

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.11
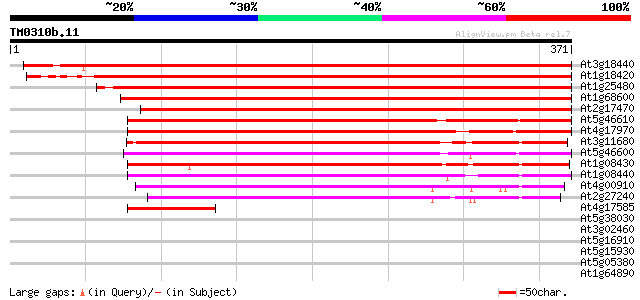
(371 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g18440 unknown protein 481 e-136
At1g18420 unknown protein 453 e-128
At1g25480 hypothetical protein 419 e-117
At1g68600 unknown protein 394 e-110
At2g17470 unknown protein 381 e-106
At5g46610 putative protein 231 4e-61
At4g17970 unknown protein 228 5e-60
At3g11680 unknown protein 219 2e-57
At5g46600 putative protein 214 5e-56
At1g08430 hypothetical protein 206 1e-53
At1g08440 hypothetical protein 199 2e-51
At4g00910 191 4e-49
At2g27240 hypothetical protein 185 4e-47
At4g17585 putative protein 61 9e-10
At5g38030 putative transmembrane protein 32 0.56
At3g02460 putative plant adhesion molecule 31 0.95
At5g16910 cellulose synthase catalytic subunit -like protein 31 1.2
At5g15930 plant adhesion molecule 1 (PAM1) 31 1.2
At5g05380 putative protein 31 1.2
At1g64890 unknown protein 31 1.2
>At3g18440 unknown protein
Length = 598
Score = 481 bits (1237), Expect = e-136
Identities = 236/364 (64%), Positives = 286/364 (77%), Gaps = 6/364 (1%)
Query: 10 KLGSFRYSFKERKEKLLSMKSSLIGYSQIGIPLPESDD--DNDGPPGSKNCCRFCSYRAL 67
K GSFR+ E++E+LLS G+S ES+D +N+ C CS L
Sbjct: 4 KQGSFRHGILEKRERLLSNN----GFSDFRFTDIESNDLLENENCGRRTRLCCCCSCGNL 59
Query: 68 SDRLVGAWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSV 127
S+++ G + K VA +AWEMG+SDPRKIVF+AK+GLAL +++LLIF + P DL R+SV
Sbjct: 60 SEKISGVYDDAKDVARKAWEMGVSDPRKIVFSAKIGLALTIVALLIFYQEPNPDLSRYSV 119
Query: 128 WAILTVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIV 187
WAILTVV VFEF+IGATLSKG NR LGTL+AGGLA+GM LS L G WEEI +SIF +
Sbjct: 120 WAILTVVVVFEFTIGATLSKGFNRALGTLSAGGLALGMAELSTLFGDWEEIFCTLSIFCI 179
Query: 188 GFCATYAKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLG 247
GF AT+ KLYP+MKAYEYGFRVFL+TYCYI++SG++TG+FI+ A+SRFLLIALGA VSLG
Sbjct: 180 GFLATFMKLYPSMKAYEYGFRVFLLTYCYILISGFRTGQFIEVAISRFLLIALGAGVSLG 239
Query: 248 VNVCLYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVY 307
VN+ +YPIWAGEDLH+LV KNFM VA SLEG VN YL C+EYER+PSKILTYQAS+D VY
Sbjct: 240 VNMFIYPIWAGEDLHNLVVKNFMNVATSLEGCVNGYLRCLEYERIPSKILTYQASEDPVY 299
Query: 308 SGYRSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCI 367
GYRSA++STS E++LM FA+WEPPHG YK YPW NYVK+SGAL+HCAF VMA+HGCI
Sbjct: 300 KGYRSAVESTSQEESLMSFAIWEPPHGPYKSFNYPWKNYVKLSGALKHCAFTVMALHGCI 359
Query: 368 LSEI 371
LSEI
Sbjct: 360 LSEI 363
>At1g18420 unknown protein
Length = 581
Score = 453 bits (1165), Expect = e-128
Identities = 228/361 (63%), Positives = 273/361 (75%), Gaps = 20/361 (5%)
Query: 12 GSFRYSFKERKEKLLSMKSSLIGYSQIGIPLPESDDDNDGPPGSKNCCRFCSYRALSDRL 71
GSFR S ++R L ++S GYS + +DDD K R CSY SD++
Sbjct: 27 GSFRQSMRDR----LILQSR--GYSNV------NDDD-------KTSVRCCSYSYFSDKI 67
Query: 72 VGAWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAIL 131
G K K V AWEMG +DPRK++F+AKMGLAL L S+LIF K P +L H +WAIL
Sbjct: 68 TGVVKKLKDVLVTAWEMGTADPRKMIFSAKMGLALTLTSILIFFKIPGLELSGHYLWAIL 127
Query: 132 TVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCA 191
TVV +FEFSIGAT SKG NRGLGTL+AGGLA+GM +S++ G W ++ SIF+V F A
Sbjct: 128 TVVVIFEFSIGATFSKGCNRGLGTLSAGGLALGMSWISEMTGNWADVFNAASIFVVAFFA 187
Query: 192 TYAKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVC 251
TYAKLYPTMK YEYGFRVFL+TYCY+IVSGY+TG+F++TAVSRFLLIALGA+V L VN C
Sbjct: 188 TYAKLYPTMKPYEYGFRVFLLTYCYVIVSGYKTGEFMETAVSRFLLIALGASVGLIVNTC 247
Query: 252 LYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQA-SDDVVYSGY 310
+YPIWAGEDLH+LVAKNF+ VA SLEG VN YL C+ Y+ +PS+IL Y+A ++D VYSGY
Sbjct: 248 IYPIWAGEDLHNLVAKNFVNVATSLEGCVNGYLECVAYDTIPSRILVYEAVAEDPVYSGY 307
Query: 311 RSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSE 370
RSA+QSTS ED LM FA WEPPHG YK +YPW+ YVKV GALRHCA MVMA+HGCILSE
Sbjct: 308 RSAVQSTSQEDTLMSFASWEPPHGPYKSFRYPWALYVKVGGALRHCAIMVMALHGCILSE 367
Query: 371 I 371
I
Sbjct: 368 I 368
>At1g25480 hypothetical protein
Length = 548
Score = 419 bits (1077), Expect = e-117
Identities = 200/314 (63%), Positives = 244/314 (77%), Gaps = 5/314 (1%)
Query: 58 CCRFCSYRALSDRLVGAWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKT 117
C RFCS D + +WK + A+ +EMG SD RK+ F+ KMG+AL L S +I+LK
Sbjct: 35 CFRFCS-----DGITASWKALYDIGAKLYEMGRSDRRKVYFSVKMGMALALCSFVIYLKE 89
Query: 118 PFEDLGRHSVWAILTVVTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEE 177
P D +++VWAILTVV VFE+SIGATL KG NR +GTL+AGGLA+G+ LS AG++EE
Sbjct: 90 PLRDASKYAVWAILTVVVVFEYSIGATLVKGFNRAIGTLSAGGLALGIARLSVSAGEFEE 149
Query: 178 IIVMVSIFIVGFCATYAKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLL 237
+I+++SIFI GF A+Y KLYP MK+YEY FRVFL+TYC ++VSG + F TA RFLL
Sbjct: 150 LIIIISIFIAGFSASYLKLYPAMKSYEYAFRVFLLTYCIVLVSGNNSRDFFSTAYYRFLL 209
Query: 238 IALGAAVSLGVNVCLYPIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKIL 297
I +GA + LGVN+ + PIWAGEDLH LV KNF VA SLEG VN YL C+EYER+PSKIL
Sbjct: 210 ILVGAGICLGVNIFILPIWAGEDLHKLVVKNFKSVANSLEGCVNGYLQCVEYERIPSKIL 269
Query: 298 TYQASDDVVYSGYRSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCA 357
TYQASDD +YSGYRS +QSTS ED+L+ FAVWEPPHG YK +PW+NYVK+SGA+RHCA
Sbjct: 270 TYQASDDPLYSGYRSVVQSTSQEDSLLDFAVWEPPHGPYKTFHHPWANYVKLSGAVRHCA 329
Query: 358 FMVMAMHGCILSEI 371
FMVMAMHGCILSEI
Sbjct: 330 FMVMAMHGCILSEI 343
>At1g68600 unknown protein
Length = 537
Score = 394 bits (1012), Expect = e-110
Identities = 187/298 (62%), Positives = 231/298 (76%)
Query: 74 AWKTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTV 133
+W+ A+ + +G SD RK+ F+ KMG+AL L S +IFLK P +D + +VWAILTV
Sbjct: 35 SWRALYEAPAKLYALGHSDRRKLYFSIKMGIALALCSFVIFLKEPLQDASKFAVWAILTV 94
Query: 134 VTVFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCATY 193
V +FE+ +GATL KG NR LGT+ AGGLA+G+ LS LAG++EE+I+++ IF+ GF A+Y
Sbjct: 95 VLIFEYYVGATLVKGFNRALGTMLAGGLALGVAQLSVLAGEFEEVIIVICIFLAGFGASY 154
Query: 194 AKLYPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLY 253
KLY +MK YEY FRVF +TYC ++VSG + F+ TA R LLI LGA + L VNV L+
Sbjct: 155 LKLYASMKPYEYAFRVFKLTYCIVLVSGNNSRDFLSTAYYRILLIGLGATICLLVNVFLF 214
Query: 254 PIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSA 313
PIWAGEDLH LVAKNF VA SLEG VN YL C+EYER+PSKILTYQASDD +YSGYRSA
Sbjct: 215 PIWAGEDLHKLVAKNFKNVANSLEGCVNGYLQCVEYERIPSKILTYQASDDPLYSGYRSA 274
Query: 314 LQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+QSTS ED+L+ FA+WEPPHG YK +PW NYVK+SGA+RHCAF VMAMHGCILSEI
Sbjct: 275 VQSTSQEDSLLDFAIWEPPHGPYKTFNHPWKNYVKLSGAVRHCAFTVMAMHGCILSEI 332
>At2g17470 unknown protein
Length = 538
Score = 381 bits (979), Expect = e-106
Identities = 179/285 (62%), Positives = 223/285 (77%)
Query: 87 EMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGATLS 146
E+G SD R+I FA KMG+AL L S++IFLK P D ++SVW ILTVV VFE+S+GATL
Sbjct: 31 ELGHSDRRRIFFAVKMGMALALCSVVIFLKEPLHDASKYSVWGILTVVVVFEYSVGATLV 90
Query: 147 KGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCATYAKLYPTMKAYEYG 206
KG NR +GT++AGGLA+G+ LS L+ +E+ I++ IF+ GF A+Y+KL+P MK YEY
Sbjct: 91 KGFNRAIGTVSAGGLALGIARLSVLSRDFEQTIIITCIFLAGFIASYSKLHPAMKPYEYA 150
Query: 207 FRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHDLVA 266
FRVFL+T+C ++VSG TG F TA RFL I +GA L VN+ ++PIWAGEDLH LVA
Sbjct: 151 FRVFLLTFCIVLVSGNNTGDFFSTAYYRFLFIVVGATTCLVVNIFIFPIWAGEDLHKLVA 210
Query: 267 KNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQSTSTEDALMGF 326
NF VA SLEG VN YL C+EYERVPSKILTYQ SDD +YSGYRSA+QST+ E++L+ F
Sbjct: 211 NNFKSVANSLEGCVNGYLQCVEYERVPSKILTYQTSDDPLYSGYRSAIQSTNQEESLLDF 270
Query: 327 AVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
A+WEPPHG Y+ +PW NYVK+SGA+RHCAF VMA+HGCILSEI
Sbjct: 271 AIWEPPHGPYRTFNHPWKNYVKLSGAVRHCAFTVMAIHGCILSEI 315
>At5g46610 putative protein
Length = 543
Score = 231 bits (590), Expect = 4e-61
Identities = 117/295 (39%), Positives = 181/295 (60%), Gaps = 8/295 (2%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFE 138
K++ W++G DPR++ A K+G++L L+SLL ++ F+ +G ++WA++TVV V E
Sbjct: 33 KKILKNIWKVGKDDPRRVKHALKVGVSLTLVSLLYLMEPLFKGIGNSAIWAVMTVVVVLE 92
Query: 139 FSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYAKLY 197
FS GATL KGLNRGLGTL AG LA + ++ +G+ + I + ++FI+G TY +
Sbjct: 93 FSAGATLCKGLNRGLGTLIAGSLAFFIEFVANDSGKIFRAIFIGAAVFIIGALITYLRFI 152
Query: 198 PTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIW 256
P +K Y+YG +FL+T+ I VS Y+ I A RF IA+G + L +++ ++PIW
Sbjct: 153 PYIKKNYDYGMLIFLLTFNLITVSSYRVDTVIKIAHERFYTIAMGVGICLLMSLLVFPIW 212
Query: 257 AGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQS 316
+GEDLH G++ S+E VN Y +E T S+D +Y+GY++ L S
Sbjct: 213 SGEDLHKSTVAKLQGLSYSIEACVNEY-----FEEEEKDEETSDLSEDTIYNGYKTVLDS 267
Query: 317 TSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
S ++AL +A WEP H R+ ++PW +YVKV LR + V+A+HGC+ +EI
Sbjct: 268 KSADEALAMYASWEPRHTRH-CHRFPWKHYVKVGSVLRQFGYTVVALHGCLKTEI 321
>At4g17970 unknown protein
Length = 560
Score = 228 bits (580), Expect = 5e-60
Identities = 118/295 (40%), Positives = 180/295 (61%), Gaps = 11/295 (3%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFE 138
K++ R W +G DPR+++ A K+GL+L L+SLL ++ F+ +G +++WA++TVV V E
Sbjct: 31 KKIPKRLWNVGKEDPRRVIHALKVGLSLTLVSLLYLMEPLFKGIGSNAIWAVMTVVVVLE 90
Query: 139 FSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYAKLY 197
FS GATL KGLNRGLGTL AG LA + ++ +G+ I + ++FI+G ATY +
Sbjct: 91 FSAGATLCKGLNRGLGTLIAGSLAFFIEFVANDSGKVLRAIFIGTAVFIIGAAATYIRFI 150
Query: 198 PTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIW 256
P +K Y+YG +FL+T+ I VS Y+ I+ A RF IA+G + L +++ ++PIW
Sbjct: 151 PYIKKNYDYGVVIFLLTFNLITVSSYRVDSVINIAHDRFYTIAVGCGICLFMSLLVFPIW 210
Query: 257 AGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQS 316
+GEDLH G++ S+E V+ Y E E+ SK D +Y GY++ L S
Sbjct: 211 SGEDLHKTTVGKLQGLSRSIEACVDEYFEEKEKEKTDSK--------DRIYEGYQAVLDS 262
Query: 317 TSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
ST++ L +A WEP H + ++P YVKV LR + V+A+HGC+ +EI
Sbjct: 263 KSTDETLALYANWEPRH-TLRCHRFPCQQYVKVGAVLRQFGYTVVALHGCLQTEI 316
>At3g11680 unknown protein
Length = 331
Score = 219 bits (557), Expect = 2e-57
Identities = 117/294 (39%), Positives = 179/294 (60%), Gaps = 14/294 (4%)
Query: 78 TKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVF 137
TKRV + DPR+I+ + K+G+AL L+SLL +++ + G +WAILTVV VF
Sbjct: 30 TKRVK-NVQKFAKDDPRRIIHSMKVGVALTLVSLLYYVRPLYISFGVTGMWAILTVVVVF 88
Query: 138 EFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAG-QWEEIIVMVSIFIVGFCATYAKL 196
EF++G TLSKGLNRG TL AG L VG L++ G Q E I++ + +F +G AT+++
Sbjct: 89 EFTVGGTLSKGLNRGFATLIAGALGVGAVHLARFFGHQGEPIVLGILVFSLGAAATFSRF 148
Query: 197 YPTMK-AYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPI 255
+P +K Y+YG +F++T+ ++ +SGY+T + + A R I +G + + V++ + P+
Sbjct: 149 FPRIKQRYDYGALIFILTFSFVAISGYRTDEILIMAYQRLSTILIGGTICILVSIFICPV 208
Query: 256 WAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQ 315
WAGEDLH ++A N +A LEG Y P KI +S Y+S L
Sbjct: 209 WAGEDLHKMIANNINKLAKYLEGFEGEYFQ-------PEKISKETSS---CVREYKSILT 258
Query: 316 STSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILS 369
S STED+L A WEP HGR++ L++PW Y+K++G +R CA + ++G +LS
Sbjct: 259 SKSTEDSLANLARWEPGHGRFR-LRHPWKKYLKIAGLVRQCAVHLEILNGYVLS 311
>At5g46600 putative protein
Length = 539
Score = 214 bits (546), Expect = 5e-56
Identities = 115/300 (38%), Positives = 177/300 (58%), Gaps = 9/300 (3%)
Query: 76 KTTKRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVT 135
K K++ W +G DPR+++ A K+G+AL L+SLL ++ FE +G++++WA++TVV
Sbjct: 31 KKMKKILRNLWNVGKEDPRRVIHALKVGVALTLVSLLYLMEPFFEGVGKNALWAVMTVVV 90
Query: 136 VFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQ-WEEIIVMVSIFIVGFCATYA 194
V EFS GATL KGLNRGLGTL AG LA + ++ +G+ I + S+F +G TY
Sbjct: 91 VLEFSAGATLRKGLNRGLGTLIAGSLAFFIEWVAIHSGKILGGIFIGTSVFTIGSMITYM 150
Query: 195 KLYPTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLY 253
+ P +K Y+YG VFL+T+ I VS Y+ I A R I +G + L +++ +
Sbjct: 151 RFIPYIKKNYDYGMLVFLLTFNLITVSSYRVDTVIKIAHERLYTIGMGIGICLFMSLLFF 210
Query: 254 PIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASD--DVVYSGYR 311
PIW+G+DLH G++ +E V+ Y E++ + SD D++Y+GY
Sbjct: 211 PIWSGDDLHKSTITKLQGLSRCIEACVSEYFE----EKLKDNETSDSESDDEDLIYNGYN 266
Query: 312 SALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+ L S S ++AL +A WEP H R + K+P Y+KV LR + V+A+HGC+ +EI
Sbjct: 267 TVLDSKSADEALAMYAKWEPRHTR-RCNKFPSQQYIKVGSVLRKFGYTVVALHGCLQTEI 325
>At1g08430 hypothetical protein
Length = 493
Score = 206 bits (525), Expect = 1e-53
Identities = 107/297 (36%), Positives = 182/297 (61%), Gaps = 11/297 (3%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKT--PFED-LGRHSVWAILTVVT 135
+ + +G DPR+I+ A K+GLAL+L+S + + PF D G +++WA++TVV
Sbjct: 5 REIVREGIRVGNEDPRRIIHAFKVGLALVLVSSFYYYQPFGPFTDYFGINAMWAVMTVVV 64
Query: 136 VFEFSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQW-EEIIVMVSIFIVGFCATYA 194
VFEFS+GATL KGLNRG+ TL AGGL +G L++L+G E I++++ +F+ +T+
Sbjct: 65 VFEFSVGATLGKGLNRGVATLVAGGLGIGAHQLARLSGATVEPILLVMLVFVQAALSTFV 124
Query: 195 KLYPTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLY 253
+ +P +K ++YG +F++T+ I +SG++ + +D A SR + +G + +++ +
Sbjct: 125 RFFPWVKTKFDYGILIFILTFALISLSGFRDEEIMDLAESRLSTVVIGGVSCILISIFVC 184
Query: 254 PIWAGEDLHDLVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSA 313
P+WAG+DLH L+A NF ++ L+ + Y E+ K++ + + Y+S
Sbjct: 185 PVWAGQDLHSLLASNFDTLSHFLQDFGDEYFEA--REKGDYKVVEKRKKN---LERYKSV 239
Query: 314 LQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSE 370
L S S E+AL +A WEPPHG+++ ++PW YV V LR CA+ + A++ I S+
Sbjct: 240 LDSKSDEEALANYAEWEPPHGQFR-FRHPWKQYVAVGALLRQCAYRIDALNSYINSD 295
>At1g08440 hypothetical protein
Length = 501
Score = 199 bits (507), Expect = 2e-51
Identities = 103/298 (34%), Positives = 176/298 (58%), Gaps = 14/298 (4%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFE 138
+ + +G DPR++V A K+GLAL L+S + + +++ G +++WA++TVV VFE
Sbjct: 5 REIVREGRRVGKEDPRRVVHAFKVGLALALVSSFYYYQPLYDNFGVNAMWAVMTVVVVFE 64
Query: 139 FSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQW-EEIIVMVSIFIVGFCATYAKLY 197
FS+GATL KGLNR + TL AGGL +G L+ L+G E I++ + +F++ +T+ + +
Sbjct: 65 FSVGATLGKGLNRAVATLVAGGLGIGAHHLASLSGPTVEPILLAIFVFVLAALSTFVRFF 124
Query: 198 PTMKA-YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIW 256
P +KA Y+YG +F++T+ I VSG++ + +D A R + +G + +++ + P+W
Sbjct: 125 PRVKARYDYGVLIFILTFALISVSGFREDEILDLAHKRLSTVIMGGVSCVLISIFVCPVW 184
Query: 257 AGEDLHDLVAKNFMGVAASLEGVVNNYLNCIE---YERVPSKILTYQASDDVVYSGYRSA 313
AG+DLH L+A NF ++ L+ + Y E + V + + Y+S
Sbjct: 185 AGQDLHSLLASNFDTLSHFLQEFGDEYFEATEDGDIKEVEKRRRNLER--------YKSV 236
Query: 314 LQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
L S S E+AL FA WEP HG+++ ++PW Y+ V LR A+ + A++ I S++
Sbjct: 237 LNSKSNEEALANFAKWEPRHGQFR-FRHPWRQYLAVGALLRQSAYRIDALNSNINSDM 293
>At4g00910
Length = 507
Score = 191 bits (486), Expect = 4e-49
Identities = 100/303 (33%), Positives = 172/303 (56%), Gaps = 20/303 (6%)
Query: 84 RAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGA 143
+AW +G DP K+V K+GLAL L+S+ +++ ++ +G +++WAI+TVV VFE ++GA
Sbjct: 55 KAWRIGADDPAKVVHCLKVGLALSLVSIFYYMRPLYDGVGGNAMWAIMTVVVVFESNVGA 114
Query: 144 TLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVGFCATYAKLYPTMKA- 202
T K +NR + T+ AG L + + ++ +G+ E ++ S+F+ F ATY++ P+ KA
Sbjct: 115 TFCKCVNRVVATILAGSLGIAVHWVATQSGKAEVFVIGCSVFLFAFAATYSRFVPSFKAR 174
Query: 203 YEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLH 262
++YG +F++T+ + V GY+ K ++ A R IA+G ++ + + V PIWAG LH
Sbjct: 175 FDYGAMIFILTFSLVSVGGYRVDKLVELAQQRVSTIAIGTSICIIITVFFCPIWAGSQLH 234
Query: 263 DLVAKNFMGVAASLEG---VVNNYLNCIEYERVPSKILTYQASDD---VVYSGYRSALQS 316
L+ +N +A SL+ V C+ +++ T + D+ + G++ L S
Sbjct: 235 RLIERNLEKLADSLDARVLTVTKTAGCVAEYFKENEVSTNRNEDENTNMKLQGFKCVLNS 294
Query: 317 TSTEDAL-----MGF-------AVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMH 364
TE+A+ GF A WEP HG + ++PW YVK+ A+R CA+ + +
Sbjct: 295 KGTEEAMPLIRFSGFSFSQANLARWEPAHGSFN-FRHPWKLYVKIGAAMRRCAYCLENLS 353
Query: 365 GCI 367
CI
Sbjct: 354 ICI 356
>At2g27240 hypothetical protein
Length = 506
Score = 185 bits (469), Expect = 4e-47
Identities = 102/299 (34%), Positives = 173/299 (57%), Gaps = 30/299 (10%)
Query: 92 DPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGATLSKGLNR 151
DPR++V + K+GL L L+S + + ++ G +++WA++TVV VFEFS+GATL KGLNR
Sbjct: 18 DPRRVVHSFKVGLVLALVSSFYYYQPLYDSFGVNAMWAVMTVVVVFEFSVGATLGKGLNR 77
Query: 152 GLGTLTAGGLAVGMGVLSKLAG-QWEEIIVMVSIFIVGFCATYAKLYPTMKA-YEYGFRV 209
TL AGGL +G L+ ++G E I++ V +F+ +T+ + +P +KA Y+Y +
Sbjct: 78 VAATLFAGGLGIGAHHLASMSGPTGEPILLAVFVFVQAALSTFVRFFPRVKARYDYSLLI 137
Query: 210 FLITYCYIIVSGYQTGKFIDTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHDLVAKNF 269
F++T+ I VSG++ + + R + +G + +++ + P+WAG+DLH L+A NF
Sbjct: 138 FILTFALISVSGFREEQVVKLTHKRISTVIIGGLSCVIISIFVCPVWAGQDLHSLIASNF 197
Query: 270 MGVAASLEG----VVNNYLNCIEYERVPSKILTYQASD------DVV------------- 306
++ L G V++ LN I R KI +++ +D +VV
Sbjct: 198 EKLSFFLLGNSFHYVSSDLNSITLLR---KIKSWRLADFGDKYCEVVENDGAKEVDKRKK 254
Query: 307 -YSGYRSALQSTSTEDALMGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMH 364
+ Y+S L S S E++L FA WEP HG+++ ++PW Y+ V +R CA+ + A++
Sbjct: 255 DFDNYKSVLNSKSNEESLANFAKWEPGHGQFR-FRHPWKQYLAVGELIRQCAYRIHALN 312
>At4g17585 putative protein
Length = 152
Score = 61.2 bits (147), Expect = 9e-10
Identities = 24/58 (41%), Positives = 42/58 (72%)
Query: 79 KRVAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDLGRHSVWAILTVVTV 136
K++ R W +G DPR+++ A K+G +L L+SLL F++ F+ +G +++WA++TVV V
Sbjct: 31 KKIPKRLWSVGKEDPRRVIHAFKVGHSLTLVSLLYFMENLFKGIGSNAIWAVMTVVAV 88
>At5g38030 putative transmembrane protein
Length = 498
Score = 32.0 bits (71), Expect = 0.56
Identities = 36/133 (27%), Positives = 63/133 (47%), Gaps = 13/133 (9%)
Query: 131 LTVVTVFEFSIGATLSKGLNRGLGTL--TAGGLAVGMGVLSKLAGQWEE--IIVMVSIFI 186
L V+V E S+ A S G+ G+G+ T G A G G LS L + +I+ V+ I
Sbjct: 82 LAAVSV-ENSVIAGFSFGVMLGMGSALETLCGQAFGAGKLSMLGVYLQRSWVILNVTAVI 140
Query: 187 VGFCATYAKLY-------PTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLIA 239
+ +A P + + F +++I + Y T KF+ + S+ +++A
Sbjct: 141 LSLLYIFAAPILAFIGQTPAISSATGIFSIYMIPQIFAYAVNYPTAKFLQSQ-SKIMVMA 199
Query: 240 LGAAVSLGVNVCL 252
+AV+L ++V L
Sbjct: 200 AISAVALVLHVLL 212
>At3g02460 putative plant adhesion molecule
Length = 353
Score = 31.2 bits (69), Expect = 0.95
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query: 81 VAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDL 122
+A R W++ LS+ KIVF K+GLAL+ +K PFE L
Sbjct: 267 LALRIWDVFLSEGVKIVF--KVGLALLKYCQDELVKLPFEKL 306
>At5g16910 cellulose synthase catalytic subunit -like protein
Length = 1145
Score = 30.8 bits (68), Expect = 1.2
Identities = 24/102 (23%), Positives = 43/102 (41%), Gaps = 5/102 (4%)
Query: 209 VFLITYCYIIVSGYQTGKFI-DTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHDLVAK 267
+FLI YC++ +G+FI T FL+ L +++L + L W+G L +
Sbjct: 930 IFLIVYCFLPALSLFSGQFIVQTLNVTFLVYLLIISITLCLLALLEIKWSGISLEEWWRN 989
Query: 268 N----FMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDV 305
G +A L V+ L + + + + DD+
Sbjct: 990 EQFWLIGGTSAHLAAVLQGLLKVVAGVEISFTLTSKSGGDDI 1031
>At5g15930 plant adhesion molecule 1 (PAM1)
Length = 356
Score = 30.8 bits (68), Expect = 1.2
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 2/41 (4%)
Query: 82 AARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDL 122
A R W++ L++ KIVF K+GLAL+ LK PFE+L
Sbjct: 265 ALRIWDVFLAEGVKIVF--KVGLALLKHCHDDLLKLPFEEL 303
>At5g05380 putative protein
Length = 217
Score = 30.8 bits (68), Expect = 1.2
Identities = 18/49 (36%), Positives = 24/49 (48%)
Query: 112 LIFLKTPFEDLGRHSVWAILTVVTVFEFSIGATLSKGLNRGLGTLTAGG 160
L+ L F D V ILT+V VF S+G+ L+ L G G + G
Sbjct: 125 LVVLGRTFSDRETLGVLVILTIVVVFLTSVGSLLTSALMIGFGIVCLHG 173
>At1g64890 unknown protein
Length = 442
Score = 30.8 bits (68), Expect = 1.2
Identities = 24/81 (29%), Positives = 41/81 (49%), Gaps = 10/81 (12%)
Query: 71 LVGAWKTTKRVAARAWEMGLSD------PRKIVFAAKMGLALILISLLIFLKTPFEDLG- 123
L+G K ++A W + PRK++ A ++ +A +IS L+F+K + DLG
Sbjct: 272 LLGISKVFGQIAMLLWGFAYNRWLKAMRPRKLLTAIQVTIAFFVISDLLFVKGVYRDLGV 331
Query: 124 RHSVWAIL---TVVTVFEFSI 141
SV+ + + T+F F I
Sbjct: 332 SDSVYVLFFSGFLETLFYFKI 352
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,231,088
Number of Sequences: 26719
Number of extensions: 343738
Number of successful extensions: 1079
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1029
Number of HSP's gapped (non-prelim): 33
length of query: 371
length of database: 11,318,596
effective HSP length: 101
effective length of query: 270
effective length of database: 8,619,977
effective search space: 2327393790
effective search space used: 2327393790
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0310b.11


