

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0307.3
(443 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
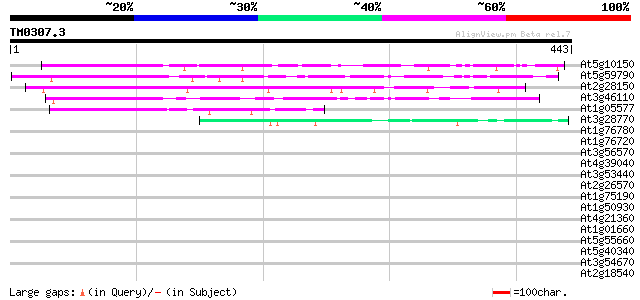
Score E
Sequences producing significant alignments: (bits) Value
At5g10150 unknown protein 216 3e-56
At5g59790 unknown protein 182 2e-46
At2g28150 unknown protein 167 1e-41
At3g46110 unknown protein 153 2e-37
At1g05577 hypothetical protein 92 4e-19
At3g28770 hypothetical protein 46 4e-05
At1g76780 putative heat shock protein 39 0.006
At1g76720 putative translation initiation factor IF-2 38 0.010
At3g56570 putative protein 38 0.013
At4g39040 unknown protein 37 0.017
At3g53440 unknown protein 37 0.017
At2g26570 unknown protein 36 0.037
At1g75190 unknown protein 36 0.037
At1g50930 hypothetical protein 36 0.037
At4g21360 putative transposable element 35 0.083
At1g01660 hypothetical protein 35 0.083
At5g55660 putative protein 35 0.11
At5g40340 unknown protein 35 0.11
At3g54670 structural maintenance of chromosomes (SMC) - like pro... 35 0.11
At2g18540 putative vicilin storage protein (globulin-like) 35 0.11
>At5g10150 unknown protein
Length = 414
Score = 216 bits (549), Expect = 3e-56
Identities = 168/431 (38%), Positives = 232/431 (52%), Gaps = 74/431 (17%)
Query: 26 KVIKPA-RKVQVVYYLFRNGLLEHPHFMELTLLPNQPLRLKDVFDRLMVLRGSGMPLQYS 84
K KP R+VQVVYYL RNG LEHPHF+E+ NQPLRL+DV +RL +LRG M QY+
Sbjct: 37 KTKKPIFRRVQVVYYLTRNGHLEHPHFIEVISPVNQPLRLRDVMNRLTILRGKCMTSQYA 96
Query: 85 WSSKRNYKSGYVWFDLALKDVIYPSEGGEYVLKGSELVEGCSERFQQLNVSN--KNGIHE 142
WS KR+Y++G+VW DLA DVIYPS+ EYVLKGSE+ +++FQ+++V+ I E
Sbjct: 97 WSCKRSYRNGFVWNDLAENDVIYPSDCAEYVLKGSEI----TDKFQEVHVNRPLSGSIQE 152
Query: 143 PPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFT--------SSTTTPHSRCSR 194
P++ +K K + +AE Y EE+EE+ E+ +S+TTP SRCSR
Sbjct: 153 APKSRL-LRSKLKPQNRTASFDDAELYVGEEEEEEDGEYELYEEKTSYTSSTTPQSRCSR 211
Query: 195 GVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLN 254
GVSTE + E + N TKTE+ + + SS ++T P +K ++ E +
Sbjct: 212 GVSTETMESTEQK----PNLTKTEQDLQVRSDSS--DLTRSNPVVKPRRHEVSTRVEDGD 265
Query: 255 RRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVG 314
PV+ R S LQ+I+CG ++ A + PR
Sbjct: 266 -----PVE----------------PGSGRGSMWLQMISCGHIATKYYAPSVMNPR----- 299
Query: 315 TTASKQRQSVDRRDQ--KAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYF 372
Q++ R+ K + ++E E + M SENPR GN Q AEEKEYF
Sbjct: 300 -----QKEENLRKGVLCKNIVKKTVVDDEREMIRFM----SENPR--FGNPQ-AEEKEYF 347
Query: 373 SGSLVEMESMK--ANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCI 430
SGS+VE S + P L+RSNS+NEE RSK+ VE K E + +E + K KCI
Sbjct: 348 SGSIVESVSQERVTAEPSLRRSNSFNEE-RSKI-VEMAK-----ETKKKEERSMAKVKCI 400
Query: 431 P---IKKSSKE 438
P + SSK+
Sbjct: 401 PRTCLMSSSKQ 411
>At5g59790 unknown protein
Length = 423
Score = 182 bits (463), Expect = 2e-46
Identities = 158/457 (34%), Positives = 224/457 (48%), Gaps = 82/457 (17%)
Query: 2 DQHQVRPHRCSSR-DTSPDRAKICRKV-IKPA--RKVQVVYYLFRNGLLEHPHFMELTLL 57
D + + P R + DTSPDR +I + +KP RKV VVYYL RNG L+HPHF+E+TL
Sbjct: 11 DNNYLVPRRSKDQQDTSPDRNRIWSEPRLKPVVNRKVPVVYYLCRNGQLDHPHFIEVTLS 70
Query: 58 PNQPLRLKDVFDRLMVLRGSGMPLQYSWSSKRNYKSGYVWFDLALKDVIYPSEGGEYVLK 117
+ L LKDV +RL LRG GM YSWSSKR+YK+G+VW DL+ D I+P +G EYVLK
Sbjct: 71 SHDGLYLKDVINRLNDLRGKGMASLYSWSSKRSYKNGFVWHDLSEDDFIFPVQGQEYVLK 130
Query: 118 GSELVEGCSERFQQLNVSNKNGIHEP----------PETNYGYNAKSKVLSNRQQQK--- 164
GSE+++ C +SN + E P+ N G + + V++ R+ Q
Sbjct: 131 GSEVLDSCL-------ISNPRSLLETSSFRDPRSLNPDKNSGDDIPA-VINRRRNQSWSS 182
Query: 165 -EAEEYEEYEEQEEEKEFT------SSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKT 217
+ EY+ Y+ E E T +ST T R R + EE+ +E + A N T
Sbjct: 183 IDLSEYKVYKATESSAESTQRLAADASTQTDDRRRRRRPAKEEI--EEVKSPASYENQST 240
Query: 218 EKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESAT 277
E SR ++ PPP + +L + R ++ P +S ES +
Sbjct: 241 E--------LSRDEIS--PPPSDSSPETLENLIKADGRLILRPSESST---DHRTVESLS 287
Query: 278 SAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASK-QRQSVDRRDQKAFMMSC 336
S + L+QLI+CG+ FK G K Q +++ R
Sbjct: 288 SGRMRASAVLMQLISCGTM--SFK----------ECGPVLLKDQGLALNGRSGCTITRGA 335
Query: 337 EEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYN 396
E+ EE RV + + G +Q E+KEYFSGSL+ E+ K P LKRS+SYN
Sbjct: 336 EDNGEE--------RVDKELK-SFGRVQ-LEDKEYFSGSLI--ETKKELVPALKRSSSYN 383
Query: 397 EERRSKLGVEEVKLKKEMEEENRERKGGVKEKCIPIK 433
+R S++G K ++E V+ KCIP K
Sbjct: 384 ADRCSRMGPTTEKDEEE----------AVRAKCIPRK 410
>At2g28150 unknown protein
Length = 540
Score = 167 bits (423), Expect = 1e-41
Identities = 150/455 (32%), Positives = 219/455 (47%), Gaps = 82/455 (18%)
Query: 13 SRDTSPDRAKICR----KVIKPARKVQVVYYLFRNGLLEHPHFMELTLLPNQPLRLKDVF 68
SR+ SP+RAK+ K + +KVQ+VYYL +N LEHPHFME+ + L L+DV
Sbjct: 9 SREVSPERAKVWTEKSPKYHQKIKKVQIVYYLSKNRQLEHPHFMEVLISSPNGLYLRDVI 68
Query: 69 DRLMVLRGSGMPLQYSWSSKRNYKSGYVWFDLALKDVIYPSEGGEYVLKGSELV-EGCSE 127
+RL VLRG GM YSWSSKR+Y++G+VW DL+ D+I P+ G EYVLKGSEL E S+
Sbjct: 69 ERLNVLRGRGMASMYSWSSKRSYRNGFVWHDLSEDDLILPANGNEYVLKGSELFDESNSD 128
Query: 128 RFQQL-NVSNKNG---IHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYE-EQEEEKEFT 182
F + N++ +N + EPP + ++ S N + +E+ E + +
Sbjct: 129 HFSPIVNLATQNMKQIVVEPPSSRSMDDSSSSSSMNNGKGTNKHSHEDDELSPPALRSVS 188
Query: 183 SSTTTPHSRCSRGVSTEELGE------------DEDRHEALQNNTKTEKVVVTNNVSSRG 230
SS +P SR ++ S+ L E E + +KT + VS+
Sbjct: 189 SSGVSPDSRDAKNSSSWCLAEYKVYKSEGLADASTQTDETVSGRSKTPIETFSRGVSTDE 248
Query: 231 NVTNVPPPMKNHSAAAASLAEK------LNRRVVVP--VQSKQRFGGELGT-ESATSAAP 281
+V++ P +N+ + AS A K ++R V P S GG+ T ES A
Sbjct: 249 DVSSEPETSENNLVSEASCAGKERESAEISRNSVSPPFSNSASSLGGKTDTLESLIRADV 308
Query: 282 SRYSA----------------------LLQLIACGSSGPEFKARTQQEPRVSSVGTTASK 319
S+ ++ L+QLI+CGS + ++ V T K
Sbjct: 309 SKMNSFRILEQEDVRMPAIPRLRASNMLMQLISCGSI-------SVKDNNFGLVPTYKPK 361
Query: 320 QRQSVDRRD--QKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLV 377
S +FMM ++R+SE P LM + EEKEYFSGSLV
Sbjct: 362 FGHSKFPSPFFSSSFMMG------------DLDRLSETPSLMGLRM---EEKEYFSGSLV 406
Query: 378 EMESMKA----NHPVLKRSNSYNEERRS-KLGVEE 407
E + K ++ LKRS+SYN +R S ++GV E
Sbjct: 407 ETKLQKKDAADSNASLKRSSSYNGDRASNQMGVAE 441
>At3g46110 unknown protein
Length = 343
Score = 153 bits (387), Expect = 2e-37
Identities = 122/392 (31%), Positives = 186/392 (47%), Gaps = 76/392 (19%)
Query: 29 KPARK--VQVVYYLFRNGLLEHPHFMELTLLPNQPLRLKDVFDRLMVLRGSGMPLQYSWS 86
KP+R+ V VVYYL RNG L+HPHF+E+ L + L LKDV +RL LRG+GM YSWS
Sbjct: 13 KPSRERIVPVVYYLSRNGRLDHPHFIEVPLSSHNGLYLKDVINRLNDLRGNGMACLYSWS 72
Query: 87 SKRNYKSGYVWFDLALKDVIYPSEGGEYVLKGSELVEGCSERFQQLNVSNKNGIHEPPET 146
SKR YK+G+VW+DL+ +D I+P G EYVLKGS++ L++ N +G
Sbjct: 73 SKRTYKNGFVWYDLSDEDFIFPVHGQEYVLKGSQI----------LDLDNNSG------- 115
Query: 147 NYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDED 206
N + Q + Y+ Y+ E E T R +S + + +D
Sbjct: 116 ----NFSAVTHRRNQSWSSVDHYKVYKASELNAEAT-----------RKLSMDASTQTDD 160
Query: 207 RHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQR 266
R + V N V+ PP + S + + R+++ +Q Q
Sbjct: 161 RRR------RKSPVDEVNEVTELSREEITSPPQSDSSPETLESLMRADGRLIL-LQEDQE 213
Query: 267 FGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDR 326
+ PS + L+QLI+CG+ FK + P + + T ++ R + +
Sbjct: 214 L-----NRTVEKMRPS--AVLMQLISCGAM--SFK---KCGPTLMNGNTRSTAVRGTGNY 261
Query: 327 RDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANH 386
R ++A E+E ++ + EEKEYFSGSL++ S K
Sbjct: 262 RLERA-------EKELKSFGRV----------------KLEEKEYFSGSLIDESSKKELV 298
Query: 387 PVLKRSNSYNEERRSKLGVEEVKLKKEMEEEN 418
P LKRS+SYN +R S++G+ + K +E+ N
Sbjct: 299 PALKRSSSYNIDRSSRMGLTKEKEGEELARAN 330
>At1g05577 hypothetical protein
Length = 372
Score = 92.4 bits (228), Expect = 4e-19
Identities = 71/227 (31%), Positives = 111/227 (48%), Gaps = 23/227 (10%)
Query: 32 RKVQVVYYLFRNGLLEHPHFMELTLLPNQPLRLKDVFDRLMVLRGSGMPLQYSWSSKRNY 91
R+V +VY+L R+G ++HPH + + L + L+DV L RG MP +SWS KR Y
Sbjct: 11 RRVNLVYFLSRSGHVDHPHLLRVHHLSRNGVFLRDVKKWLADARGDAMPDAFSWSCKRRY 70
Query: 92 KSGYVWFDLALKDVIYPSEGGEYVLKGSELVEGCSERFQQLNVSNKNGIHEPPETNYGYN 151
K+GYVW DL D+I P EYVLKGSE++ S + NV K + N G +
Sbjct: 71 KNGYVWQDLLDDDLITPISDNEYVLKGSEILLS-SPKEDYPNVEKKAWV----TRNGGID 125
Query: 152 AKSKV----LSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPH------SRCSRGVSTEEL 201
A+ K+ L++ + QKE+ + T +TT + + VS +
Sbjct: 126 AEEKLQKLKLTSEKIQKESPVFCSQRSTATTSTVTEESTTNEEGFVLKKQDPKTVSGQRD 185
Query: 202 GEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAAS 248
G E+ N+ ++ + V++ SS + N K++S+ AS
Sbjct: 186 GSTEN---GSGNDVESGRPSVSSTTSSSSYIKN-----KSYSSVRAS 224
>At3g28770 hypothetical protein
Length = 2081
Score = 46.2 bits (108), Expect = 4e-05
Identities = 67/316 (21%), Positives = 123/316 (38%), Gaps = 54/316 (17%)
Query: 151 NAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGED--EDRH 208
N + V N+++ ++ E+ E+ E ++ + T S +R + E GED ED+
Sbjct: 770 NKEENVQGNKKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKE 829
Query: 209 EA-----LQNNTKTEKVVVTNNVSSRGNVTNVPPPMK-----NHSAAAASLAEKLNRRVV 258
E+ ++ K E V NV ++ + ++ N + E++ R
Sbjct: 830 ESKDYQSVEAKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDK 889
Query: 259 VPVQSKQRFGGELGTESATSAAPS-RYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTA 317
+ + F + + + S +Y + K +E ++ T+
Sbjct: 890 SSTKEVRDFANNMDIDVQKGSGESVKYKK------------DEKKEGNKEENKDTINTS- 936
Query: 318 SKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRV------------SENPRLMLGNLQN 365
SKQ+ ++ +K S +++EE+ + N + SEN +L N N
Sbjct: 937 SKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDN 996
Query: 366 AEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGV 425
E+KE +S N R EE++SK E K KK+ +++ RE K
Sbjct: 997 KEKKE-------SEDSASKN-----REKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSE 1044
Query: 426 KEKCIPIKKSSKESSR 441
+ K K KE SR
Sbjct: 1045 ERK----SKKEKEESR 1056
Score = 37.4 bits (85), Expect = 0.017
Identities = 73/376 (19%), Positives = 143/376 (37%), Gaps = 30/376 (7%)
Query: 73 VLRGSGMPLQYSWSSKR--NYKSGYVWFDLALKDVIYPSEGGEYVLKGSELVEGCSERFQ 130
V +GSG ++Y K+ N + + + K + + K S + + ++ +
Sbjct: 906 VQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKE 965
Query: 131 QLNVSNKNGIHEPPETNYGYNAKSKVLS-NRQQQKEAEEYEEYEEQEEEKEFTSSTTTPH 189
+N K ET N+K K + + +++KE+E+ +++E E S T
Sbjct: 966 YVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEE 1025
Query: 190 SRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNH-SAAAAS 248
++ + S ++ E++D E K E + + T +NH S
Sbjct: 1026 AKKEKKKSQDKKREEKDSEERKSKKEKEESRDL--KAKKKEEETKEKKESENHKSKKKED 1083
Query: 249 LAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEP 308
E + + + + K+ ++S + L+ K + E
Sbjct: 1084 KKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLE-----DQNSNKKKEDKNEK 1138
Query: 309 RVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEE 368
+ S K+ ++++ + E +E E+ N V + + + Q +E
Sbjct: 1139 KKSQHVKLVKKESDKKEKKENE----EKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKE 1194
Query: 369 KEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKE- 427
KE + E E K LK++ E+R+ + VEE K +KE ++E + K K
Sbjct: 1195 KE-----MKESEEKK-----LKKNE---EDRKKQTSVEENKKQKETKKEKNKPKDDKKNT 1241
Query: 428 -KCIPIKKSSKESSRR 442
K KK S ES +
Sbjct: 1242 TKQSGGKKESMESESK 1257
Score = 35.8 bits (81), Expect = 0.049
Identities = 58/311 (18%), Positives = 125/311 (39%), Gaps = 49/311 (15%)
Query: 153 KSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQ 212
+ K+ N + +K+ EE ++Q+E K+ + ++ ++ + + EA +
Sbjct: 1202 EKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEA-E 1260
Query: 213 NNTKTEKVVVTNNVSSRGNVT-NVPPPMKNHSAAAASLAEKLNRRVV-VPVQSKQRFGGE 270
N K++ ++ S+ + +HS + A E N ++ Q+ + E
Sbjct: 1261 NQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNE 1320
Query: 271 LGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASK--QRQSVDRRD 328
+ TS A ++ + + +P+ TT +++S++
Sbjct: 1321 EDRKKQTSVAENKKQKETK-------------EEKNKPKDDKKNTTKQSGGKKESMESES 1367
Query: 329 QKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQ-------NAEEKEYFSGSLVEMES 381
++A E +++ +A + S+N LM + Q A+ E + L++ +S
Sbjct: 1368 KEA-----ENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADS 1422
Query: 382 MKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENR----------ERKGGVKEKCIP 431
+R+N E+R+ + V E K +KE +EE E+ GG KE
Sbjct: 1423 QATT----QRNNE--EDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTEQSGGKKES--- 1473
Query: 432 IKKSSKESSRR 442
++ SKE+ +
Sbjct: 1474 MESESKEAENQ 1484
Score = 35.0 bits (79), Expect = 0.083
Identities = 59/307 (19%), Positives = 112/307 (36%), Gaps = 35/307 (11%)
Query: 126 SERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSN-RQQQKEAEEYEEYEEQEEEKEFTSS 184
S+ +Q + KN + +TN G SK L + R + +A + E +++ EE +
Sbjct: 831 SKDYQSVEAKEKNE-NGGVDTNVGNKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDK 889
Query: 185 TTTPHSR---------CSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNV 235
++T R +G + +++ E + K T N SS+ +
Sbjct: 890 SSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKD-----TINTSSKQKGKDK 944
Query: 236 PPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGS 295
K + E+ + V KQ + T+S S +
Sbjct: 945 KKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESED 1004
Query: 296 SGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSEN 355
S + + + + E + S A K+++ +D+K EE++ EE + S +
Sbjct: 1005 SASKNREKKEYEEKKSKTKEEAKKEKKK--SQDKKR-----EEKDSEERKSKKEKEESRD 1057
Query: 356 PRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEME 415
+ + E+KE NH K+ + E + EE K +K+
Sbjct: 1058 LKAKKKEEETKEKKE------------SENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKH 1105
Query: 416 EENRERK 422
EE++ RK
Sbjct: 1106 EESKSRK 1112
Score = 35.0 bits (79), Expect = 0.083
Identities = 60/339 (17%), Positives = 136/339 (39%), Gaps = 24/339 (7%)
Query: 117 KGSELVEGCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQE 176
K SE + E+ + ++ K E E N KSK ++++ ++ + ++ E+++
Sbjct: 1041 KDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKK 1100
Query: 177 EEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKV-VVTNNVSSRGNVTNV 235
E+K+ S + + + E + E K++ V +V + N
Sbjct: 1101 EKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENE 1160
Query: 236 PPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGS 295
++ S +++++ + +Q+ + ES + +
Sbjct: 1161 EKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEE 1220
Query: 296 SGPEFKARTQQ-EPRVSSVGTTASK--QRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRV 352
+ + + + ++ +P+ TT +++S++ ++A E +++ +A +
Sbjct: 1221 NKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEA-----ENQQKSQATTQADSDE 1275
Query: 353 SENPRLMLGNLQ-------NAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGV 405
S+N LM + Q A+ E + L++ +S +R+N E+R+ + V
Sbjct: 1276 SKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATT----QRNNE--EDRKKQTSV 1329
Query: 406 EEVKLKKEMEEENRERKGGVKE--KCIPIKKSSKESSRR 442
E K +KE +EE + K K K KK S ES +
Sbjct: 1330 AENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESK 1368
>At1g76780 putative heat shock protein
Length = 1871
Score = 38.9 bits (89), Expect = 0.006
Identities = 61/283 (21%), Positives = 119/283 (41%), Gaps = 36/283 (12%)
Query: 168 EYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDE-DRHEALQNNTKTEKVVVTNNV 226
E EE EE+EEE+ S +G EL +D ++ + + EK + +N
Sbjct: 61 EEEEEEEEEEEERVDVSEAEHKEETEKG----ELKDDYLEKSHQIDERIEEEKGLADSNK 116
Query: 227 SSRGNVTNVPPPMKNHSAAAASLAEKL-NRRVVVPVQSKQR--FGGELGTE--------- 274
S + PP ++ + E+ N + +V +S FG E
Sbjct: 117 ESVDSSLRKPPDIEGRECHEQTRHEEQENNKQLVQAESDDSDDFGSRAFEEIEEQESDVL 176
Query: 275 SATSAAPSRYSALLQLIACGS---SGPEFKARTQQEPRVSSV--GTTASKQRQSVDRRDQ 329
TS + + + + G G E R +E ++S + G T+ +++ V + D+
Sbjct: 177 DRTSTSGAMEKEMTDDVGDGLRKVQGIEEPERHNEESKISEMVDGETSGHEKKKVVKMDK 236
Query: 330 KAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAE----EKEYFSGSLVEMESMKAN 385
K + + +EE + R + + ++G+ + AE ++E+ S L E K N
Sbjct: 237 K----NRDVKEEVDGAMGEGFRPNIDRTQVVGDDEIAETEKNDEEFESDKLEADEVDKIN 292
Query: 386 HPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEK 428
+ ++E+R +KL+++ E+ ++E+KG KE+
Sbjct: 293 EGGNTKVRRHSEDRNL------IKLQEKEEQHSKEQKGHSKEE 329
>At1g76720 putative translation initiation factor IF-2
Length = 1224
Score = 38.1 bits (87), Expect = 0.010
Identities = 67/310 (21%), Positives = 119/310 (37%), Gaps = 46/310 (14%)
Query: 144 PETNYGYNAKSKVLSNRQQQKEA--EEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEEL 201
P T G +S S + K A +E E EE E T S S + L
Sbjct: 175 PITFSGKKTRSSKSSKKTTNKVALPDEEEGTMGDEESLEITFSGKKKGS-----IVLASL 229
Query: 202 GED---EDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVV 258
G+D ++ +A +TK+ +V+ T + + KN + A +L E+
Sbjct: 230 GDDSVADETSQAKTPDTKSVEVIETGKIKKK---------KKNKNKVARTLEEE------ 274
Query: 259 VPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTAS 318
+ ELG A S + ++ A + ++E +V T A+
Sbjct: 275 ---DDLDKLLAELGETPAAERPASSTPEVEKVQAQPGPVAPVENAGEKEGEKETVETAAA 331
Query: 319 KQRQSVDRRDQK-------AFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEY 371
K+++ +D++ + +EE++EE+V + P+ + AE+K
Sbjct: 332 KKKKKKKEKDKEKKAAAAATSSVEAKEEKQEESVTEPLQ-----PKKKDAKGKAAEKK-- 384
Query: 372 FSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEME----EENRERKGGVKE 427
+ EM+ A + EE + + EE + ++E+E E R+RK KE
Sbjct: 385 IPKHVREMQEALARRQEAEERKKKEEEEKLRKEEEERRRQEELEAQAEEAKRKRKEKEKE 444
Query: 428 KCIPIKKSSK 437
K + K K
Sbjct: 445 KLLRKKLEGK 454
Score = 29.6 bits (65), Expect = 3.5
Identities = 68/350 (19%), Positives = 127/350 (35%), Gaps = 54/350 (15%)
Query: 133 NVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEE-EKEFTSSTTTPHSR 191
N K G E ET K K +++++ A E +EE ++E + P +
Sbjct: 315 NAGEKEGEKETVETAAAKKKKKKKEKDKEKKAAAAATSSVEAKEEKQEESVTEPLQPKKK 374
Query: 192 CSRGVSTEE-----LGEDED----RHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNH 242
++G + E+ + E ++ R EA + K E+ + R +
Sbjct: 375 DAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEEEKLRKEEEERRR--------QEE 426
Query: 243 SAAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKA 302
A A A++ + +++ G+L T + A R + QL+A G P A
Sbjct: 427 LEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAKQKTEAQKREAFKNQLLAAGRGLP--VA 484
Query: 303 RTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLM--- 359
+ S A+K++ S + + + + E E +E A + SE+ +
Sbjct: 485 DDVGDATSSKRPIYANKKKPSRQKGNDTSVQVEDEVEPQENHAATLGEVGSEDTEKVDLL 544
Query: 360 -------------LGNLQNAEEKEYFSGSLVEMESMKAN------HPVLKRS----NSYN 396
+ EE + + + +K + PV+K+ +S +
Sbjct: 545 ESANTGEKSGPADVAQENGVEEDDEWDAKSWDNVDLKIDDKEEEAQPVVKKELKAHDSDH 604
Query: 397 EERR--------SKLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKE 438
E + SKL VK E+E+ + K K KC+ + KE
Sbjct: 605 ETEKPTAKPAGMSKLTTGAVKAISEVEDAATQTKRAKKGKCLAPSEFIKE 654
>At3g56570 putative protein
Length = 531
Score = 37.7 bits (86), Expect = 0.013
Identities = 41/148 (27%), Positives = 62/148 (41%), Gaps = 25/148 (16%)
Query: 145 ETNYGYNAKSKVLSNRQQQKEA-------EEYEEYEEQEEEKEFTSSTTTPHSRCSRGVS 197
ET SK+ S+ +Q E +E +E EE+EEE+E G
Sbjct: 225 ETTDEDEPSSKISSSPEQSFEEVPGENTDDEAKEEEEEEEEEE-------------EGEE 271
Query: 198 TEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRV 257
EE E+E+ LQN+ K+++ +VS+ V N M N A+L +
Sbjct: 272 EEEGEEEEENSSMLQNDQSGLKMIMVKDVSAGAEVFNTYGLMGN-----AALLHRYGFTE 326
Query: 258 VVPVQSKQRFGGELGTESATSAAPSRYS 285
+ EL TE +TS+ SRY+
Sbjct: 327 LDNPYDIVNIDLELVTEWSTSSFTSRYT 354
>At4g39040 unknown protein
Length = 296
Score = 37.4 bits (85), Expect = 0.017
Identities = 30/94 (31%), Positives = 44/94 (45%), Gaps = 11/94 (11%)
Query: 166 AEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNN 225
AEEYE YE++EEE E S ++EE+ E D+ E +N K +
Sbjct: 94 AEEYE-YEDEEEEDEQDSGVVVSERGIEDSEASEEVSEIGDKEEKTENTKKKK------- 145
Query: 226 VSSRGNVTNVPPPMKNHSAA-AASLAEKLNRRVV 258
SRG+ + K A+ A SL +KL ++V
Sbjct: 146 --SRGSALKLSIKEKKELASYAHSLGDKLKCQLV 177
>At3g53440 unknown protein
Length = 512
Score = 37.4 bits (85), Expect = 0.017
Identities = 35/136 (25%), Positives = 57/136 (41%), Gaps = 20/136 (14%)
Query: 305 QQEPRVSSVG-TTASKQRQSVDRRDQKAFMMSCEEEE-EEEAVA---------MMMNRVS 353
++ PR SS G + + S+ ++ + + SC + E V +RV
Sbjct: 126 RRSPRFSSGGGESVDQSSSSIGKKSGNSVLSSCSTKSISSEKVKGGDGLKSKDRSRSRVR 185
Query: 354 ENPRLMLGNLQ-NAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKK 412
+ M N EE+E S L E + M+ P NEE ++ +EVK K
Sbjct: 186 PKTKKMFSGCDDNEEEEEEVSVCLTERKRMRIAKP--------NEEENAEAKADEVKRKG 237
Query: 413 EMEEENRERKGGVKEK 428
+ EEE + + G+K K
Sbjct: 238 KEEEEEEDEEEGLKAK 253
>At2g26570 unknown protein
Length = 807
Score = 36.2 bits (82), Expect = 0.037
Identities = 68/279 (24%), Positives = 108/279 (38%), Gaps = 43/279 (15%)
Query: 174 EQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVT 233
E E+EK +S S V++ E D R E +K + +R +
Sbjct: 496 ELEKEKSTLASIKQREGMASIAVASIEAEIDRTRSEIASVQSKEK--------DAREKMV 547
Query: 234 NVPPPMKN---HSAAAASLAEKLNRRV-VVPVQSKQRFGGELGTESATSAAP-------- 281
+P ++ + A SLAE + +++Q G ES AA
Sbjct: 548 ELPKQLQQAAEEADEAKSLAEVAREELRKAKEEAEQAKAGASTMESRLFAAQKEIEAAKA 607
Query: 282 SRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEE 341
S AL + A S KA PR SV T + ++ + +R +A EE
Sbjct: 608 SERLALAAIKALEESESTLKANDTDSPR--SV-TLSLEEYYELSKRAHEA------EELA 658
Query: 342 EEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEE-RR 400
VA ++R+ E + +L+ EE M A LK + E+ +
Sbjct: 659 NARVAAAVSRIEEAKETEMRSLEKLEEVN---------RDMDARKKALKEATEKAEKAKE 709
Query: 401 SKLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKES 439
KLGVE+ +L+K E ++RK G + +K+ KES
Sbjct: 710 GKLGVEQ-ELRKWRAEHEQKRKAG---DGVNTEKNLKES 744
>At1g75190 unknown protein
Length = 131
Score = 36.2 bits (82), Expect = 0.037
Identities = 26/82 (31%), Positives = 38/82 (45%), Gaps = 5/82 (6%)
Query: 160 RQQQKEAEEYEEYEEQEEEKEFTSSTTTPH--SRCSRGVSTEELGEDEDRHEALQNNTKT 217
+++ +E E+YEEYE++EEE E + R S E + +HE L+ K
Sbjct: 46 KEESEEDEDYEEYEDEEEEDEEAEVVINREKLKKKVRSSSGSMEKEQKMKHEELEEEEKP 105
Query: 218 E---KVVVTNNVSSRGNVTNVP 236
E KVVV S R + P
Sbjct: 106 EMTVKVVVPPPSSRRSRRKSTP 127
>At1g50930 hypothetical protein
Length = 196
Score = 36.2 bits (82), Expect = 0.037
Identities = 22/104 (21%), Positives = 50/104 (47%), Gaps = 3/104 (2%)
Query: 126 SERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSST 185
S+ ++ + N + N N K + + ++ +++E EE ++ EE+EE++E T+S+
Sbjct: 65 SDAASPVHTTKINNVVRRKANNINTNPKKRRIIHQHKEEEEEELQKGEEEEEDEEDTASS 124
Query: 186 TTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSR 229
S ++ S + D R+ +N +E++ S+
Sbjct: 125 P---SNKTKIFSVLDHANDNTRYGKTMDNVTSEEIGCITETGSK 165
>At4g21360 putative transposable element
Length = 1308
Score = 35.0 bits (79), Expect = 0.083
Identities = 29/104 (27%), Positives = 46/104 (43%), Gaps = 16/104 (15%)
Query: 158 SNRQQQKEAEEYEEYEE-QEEEKEFTSSTTTPHSRCSR--GVSTEELGEDEDRHEALQNN 214
S+ Q K+ + E Y E Q KE H CS+ GV+ EE+ ++ D ++
Sbjct: 720 SSEQSVKDISDLEGYNEFQVSVKE--------HGECSKTGGVTIEEIDQESDSENSVTQE 771
Query: 215 TKTEKVVVTNNVSSRGNVTNVP-PPMK----NHSAAAASLAEKL 253
+ ++N S+R P PP K H A A +AE++
Sbjct: 772 PLIASIDLSNYQSARDRERRAPNPPQKLADYTHFALALVMAEEI 815
>At1g01660 hypothetical protein
Length = 568
Score = 35.0 bits (79), Expect = 0.083
Identities = 41/158 (25%), Positives = 72/158 (44%), Gaps = 30/158 (18%)
Query: 283 RYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTA-SKQRQSVDRRDQKAFMMSCEEEEE 341
R+ A + S E++A +E ++ G A +KQR+ V++ Q E
Sbjct: 251 RFKAENTAVEAIRSAREYEAMYNEEAKLRKEGKEALAKQRKMVEKTKQ----------ER 300
Query: 342 EEAVAMMMN-RVSENPRL--------MLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRS 392
++A+ +++N R N L MLG +EKE + E+E ++A +++
Sbjct: 301 DDALIIILNGRKLYNEELRRRVEAEEMLG-----KEKEEHERTKKEIEEVRA---IVQDG 352
Query: 393 NSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCI 430
YNE+ R + +EE K EEE + K +E C+
Sbjct: 353 TLYNEQLRHRKEMEE--SMKRQEEELEKTKKEKEEACM 388
>At5g55660 putative protein
Length = 759
Score = 34.7 bits (78), Expect = 0.11
Identities = 55/273 (20%), Positives = 103/273 (37%), Gaps = 43/273 (15%)
Query: 161 QQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKV 220
+++KE +E EE E++ EE+E + P E E+ + E + TK +K
Sbjct: 511 EEEKEDDEEEEKEQEVEEEEEENENGIPDKSEDEAPQLSESEENVESEEESEEETKKKK- 569
Query: 221 VVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQS--------KQRFGGELG 272
RG+ T+ S S + +++ VP +S K+ G
Sbjct: 570 --------RGSRTS--------SDKKESAGKSRSKKTAVPTKSSPPKKATQKRSAGKRKK 613
Query: 273 TESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAF 332
++ + +P +S K + + ++ + SK++ + +R K
Sbjct: 614 SDDDSDTSPK------------ASSKRKKTEKPAKEQAAAPLKSVSKEKPVIGKRGGKGK 661
Query: 333 MMSCEEEEEE--EAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLK 390
+ E +EE A+ ++ V N L+ + K F+ SL +S +
Sbjct: 662 DKNKEPSDEELKTAIIDILKGVDFNTATFTDILKRLDAK--FNISLASKKS-SIKRMIQD 718
Query: 391 RSNSYNEERRSKLGVEEVKLKKEMEEENRERKG 423
+E + G EE + E EEE + KG
Sbjct: 719 ELTKLADEAEDEEG-EEEDAEHEEEEEKEKAKG 750
Score = 33.9 bits (76), Expect = 0.18
Identities = 35/149 (23%), Positives = 62/149 (41%), Gaps = 17/149 (11%)
Query: 292 ACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNR 351
A GSS + A++Q++ ++ S + ++K E++EEEE +
Sbjct: 476 AAGSSSSKRSAKSQKKTEEATRTNKKSVAHSDDESEEEK------EDDEEEEKEQEVEEE 529
Query: 352 VSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLK 411
EN + ++ +E S S +ES S E ++ K G K
Sbjct: 530 EEENENGIPD--KSEDEAPQLSESEENVES---------EEESEEETKKKKRGSRTSSDK 578
Query: 412 KEMEEENRERKGGVKEKCIPIKKSSKESS 440
KE ++R +K V K P KK++++ S
Sbjct: 579 KESAGKSRSKKTAVPTKSSPPKKATQKRS 607
Score = 33.5 bits (75), Expect = 0.24
Identities = 71/368 (19%), Positives = 124/368 (33%), Gaps = 65/368 (17%)
Query: 119 SELVEGCSE--RFQQLNVSNKNGIHEPPETNYGYNAK-----------SKVLSNRQQQKE 165
+E VE E + + L N+ + E ETN G K +KV + K+
Sbjct: 180 AEKVENVDEDDKEEALKEKNEAELAEEEETNKGEEVKEANKEDDVEADTKVAEPEVEDKK 239
Query: 166 AEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNN 225
E +E E++EEEKE S + E++ + R + T+ + T +
Sbjct: 240 TESKDENEDKEEEKEDEKEDEKEESNDDKEDKKEDIKKSNKRGKGKTEKTRGK----TKS 295
Query: 226 VSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPS--- 282
+ ++ P + E+L VV S + F E G + P+
Sbjct: 296 DEEKKDIEPKTPFFSDRPVRERKSVERL--VAVVDKDSSREFHVEKGKGTPLKDIPNVAY 353
Query: 283 ------RYSALLQLIACGSSGPEFKARTQQEPRV---------SSVGTTASKQRQSVDRR 327
QL G KA TQ + + K ++ ++
Sbjct: 354 KVSRKKSDEVFKQLHTILFGGKRVKA-TQLKAHILRFSGYKWQGDEEKAKLKVKEKFEKI 412
Query: 328 DQKAFMMSCE--------EEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEM 379
+++ + C+ ++E + + E P L N +EK +
Sbjct: 413 NKEKLLEFCDLFDISVAKATTKKEDIVTKLVEFLEKPHATTDVLVNEKEKGVKRKRTPKK 472
Query: 380 ESMKANHPVLKRS-------------------NSYNEERRSKLGVEEVKLKKEMEEENRE 420
S A KRS +S +E K EE + ++E+EEE E
Sbjct: 473 SSPAAGSSSSKRSAKSQKKTEEATRTNKKSVAHSDDESEEEKEDDEEEEKEQEVEEEEEE 532
Query: 421 RKGGVKEK 428
+ G+ +K
Sbjct: 533 NENGIPDK 540
Score = 32.7 bits (73), Expect = 0.41
Identities = 34/122 (27%), Positives = 50/122 (40%), Gaps = 13/122 (10%)
Query: 142 EPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKE------FTSSTTTPH-SRCSR 194
E TN A S S +++ + EE +E E +EEE+E S P S
Sbjct: 494 EATRTNKKSVAHSDDESEEEKEDDEEEEKEQEVEEEEEENENGIPDKSEDEAPQLSESEE 553
Query: 195 GVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVP----PPMKNHSAAAASLA 250
V +EE E+E + + + T ++K SR T VP PP K +A
Sbjct: 554 NVESEEESEEETKKKKRGSRTSSDKKESAG--KSRSKKTAVPTKSSPPKKATQKRSAGKR 611
Query: 251 EK 252
+K
Sbjct: 612 KK 613
>At5g40340 unknown protein
Length = 1008
Score = 34.7 bits (78), Expect = 0.11
Identities = 30/122 (24%), Positives = 50/122 (40%), Gaps = 5/122 (4%)
Query: 98 FDLALKDVIYPSEGGEYVLKGSELVEGCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVL 157
+D + K+ SE G+ V KG E + + Q+ G E +G K+
Sbjct: 675 YDSSDKEKEELSEMGKPVTKGKEKKDKKGKAKQKAEEIEVTGKEENETDKHG-----KMK 729
Query: 158 SNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKT 217
R+++K + E E +E +KE ST R + GE+E + E ++ K
Sbjct: 730 KERKRKKSESKKEGGEGEETQKEANESTKKERKRKKSESKKQSDGEEETQKEPSESTKKE 789
Query: 218 EK 219
K
Sbjct: 790 RK 791
Score = 29.3 bits (64), Expect = 4.6
Identities = 27/90 (30%), Positives = 47/90 (52%), Gaps = 12/90 (13%)
Query: 109 SEGGEYVLKGSELVEGCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEE 168
S+ GE K S++V E+ ++ N+ N++G+ E N G ++ K+ + ++++E EE
Sbjct: 43 SDNGE---KKSDVVVDVDEKNEK-NL-NESGVIEDCVMN-GVSSLLKLKEDVEEEEEEEE 96
Query: 169 YEEYEEQ------EEEKEFTSSTTTPHSRC 192
EE EE+ EEE+E H C
Sbjct: 97 EEEEEEEDGEDEEEEEEEEEEEEEEEHGYC 126
Score = 28.5 bits (62), Expect = 7.8
Identities = 24/110 (21%), Positives = 41/110 (36%)
Query: 110 EGGEYVLKGSELVEGCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEY 169
EGGE E E + ++ +K ET + +K R+ + ++
Sbjct: 742 EGGEGEETQKEANESTKKERKRKKSESKKQSDGEEETQKEPSESTKKERKRKNPESKKKA 801
Query: 170 EEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEK 219
E EE+E KE ST R EE+ + ++ E + + K
Sbjct: 802 EAVEEEETRKESVESTKKERKRKKPKHDEEEVPNETEKPEKKKKKKREGK 851
>At3g54670 structural maintenance of chromosomes (SMC) - like
protein
Length = 1265
Score = 34.7 bits (78), Expect = 0.11
Identities = 39/148 (26%), Positives = 66/148 (44%), Gaps = 17/148 (11%)
Query: 46 LEHPHFMELTLLPNQPLRLKDVFDRLMVLRGSG--------MPLQYSWSSK-RNYKSGYV 96
L+ +T +P Q +R+K VF+RL L G+ PL S SK + Y
Sbjct: 573 LKEQRLPPMTFIPLQSVRVKQVFERLRNLGGTAKLVFDVIQYPLHISEVSKIYIFVLNYS 632
Query: 97 WFDLALKDVIYPSEGGEYV---LKGSELVEGCSERFQQLNVS----NKNGIHEPPETNYG 149
FD L+ + + G V L+ ++++ ERF+ + V K G T+ G
Sbjct: 633 TFDPELEKAVLYAVGNTLVCDELEEAKVLSWSGERFKVVTVDGILLTKAGT-MTGGTSGG 691
Query: 150 YNAKSKVLSNRQQQKEAEEYEEYEEQEE 177
AKS +++ + + E++E+Q E
Sbjct: 692 MEAKSNKWDDKKIEGLKKNKEDFEQQLE 719
Score = 29.3 bits (64), Expect = 4.6
Identities = 39/168 (23%), Positives = 75/168 (44%), Gaps = 12/168 (7%)
Query: 257 VVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTT 316
++V ++ F G++ ES S P + LL+ I+ GS + + +E + S+
Sbjct: 136 ILVKARNFLVFQGDV--ESIASKNPKELTGLLEEIS-GSEELKKEYEGLEEKKASAEEKA 192
Query: 317 ASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLM--LGNLQNAEEKEYFSG 374
A ++ ++K + +EE E+ + + R + L N++N EK
Sbjct: 193 ALIYQKKKTIGNEKKLKKAQKEEAEKHLRLQEELKALKRERFLWQLYNIENDIEK----- 247
Query: 375 SLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERK 422
+ +++S K+N + R E K VE+ K KE+ + RE+K
Sbjct: 248 ANEDVDSEKSNRKDVMRELEKFEREAGKRKVEQAKYLKEIAQ--REKK 293
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 34.7 bits (78), Expect = 0.11
Identities = 27/122 (22%), Positives = 48/122 (39%), Gaps = 1/122 (0%)
Query: 299 EFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEE-EEEEAVAMMMNRVSENPR 357
E R +QE + K R+ +R+ ++ E+E +++E M + E R
Sbjct: 566 EMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKKEREEMERKKREEEAR 625
Query: 358 LMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEE 417
+ E+E ++E + ++R E + EE + KKE EEE
Sbjct: 626 KREEEMAKIREEERQRKEREDVERKRREEEAMRREEERKREEEAAKRAEEERRKKEEEEE 685
Query: 418 NR 419
R
Sbjct: 686 KR 687
Score = 33.9 bits (76), Expect = 0.18
Identities = 33/150 (22%), Positives = 61/150 (40%), Gaps = 10/150 (6%)
Query: 299 EFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEE------EEEEAVAMMMNRV 352
E K R ++E +++R+ RR+++ EEE EEEEA R
Sbjct: 426 ERKRREEEEIERRRKEEEEARKREEAKRREEEEAKRREEEETERKKREEEEARKREEERK 485
Query: 353 SENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKK 412
E + + +E + E + + K+ EE R + EEV+ K+
Sbjct: 486 REEEEAKRREEERKKREEEAEQARKREEEREKEEEMAKK----REEERQRKEREEVERKR 541
Query: 413 EMEEENRERKGGVKEKCIPIKKSSKESSRR 442
E+E + R+ +++ K+ + + RR
Sbjct: 542 REEQERKRREEEARKREEERKREEEMAKRR 571
Score = 30.8 bits (68), Expect = 1.6
Identities = 33/154 (21%), Positives = 61/154 (39%), Gaps = 11/154 (7%)
Query: 299 EFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRL 358
E +AR ++E R A ++ + +R+++A EEE E+ M R E R
Sbjct: 474 EEEARKREEERKREE-EEAKRREEERKKREEEAEQARKREEEREKEEEMAKKREEERQRK 532
Query: 359 MLGNLQNAEEKEYFSGSLVE----------MESMKANHPVLKRSNSYNEERRSKLGVEEV 408
++ +E E E A +R EE K+ E+
Sbjct: 533 EREEVERKRREEQERKRREEEARKREEERKREEEMAKRREQERQRKEREEVERKIREEQE 592
Query: 409 KLKKEMEEENRERKGGVKEKCIPIKKSSKESSRR 442
+ ++E + RE++ KE+ +K +E +R+
Sbjct: 593 RKREEEMAKRREQERQKKEREEMERKKREEEARK 626
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,121,835
Number of Sequences: 26719
Number of extensions: 456197
Number of successful extensions: 3464
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 128
Number of HSP's that attempted gapping in prelim test: 2678
Number of HSP's gapped (non-prelim): 507
length of query: 443
length of database: 11,318,596
effective HSP length: 102
effective length of query: 341
effective length of database: 8,593,258
effective search space: 2930300978
effective search space used: 2930300978
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0307.3


