSQSLLP---AIVELAEDRH-WRVRLAIIEYIPLLASQLGVGF--FDDKLGALC 441
V L + ++P ++ + DR W+V+ AI+ + +L + G+ F +L
Sbjct: 2059 VTSEQALKEFVIPITGPLIRIIGDRFPWQVKSAILATLIILIQRGGMALKPFLPQLQTTF 2118
Query: 442 MQWLKDKVYSIRDAAANNVKRLA 464
++ L+D +IR +AA + +L+
Sbjct: 2119 VKCLQDSTRTIRSSAAVALGKLS 2141
>At2g01690 unknown protein
Length = 743
Score = 51.2 bits (121), Expect = 2e-06
Identities = 79/308 (25%), Positives = 132/308 (42%), Gaps = 32/308 (10%)
Query: 180 RRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQD----DQDSVRLLAVEGCAAL--GKLL 233
R++AA L + ++ I V + L ++ Q + R + G AA+ G
Sbjct: 24 RKNAALELENIVKNLTSSGDHDKISKVIEMLIKEFAKSPQANHRKGGLIGLAAVTVGLST 83
Query: 234 EPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPE--PTRTELVPAYVRLLRD 291
E + + I+P ++N D+ RVRY LY + + V + ++ A +L D
Sbjct: 84 EAAQYLEQIVPPVINSFSDQDSRVRYYACEALYNIAKVVRGDFIIFFNKIFDALCKLSAD 143
Query: 292 NEAEVRIAA--AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPV 349
++A V+ AA ++ K S + +IE +P +KE + +VR L +G V
Sbjct: 144 SDANVQSAAHLLDRLVKDIVTESDQFSIEEFIPLLKERMNVLNPYVRQFL----VGWITV 199
Query: 350 LGKDATIEQL--LPIFL----SLLKDEFPDVRLNIISKLDQVNQVI----GIDLLSQSLL 399
L I+ L LP FL ++L D ++R S L + Q I +D +
Sbjct: 200 LDSVPDIDMLGFLPDFLDGLFNMLSDSSHEIRQQADSALSEFLQEIKNSPSVDY--GRMA 257
Query: 400 PAIVELAEDRHWRVRLAII----EYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDA 455
+V+ A RL I E++ L QL V ++ D LGA+ + + DK IR
Sbjct: 258 EILVQRAASPDEFTRLTAITWINEFVKLGGDQL-VRYYADILGAI-LPCISDKEEKIRVV 315
Query: 456 AANNVKRL 463
A + L
Sbjct: 316 ARETNEEL 323
>At5g64270 nuclear protein-like
Length = 1269
Score = 50.1 bits (118), Expect = 3e-06
Identities = 79/360 (21%), Positives = 154/360 (41%), Gaps = 31/360 (8%)
Query: 239 VAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLL-------RD 291
VA I+ +V +D+S + R MV + ++ +G L + +
Sbjct: 810 VADIVGRVVEDLKDESEQYRRMVMETIDKVVTNLGASDIDARLEELLIDGILYAFQEQTS 869
Query: 292 NEAEVRIAAAGKVTK-FSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVL 350
++A V + G V + + P L I +K + S VR A +I +A V+
Sbjct: 870 DDANVMLNGFGAVVNALGQRVKPYLP--QICGTIKWRLNNKSAKVRQQAADLISRIAVVM 927
Query: 351 ---GKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLS---QSLLPAIVE 404
G++ + L + L +E+P+V +I+ L + VIG+ ++ + LLP +
Sbjct: 928 KQCGEEQLMGHLGVVLYEYLGEEYPEVLGSILGALKAIVNVIGMTKMTPPIKDLLPRLTP 987
Query: 405 LAEDRHWRVRLAIIEYIPLLASQLGVGFFDDK-LGALC---MQWLKDKVYSIRDAAANNV 460
+ ++RH +V+ I+ + +A + G F + +C ++ LK IR A N
Sbjct: 988 ILKNRHEKVQENCIDLVGRIADR-GAEFVPAREWMRICFELLEMLKAHKKGIRRATVNTF 1046
Query: 461 KRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVV 520
+A+ GP Q ++ +L+ + R+ AI+++A + +LP ++
Sbjct: 1047 GYIAKAIGP----QDVLATLLNNLKVQERQNRVCTTVAIAIVA----ETCSPFTVLPALM 1098
Query: 521 NASKDRVPNIKFNVAKVLQSLIPIVEESVVE--NTLRPCLVELSEDPDVDVRFFASQALQ 578
N + N++ V K L L + E + + P L + D D+ R A+ A++
Sbjct: 1099 NEYRVPELNVQNGVLKSLSFLFEYIGEMGKDYIYAVTPLLEDALMDRDLVHRQTAASAVK 1158
>At4g29380 unknown protein
Length = 1494
Score = 45.4 bits (106), Expect = 9e-05
Identities = 68/331 (20%), Positives = 130/331 (38%), Gaps = 14/331 (4%)
Query: 233 LEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGP-EPTRTELVPAYV----- 286
++ + + +LP +V D + VR L ++ V P+ ++ P Y+
Sbjct: 476 IDDDDRLQRVLPYVVALLSDPTAIVRCAAMETLCDILPLVRDFPPSDAKIFPEYIFPMLS 535
Query: 287 RLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGM 346
L D E VRI A + K + L L V L+ +SQ + + AS
Sbjct: 536 MLPEDTEESVRICYASNIAKLALTAYGFLIHSFQLSDVGVLNELNSQQISTTPASETPSH 595
Query: 347 APVLGKDATIEQLLPIFLSLLKD------EFPDVRLNIISKLDQVNQVIGIDLLSQSLLP 400
+A ++QL ++++ + P+VR ++ + ++ G + LLP
Sbjct: 596 LQKANGNAQLQQLRKTIAEVVQELVMGPKQTPNVRRALLQDIGELCFFFGQRQSNDFLLP 655
Query: 401 AIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNV 460
+ DR ++R E I + +G ++ L Q L D+ ++ A +
Sbjct: 656 ILPAFLNDRDEQLRSVFFEKIVYVCFFVGQRSVEEYLLPYIDQALSDQTEAVIVNALECL 715
Query: 461 KRLAE-EFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLV 519
L + F A+ +I V ++ P R ++ I+ + LG+ + S + V
Sbjct: 716 STLCKSSFLRKRALLQMIECVYPLLCYPSQWVRRAVVTFIAASSECLGA-VDSYAFIAPV 774
Query: 520 VNASKDRVPNIKFNVAKVLQSLIPIVEESVV 550
+ + R+P + +L L P V VV
Sbjct: 775 IRSYLSRLPASIASEEGLLSCLKPPVTREVV 805
Score = 37.7 bits (86), Expect = 0.018
Identities = 40/186 (21%), Positives = 74/186 (39%), Gaps = 3/186 (1%)
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDS--VRLLAVEGCAALGKLLEPQECVAHI 242
++L K + L+ I V +L + + VR ++ L ++ +
Sbjct: 594 SHLQKANGNAQLQQLRKTIAEVVQELVMGPKQTPNVRRALLQDIGELCFFFGQRQSNDFL 653
Query: 243 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRD-NEAEVRIAAA 301
LP++ F D+ ++R + ++ +C VG L+P + L D EA + A
Sbjct: 654 LPILPAFLNDRDEQLRSVFFEKIVYVCFFVGQRSVEEYLLPYIDQALSDQTEAVIVNALE 713
Query: 302 GKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLP 361
T + A+ ++ CV L SQ VR A+ + I + LG + + P
Sbjct: 714 CLSTLCKSSFLRKRALLQMIECVYPLLCYPSQWVRRAVVTFIAASSECLGAVDSYAFIAP 773
Query: 362 IFLSLL 367
+ S L
Sbjct: 774 VIRSYL 779
Score = 37.7 bits (86), Expect = 0.018
Identities = 62/315 (19%), Positives = 122/315 (38%), Gaps = 29/315 (9%)
Query: 46 RKELIPFLSENNDDDDEVLLAMAEELG-----VFIPYVGGVE-HANVLLPPLETLC--TV 97
+K PFL + DD L+++ + F+P G + VL+ + C +
Sbjct: 397 KKNRHPFLKKITMDDLGTLMSLYDSRSDTYGTPFLPVEGNMRCEGMVLIASMLCSCIRNI 456
Query: 98 EETCVRDKSVESLCRIGAQMREQDLVEHFIPLVKRLAAGEWFTARVSS----CGLFHIAY 153
+ +R +++ L + + D ++ +P V L + R ++ C + +
Sbjct: 457 KLPHLRREAILLLRSCSLYIDDDDRLQRVLPYVVALLSDPTAIVRCAAMETLCDILPLVR 516
Query: 154 PSAPEAQKA---ELRAMYGQLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMS---VF 207
P K + M L +D VR A+N+ K A T + S +S V
Sbjct: 517 DFPPSDAKIFPEYIFPMLSMLPEDTEESVRICYASNIAKLALTAYGFLIHSFQLSDVGVL 576
Query: 208 DDL-------TQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYM 260
++L T + L G A L +L ++ +A ++ +V ++ VR
Sbjct: 577 NELNSQQISTTPASETPSHLQKANGNAQLQQL---RKTIAEVVQELV-MGPKQTPNVRRA 632
Query: 261 VANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHI 320
+ + ELC G + L+P L D + ++R K+ + E++
Sbjct: 633 LLQDIGELCFFFGQRQSNDFLLPILPAFLNDRDEQLRSVFFEKIVYVCFFVGQRSVEEYL 692
Query: 321 LPCVKELSADSSQHV 335
LP + + +D ++ V
Sbjct: 693 LPYIDQALSDQTEAV 707
>At2g40730 unknown protein
Length = 604
Score = 42.0 bits (97), Expect = 0.001
Identities = 45/196 (22%), Positives = 80/196 (39%), Gaps = 18/196 (9%)
Query: 235 PQECVAH-ILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNE 293
P+E V +LP++ + + S + A L ++ + E +++P V+L N+
Sbjct: 313 PREIVLKKLLPLLASSLEFGSAAAPALTA--LLKMGSWLSTEDFSVKVLPTIVKLFASND 370
Query: 294 AEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKD 353
+R++ V +F +S ++ E + P V AD+S +R ++ +AP L +
Sbjct: 371 RAIRVSLLQHVDQFGESMSGQIVDEQVYPHVATGFADTSAFLRELTLKSMLVLAPKLSQR 430
Query: 354 ATIEQLLPIFLSL---------------LKDEFPDVRLNIISKLDQVNQVIGIDLLSQSL 398
LL L L+D FP R I L + ++ +
Sbjct: 431 TLSGSLLKYLSKLQTRKRVLINAFTVRALRDTFPPARGAGIVALCATSTTYDDTEIATRI 490
Query: 399 LPAIVELAEDRHWRVR 414
LP IV L D+ VR
Sbjct: 491 LPNIVVLTIDQDSDVR 506
Score = 36.2 bits (82), Expect = 0.052
Identities = 47/253 (18%), Positives = 91/253 (35%), Gaps = 56/253 (22%)
Query: 346 MAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVEL 405
+A L ++ +++LLP+ S L EF ++ L ++ + + S +LP IV+L
Sbjct: 308 VAEQLPREIVLKKLLPLLASSL--EFGSAAAPALTALLKMGSWLSTEDFSVKVLPTIVKL 365
Query: 406 AEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAE 465
+R+++++++ + D++
Sbjct: 366 FASNDRAIRVSLLQHVDQFGESMSGQIVDEQ----------------------------- 396
Query: 466 EFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLL--------- 516
+ P V D R L+++ +LAP L S LL
Sbjct: 397 ----------VYPHVATGFADTSAFLRELTLKSMLVLAPKLSQRTLSGSLLKYLSKLQTR 446
Query: 517 ------PLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVR 570
V A +D P + L + +++ + + P +V L+ D D DVR
Sbjct: 447 KRVLINAFTVRALRDTFPPARGAGIVALCATSTTYDDTEIATRILPNIVVLTIDQDSDVR 506
Query: 571 FFASQALQSSDQV 583
A QA++ Q+
Sbjct: 507 SKAFQAVEQFLQI 519
>At5g16210 unknown protein
Length = 1180
Score = 38.5 bits (88), Expect = 0.011
Identities = 20/88 (22%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Query: 495 ILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVENTL 554
+L A+ L E+ + + ++ D +K N AK+L++++P ++ V +
Sbjct: 877 VLDAVRFLCTF---EVHHNMIFGILWEMVVDSTAELKINAAKLLKTIVPYIDAKVASANV 933
Query: 555 RPCLVELSEDPDVDVRFFASQALQSSDQ 582
P L+ L D +++V++ + A S Q
Sbjct: 934 LPALITLGSDQNLNVKYASIDAFGSVAQ 961
Score = 37.4 bits (85), Expect = 0.023
Identities = 42/203 (20%), Positives = 82/203 (39%), Gaps = 21/203 (10%)
Query: 334 HVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDL 393
H+R+ + ++ ++ G LP+FL D+ D+R
Sbjct: 765 HLRNRITKFLLAVSERFGSSYLTHIELPVFLVAAGDDEADLRF----------------- 807
Query: 394 LSQSLLPAIVELAEDRHWRVRLAIIEYIPLL-ASQLGVGFFDDKLGALCMQWL-KDKVYS 451
L ++ P I L RLA + +PLL A LG ++L Q L + K
Sbjct: 808 LPSAIHPRIKGLKPRTAVANRLATLCILPLLLAGVLGAPSKREELTIFLRQLLVESKTKE 867
Query: 452 IRDAAANNVKRLAEEFGPDWAMQH--IIPQVLDMINDPHYLYRMTILQAISLLAPVLGSE 509
+ + NN A F + + H I + +M+ D ++ + + + P + ++
Sbjct: 868 NQSSKHNNEVLDAVRFLCTFEVHHNMIFGILWEMVVDSTAELKINAAKLLKTIVPYIDAK 927
Query: 510 ITSSKLLPLVVNASKDRVPNIKF 532
+ S+ +LP ++ D+ N+K+
Sbjct: 928 VASANVLPALITLGSDQNLNVKY 950
Score = 37.0 bits (84), Expect = 0.031
Identities = 27/106 (25%), Positives = 49/106 (45%), Gaps = 3/106 (2%)
Query: 258 RYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAI 317
R ++ + L VG T TEL+P + E R+ A + + + PE+
Sbjct: 510 RRIIMDACVSLSRNVGEMRTETELLPQCWEQINHTYEERRLLVAQSCGELAEYVRPEIRD 569
Query: 318 EHILPCVKELSADSSQHVRSALA---SVIMGMAPVLGKDATIEQLL 360
IL V++L DS+ VR A A ++++ + P K +E+++
Sbjct: 570 SLILSIVQQLIEDSATVVREAAAHNLALLLPLFPNTDKYFKVEEMM 615
Score = 33.9 bits (76), Expect = 0.26
Identities = 36/178 (20%), Positives = 73/178 (40%), Gaps = 14/178 (7%)
Query: 253 KSWRVRYMVANQLYELC------EAVGPEPTRTELVPAYVRLL------RDNEAEVRIAA 300
K + R VAN+L LC V P++ E + ++R L ++N++
Sbjct: 817 KGLKPRTAVANRLATLCILPLLLAGVLGAPSKREELTIFLRQLLVESKTKENQSSKHNNE 876
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLL 360
+F + + E+ I + E+ DS+ ++ A ++ + P + +L
Sbjct: 877 VLDAVRF--LCTFEVHHNMIFGILWEMVVDSTAELKINAAKLLKTIVPYIDAKVASANVL 934
Query: 361 PIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAII 418
P ++L D+ +V+ I V Q +D++ +L + ED +A+I
Sbjct: 935 PALITLGSDQNLNVKYASIDAFGSVAQHFKVDMIVDKILVQMDAFMEDGSHEAIIAVI 992
Score = 33.5 bits (75), Expect = 0.34
Identities = 27/98 (27%), Positives = 46/98 (46%), Gaps = 1/98 (1%)
Query: 14 LIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSEN-NDDDDEVLLAMAEELG 72
L + +K D Q R + +++R +GE RT EL+P E N +E L +A+ G
Sbjct: 498 LFNLIKRPDEQQRRIIMDACVSLSRNVGEMRTETELLPQCWEQINHTYEERRLLVAQSCG 557
Query: 73 VFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESL 110
YV +++L ++ L T VR+ + +L
Sbjct: 558 ELAEYVRPEIRDSLILSIVQQLIEDSATVVREAAAHNL 595
Score = 32.7 bits (73), Expect = 0.58
Identities = 18/76 (23%), Positives = 36/76 (46%)
Query: 362 IFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYI 421
I ++ D ++++N L + I + S ++LPA++ L D++ V+ A I+
Sbjct: 897 ILWEMVVDSTAELKINAAKLLKTIVPYIDAKVASANVLPALITLGSDQNLNVKYASIDAF 956
Query: 422 PLLASQLGVGFFDDKL 437
+A V DK+
Sbjct: 957 GSVAQHFKVDMIVDKI 972
Score = 30.4 bits (67), Expect = 2.9
Identities = 35/190 (18%), Positives = 76/190 (39%), Gaps = 12/190 (6%)
Query: 189 KFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILPVIVN 248
+F T E H + I + ++ D +++ A + + ++ + A++LP ++
Sbjct: 882 RFLCTFEVHH--NMIFGILWEMVVDSTAELKINAAKLLKTIVPYIDAKVASANVLPALIT 939
Query: 249 FSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFS 308
D++ V+Y + + + + +++ + D E IA + +
Sbjct: 940 LGSDQNLNVKYASIDAFGSVAQHFKVDMIVDKILVQMDAFMEDGSHEAIIAVI-RALLVA 998
Query: 309 RILSPELAIEHILPCVKELSADSSQHV--------RSALASVIMGM-APVLGKDATIEQL 359
+ E +++L + +LSA S +A I + A L + + E L
Sbjct: 999 IPHTTERLRDYLLSKILQLSASPSSSTDVNRRRERANAFCEAIRALDATDLSQTSVKEYL 1058
Query: 360 LPIFLSLLKD 369
LP +LLKD
Sbjct: 1059 LPAIQNLLKD 1068
Score = 30.4 bits (67), Expect = 2.9
Identities = 21/90 (23%), Positives = 40/90 (44%)
Query: 476 IIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVA 535
++PQ + IN + R+ + Q+ LA + EI S +L +V +D ++ A
Sbjct: 533 LLPQCWEQINHTYEERRLLVAQSCGELAEYVRPEIRDSLILSIVQQLIEDSATVVREAAA 592
Query: 536 KVLQSLIPIVEESVVENTLRPCLVELSEDP 565
L L+P+ + + + +L DP
Sbjct: 593 HNLALLLPLFPNTDKYFKVEEMMFQLICDP 622
Score = 30.4 bits (67), Expect = 2.9
Identities = 37/163 (22%), Positives = 66/163 (39%), Gaps = 20/163 (12%)
Query: 357 EQLLPIFL--------SLLKDEFPDVRLNIISKLDQVNQVIGIDL---LSQS-------- 397
E+LLP+ + S +D N+I + D+ + I +D LS++
Sbjct: 472 EELLPLMMCAIERHPVSSTRDSLTHTLFNLIKRPDEQQRRIIMDACVSLSRNVGEMRTET 531
Query: 398 -LLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAA 456
LLP E + RL + + LA + D + ++ Q ++D +R+AA
Sbjct: 532 ELLPQCWEQINHTYEERRLLVAQSCGELAEYVRPEIRDSLILSIVQQLIEDSATVVREAA 591
Query: 457 ANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAI 499
A+N+ L F + + +I DP L T L+ +
Sbjct: 592 AHNLALLLPLFPNTDKYFKVEEMMFQLICDPSGLVVETTLKEL 634
>At1g22870 hypothetical protein
Length = 811
Score = 37.0 bits (84), Expect = 0.031
Identities = 32/150 (21%), Positives = 67/150 (44%), Gaps = 3/150 (2%)
Query: 220 LLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRT 279
LL ++ + + V+H+LP+++ D R++ V + + + + + R
Sbjct: 341 LLLIKRAELIINKTNAEHLVSHVLPLLLRAYNDNDVRIQEEVLKRSTSVAKQLDGQVVRQ 400
Query: 280 ELVPAYVRL-LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSA-DSSQHVRS 337
++P L L+ A VR+ A + + + L +LA+ IL ++ +A D S
Sbjct: 401 AILPRVHGLALKTTVAAVRVNALLCLAELVQTLD-KLAVTEILQTIQRCTAVDRSAPTLM 459
Query: 338 ALASVIMGMAPVLGKDATIEQLLPIFLSLL 367
++ + G + T E +LP+ + LL
Sbjct: 460 CTLAIANAILKQYGVEFTSEHVLPLIIPLL 489
Score = 29.3 bits (64), Expect = 6.4
Identities = 19/84 (22%), Positives = 38/84 (44%), Gaps = 1/84 (1%)
Query: 473 MQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVN-ASKDRVPNIK 531
+ H++P +L ND + +L+ + +A L ++ +LP V A K V ++
Sbjct: 360 VSHVLPLLLRAYNDNDVRIQEEVLKRSTSVAKQLDGQVVRQAILPRVHGLALKTTVAAVR 419
Query: 532 FNVAKVLQSLIPIVEESVVENTLR 555
N L L+ +++ V L+
Sbjct: 420 VNALLCLAELVQTLDKLAVTEILQ 443
>At1g71410 hypothetical protein
Length = 859
Score = 35.8 bits (81), Expect = 0.068
Identities = 31/149 (20%), Positives = 63/149 (41%), Gaps = 1/149 (0%)
Query: 220 LLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRT 279
LL V+ + + + V+H+LP+++ D R++ V + + + + + R
Sbjct: 372 LLLVKHADLITNKTDSEHLVSHVLPLLLRAYNDNDVRIQEEVLKRSTSVAKQLDGQVVRQ 431
Query: 280 ELVPAYVRL-LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSA 338
++P L L+ A VR+ A + + + L AIE + + + D S
Sbjct: 432 AILPRVHGLALKTTVAAVRVNALLCLAELVQTLDKPAAIEILETIQRCTAVDRSAPTLMC 491
Query: 339 LASVIMGMAPVLGKDATIEQLLPIFLSLL 367
+V + G + T E +L + + LL
Sbjct: 492 TLAVANAILKQYGVEFTAEHVLTLMMPLL 520
>At1g59850 hypothetical protein
Length = 498
Score = 34.3 bits (77), Expect = 0.20
Identities = 63/260 (24%), Positives = 111/260 (42%), Gaps = 18/260 (6%)
Query: 176 MPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEP 235
MP V+ ++ + + A TV LK +++ + L+ D+D++ L A E ++ L P
Sbjct: 1 MPSVQIRSSPSHSQPAMTV--TDLKQRVIACLNRLS--DRDTLALAAAE-LDSIALNLSP 55
Query: 236 QECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPE--PTRTELVPAYVRLLRDNE 293
+ I + S KS VR + L L + G P +++V +R LRD +
Sbjct: 56 ETFSLFINCLQSTDSSAKS-PVRKHCVSLLSVLSRSHGDSLAPHLSKMVSTVLRRLRDPD 114
Query: 294 AEVRIAA-AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGK 352
+ VR A A V + I +I P ++ + D + + + A++ + A
Sbjct: 115 SSVRAACVAASVDMTTNITGQPFSIL-FGPMIETVIHDCDPNAQIS-AAMCLAAAVDAAD 172
Query: 353 DATIEQL---LPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQS----LLPAIVEL 405
+ +EQL LP LLK E + ++ + V +G ++ LLP + E
Sbjct: 173 EPDVEQLQKALPKIGKLLKSEGFKAKAELLGAIGTVIGAVGGRNSEKAVLDWLLPNVSEF 232
Query: 406 AEDRHWRVRLAIIEYIPLLA 425
WR R A E + +A
Sbjct: 233 LSSDDWRARKAAAEAMARVA 252
>At5g14790 unknown protein
Length = 346
Score = 32.3 bits (72), Expect = 0.75
Identities = 26/102 (25%), Positives = 47/102 (45%), Gaps = 11/102 (10%)
Query: 281 LVPAYVRLLRDNEAEVRI---AAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRS 337
L P +LL + + R AA +T ++ +SP L + + PC+K + + +R+
Sbjct: 166 LEPLLTQLLLKSSQDKRFVCEAAEKALTAMTKYVSPTLLLPKLQPCLK----NRNPRIRA 221
Query: 338 ALASVIMGMAPVLG----KDATIEQLLPIFLSLLKDEFPDVR 375
+ P LG K+ I++L+ S L D+ P+ R
Sbjct: 222 KASLCFSRSVPRLGVEGIKEYGIDKLVQAAASQLSDQLPESR 263
>At1g64960 hypothetical protein
Length = 1227
Score = 32.3 bits (72), Expect = 0.75
Identities = 28/102 (27%), Positives = 44/102 (42%), Gaps = 8/102 (7%)
Query: 170 QLCQDDMPMVRRSAATNLGK----FAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEG 225
+L D+ P VR A L + F + +A + + +FDD++ + VRL V G
Sbjct: 397 KLLSDECPDVRSVAVEGLSRVFYLFWEVIPSATITKVLTKIFDDMSHESCSEVRLSTVNG 456
Query: 226 CAALGKLLEPQE--CVAHILPVIVNFSQDKSWRVRYMVANQL 265
L L PQ + ILP + + D VR + + L
Sbjct: 457 ITYL--LANPQSHGILKVILPRLGHLMLDSVTSVRVAMVDLL 496
>At4g27640 putative protein
Length = 651
Score = 32.0 bits (71), Expect = 0.98
Identities = 22/81 (27%), Positives = 40/81 (49%), Gaps = 7/81 (8%)
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELA--IEHILPCVKELSADSSQHVRSALASVIMGM 346
LRD E VR AA+ + +F+ L PE+ + +LPC+ D+S+ V+ +
Sbjct: 387 LRDPELVVRGAASFAIGQFAEHLQPEILSHYQSVLPCLLIAIEDTSEEVKEKSHYALAAF 446
Query: 347 APVLGKDATIEQLLPIFLSLL 367
+G E+++P+ L+
Sbjct: 447 CENMG-----EEIVPLLDHLM 462
>At4g01660 putative ABC transporter
Length = 623
Score = 31.2 bits (69), Expect = 1.7
Identities = 23/86 (26%), Positives = 44/86 (50%), Gaps = 8/86 (9%)
Query: 344 MGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIV 403
+G+ + G + Q+L I +DE V I++ L+ V Q G D++ +S L ++
Sbjct: 230 LGLCEMRGAALKVGQMLSI-----QDESL-VPAPILNALEYVRQ--GADVMPRSQLNPVL 281
Query: 404 ELAEDRHWRVRLAIIEYIPLLASQLG 429
+ +W+ +L +Y PL A+ +G
Sbjct: 282 DAELGSNWQSKLTSFDYEPLAAASIG 307
>At1g76480 unknown protein
Length = 177
Score = 31.2 bits (69), Expect = 1.7
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 6/67 (8%)
Query: 10 PIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDDEVLLAMAE 69
P V + LKN +++L+LN +RRL + + + + KE+ NN VL+ +
Sbjct: 49 PARVSLQLLKNHEMELKLNQMRRLLNTEKLIFKAESLKEV------NNKAGLNVLVTLEP 102
Query: 70 ELGVFIP 76
E V IP
Sbjct: 103 EGIVAIP 109
>At3g20170 unknown protein
Length = 475
Score = 30.0 bits (66), Expect = 3.7
Identities = 39/191 (20%), Positives = 73/191 (37%), Gaps = 7/191 (3%)
Query: 281 LVPAYVRLLRDNEAEVRIAAA---GKVTKFSRILSPELAIEHILPCVKELSADSSQHVRS 337
++PA V L RD + + ++ A G ++ + + P I P EL + +
Sbjct: 235 VIPALVDLYRDGDDKAKLLAGNALGIISAQTEYIRPVTEAGSI-PLYVELLSGQDPMGKD 293
Query: 338 ALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQS 397
V +A G I + L L +E +++ L + + +
Sbjct: 294 IAEDVFCILAVAEGNAVLIAEQLVRILRAGDNEAKLAASDVLWDLAGYRHSVSV-IRGSG 352
Query: 398 LLPAIVELAEDRHWRVRLAIIEYIPLLASQLG--VGFFDDKLGALCMQWLKDKVYSIRDA 455
+P ++EL D R I I L+ F D + + ++WL D+ +RD
Sbjct: 353 AIPLLIELLRDGSLEFRERISGAISQLSYNENDREAFSDSGMIPILIEWLGDESEELRDN 412
Query: 456 AANNVKRLAEE 466
AA + +E+
Sbjct: 413 AAEALINFSED 423
>At5g65530 unknown protein
Length = 456
Score = 29.6 bits (65), Expect = 4.9
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 3/45 (6%)
Query: 25 LRLNSIRRLSTIARALGEERTRKELIPFL---SENNDDDDEVLLA 66
++ SIRR S I E TRK+ P L SEN+ D D++L+A
Sbjct: 81 IKKKSIRRFSVIPLLASYELTRKKKQPKLSPCSENDFDCDQILVA 125
>At5g24140 squalene monooxygenase 2 (squalene epoxidase 2) (SE 2)
(sp|O65403)
Length = 516
Score = 28.9 bits (63), Expect = 8.3
Identities = 29/106 (27%), Positives = 45/106 (42%), Gaps = 20/106 (18%)
Query: 287 RLLRDNEAEVRIAAAGKVTKFSRILSPE-----------LAIEHI----LPCVKELSADS 331
R L DN EV G VTK SR+ P I I + CV E+ ADS
Sbjct: 211 RSLVDNTEEVLSYMVGYVTKNSRLEDPHSLHLIFSKPLVCVIYQITSDEVRCVAEVPADS 270
Query: 332 SQHVRSALASVIM--GMAPVLGKDATIEQLLPIFLSLLKDEFPDVR 375
+ + S + MAP + + + + IFL +++ P+++
Sbjct: 271 IPSISNGEMSTFLKKSMAPQIPETGNLRE---IFLKGIEEGLPEIK 313
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,879,108
Number of Sequences: 26719
Number of extensions: 480359
Number of successful extensions: 1448
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 1312
Number of HSP's gapped (non-prelim): 66
length of query: 587
length of database: 11,318,596
effective HSP length: 105
effective length of query: 482
effective length of database: 8,513,101
effective search space: 4103314682
effective search space used: 4103314682
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0310b.14


