

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0307.15
(326 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
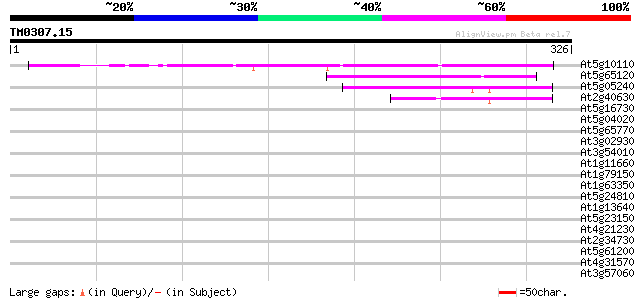
Score E
Sequences producing significant alignments: (bits) Value
At5g10110 unknown protein 125 4e-29
At5g65120 unknown protein 99 3e-21
At5g05240 unknown protein 49 3e-06
At2g40630 unknown protein 42 5e-04
At5g16730 putative protein 38 0.007
At5g04020 pathogen-induced calmodulin-binding protein (PICBP) 33 0.16
At5g65770 nuclear matrix constituent protein 1 (NMCP1)-like 33 0.27
At3g02930 unknown protein 32 0.36
At3g54010 pasticcino 1-A (PAS1-A) 31 0.80
At1g11660 heat-shock protein like 31 1.0
At1g79150 hypothetical protein 30 1.4
At1g63350 disease resistance protein, putative 30 1.4
At5g24810 unknown protein 30 2.3
At1g13640 unknown protein 30 2.3
At5g23150 transcription factor-like protein (gb|AAD31171.1) 29 3.0
At4g21230 receptor kinase - like protein 29 3.0
At2g34730 putative myosin heavy chain 29 3.0
At5g61200 putative protein 28 6.8
At4g31570 putative protein 28 6.8
At3g57060 putative protein 28 6.8
>At5g10110 unknown protein
Length = 321
Score = 125 bits (313), Expect = 4e-29
Identities = 113/330 (34%), Positives = 155/330 (46%), Gaps = 54/330 (16%)
Query: 12 VKPQPRPVLSDVTNLRAKRPFSSISADDGDSRFTKKKVQSKLGVQAQQGRSNKDKVVVSQ 71
V Q RPVLSD+TNL KR SSI D D + G++ +V S+
Sbjct: 9 VSEQTRPVLSDLTNLPKKRGISSILGDLLD----------------ESGKTVAHEV--SK 50
Query: 72 QK-GQNPCLELPFCGDDSLGNVSSQESNLQLPSGGLE-EKNLLGGAVVSGPTEVPVAVLG 129
+K + CL + D L V + L G +KN G AV V
Sbjct: 51 EKFSKRLCLVV-----DDL--VKENATPLDTIQGSCSFDKNTSGDAVDKEEYHDAVMEFS 103
Query: 130 EIGQGRPLNEG-----FIDRGIESGQRDVRAVDNLGSPKC-GGRAVELPTMSDSCDSRFP 183
G + L E + + G G R++ A N GG + L ++ +SR
Sbjct: 104 S-GDCKALKESKSIQIYFEPGDRDGARELNAAANADQTDVTGGEGLALSLLTSDTESRNL 162
Query: 184 ----------------GLERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHY 227
+ RCS ++ N + DLL+SC CSFC A++IWSDL+Y
Sbjct: 163 LKTGKELSDCQNLRSFEMSRCSNVK-NKEHVNLNTGDDLLRSCCCSFCLTASYIWSDLNY 221
Query: 228 QDVKGRLSALKKSQKDASNAVQKLSEIKD-SVMPDQQSSSESSKLELALMHQWKSLFDFM 286
QD KGRLSA+KKSQK ASN +Q +K+ S +S S+K E LM QW+SLF M
Sbjct: 222 QDSKGRLSAMKKSQKAASNLIQ--GNVKERSTDFHATGNSVSAKQESKLMAQWRSLFLSM 279
Query: 287 ENTFAEESRQLESSFETLKDLRDNCKNDLD 316
+ +EES L++SF +K LRD+CK DL+
Sbjct: 280 GDILSEESSHLQNSFVRMKKLRDDCKMDLE 309
>At5g65120 unknown protein
Length = 324
Score = 99.0 bits (245), Expect = 3e-21
Identities = 55/122 (45%), Positives = 72/122 (58%), Gaps = 1/122 (0%)
Query: 185 LERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDA 244
+ RCS + G A LKSC+CSFC AA+IWSDLHYQD+KGRLS LKKSQK+A
Sbjct: 176 MNRCSNVDGMGIVNHHMEADGELKSCSCSFCLTAAYIWSDLHYQDIKGRLSVLKKSQKEA 235
Query: 245 SNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETL 304
S +Q+ + + ++S+ S+ + M QW SLF ME A ES L SS + +
Sbjct: 236 SGLIQRNDRGTPTDIYGSENSNNSTNTDNP-MEQWTSLFRNMEGILARESNHLVSSSDLI 294
Query: 305 KD 306
D
Sbjct: 295 FD 296
>At5g05240 unknown protein
Length = 530
Score = 49.3 bits (116), Expect = 3e-06
Identities = 37/132 (28%), Positives = 64/132 (48%), Gaps = 10/132 (7%)
Query: 194 NAGPTSAAAAADLLKSCTCSF-CSKAAHIWSDLHYQDVKGRLSALKKSQKDASNAVQ-KL 251
N S+ A+DL + + + A W +L +QD+KGR+SAL++S+K V +L
Sbjct: 344 NDSSLSSEDASDLNSASVLTVNAATVASQWLELLHQDIKGRVSALRRSRKRVRAVVTIEL 403
Query: 252 SEIKDSVMPDQQSSSE------SSKLELALMH--QWKSLFDFMENTFAEESRQLESSFET 303
+ P Q + +SK +H +W +LF +E+ +EE QLES
Sbjct: 404 PHLIRKEFPADQENDPTLLLGGASKASTVDIHKSRWMTLFKQLEHKLSEEESQLESWLNQ 463
Query: 304 LKDLRDNCKNDL 315
++ ++ +C L
Sbjct: 464 VRYMQSHCDEGL 475
>At2g40630 unknown protein
Length = 535
Score = 42.0 bits (97), Expect = 5e-04
Identities = 26/111 (23%), Positives = 50/111 (44%), Gaps = 19/111 (17%)
Query: 222 WSDLHYQDVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMH---- 277
W + QD+ GRLSA++ S+ N + +E+ + SS++++ LE+ +
Sbjct: 372 WLEFLQQDLSGRLSAVQDSRNRVQNIL--TTELPLLASSRESSSNQANSLEMVTTNTSGD 429
Query: 278 -------------QWKSLFDFMENTFAEESRQLESSFETLKDLRDNCKNDL 315
+W + FD + +E R LE S +K+++ C + L
Sbjct: 430 ASSDKAATETHQKRWTAKFDQINKALYDEQRDLERSLNQVKEMQSRCNHGL 480
>At5g16730 putative protein
Length = 853
Score = 38.1 bits (87), Expect = 0.007
Identities = 22/91 (24%), Positives = 47/91 (51%), Gaps = 1/91 (1%)
Query: 233 RLSALKKSQKDASNAVQKLSEIKDSVMPD-QQSSSESSKLELALMHQWKSLFDFMENTFA 291
RLS +K+ K A+ + L + K + + +Q+ E+ ++ L L K+ EN+
Sbjct: 99 RLSQIKEDLKKANERISSLEKDKAKALDELKQAKKEAEQVTLKLDDALKAQKHVEENSEI 158
Query: 292 EESRQLESSFETLKDLRDNCKNDLDSTDNTH 322
E+ + +E+ E +++ + K +L++ N H
Sbjct: 159 EKFQAVEAGIEAVQNNEEELKKELETVKNQH 189
Score = 32.7 bits (73), Expect = 0.27
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 9/78 (11%)
Query: 236 ALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLF--------DFME 287
ALKK Q DA++ VQ+LSE K ++ D +SS E + M S + E
Sbjct: 427 ALKKEQ-DATSRVQRLSEEKSKLLSDLESSKEEEEKSKKAMESLASALHEVSSEGRELKE 485
Query: 288 NTFAEESRQLESSFETLK 305
++ + E+ + LK
Sbjct: 486 KLLSQGDHEYETQIDDLK 503
>At5g04020 pathogen-induced calmodulin-binding protein (PICBP)
Length = 1495
Score = 33.5 bits (75), Expect = 0.16
Identities = 34/131 (25%), Positives = 58/131 (43%), Gaps = 21/131 (16%)
Query: 195 AGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDASNAVQKLSEI 254
+G T +AA+ S S S AAH ++ + + ++G+L L++ K+++
Sbjct: 1243 SGSTPGSAASSRNISRQSSISSMAAHYENEANAEIIRGKLRNLQEDLKESAKL------- 1295
Query: 255 KDSVMPDQQSSSESSKLELALMHQWKSLFDFMEN-----TFAEESRQLESSFETLKDLR- 308
D V D + + S L W+ L ME+ T EE+R+ E E +D
Sbjct: 1296 -DGVSKDLEEKQQCSSL-------WRILCKQMEDNEKNQTLPEETRKEEEEEELKEDTSV 1347
Query: 309 DNCKNDLDSTD 319
D K +L T+
Sbjct: 1348 DGEKMELYQTE 1358
>At5g65770 nuclear matrix constituent protein 1 (NMCP1)-like
Length = 1042
Score = 32.7 bits (73), Expect = 0.27
Identities = 28/98 (28%), Positives = 44/98 (44%), Gaps = 9/98 (9%)
Query: 233 RLSALKKSQKDASNAVQKLSEIKDSVMP---DQQSSSESSKLELALMH-QWKSLFDFMEN 288
+L ALK + S KL E D + + + ++ K+E A +W+ + D
Sbjct: 512 KLEALKSETSELSTLEMKLKEELDDLRAQKLEMLAEADRLKVEKAKFEAEWEHI-DVKRE 570
Query: 289 TFAEES----RQLESSFETLKDLRDNCKNDLDSTDNTH 322
+E+ RQ E+ LKD RDN K + D+ N H
Sbjct: 571 ELRKEAEYITRQREAFSMYLKDERDNIKEERDALRNQH 608
>At3g02930 unknown protein
Length = 806
Score = 32.3 bits (72), Expect = 0.36
Identities = 23/95 (24%), Positives = 45/95 (47%), Gaps = 10/95 (10%)
Query: 228 QDVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFME 287
+D+K + + + + A+ +L E + +++ S KL+ AL Q KSL E
Sbjct: 94 EDLKKANELIASLENEKAKALDQLKEAR------KEAEEASEKLDEALEAQKKSL----E 143
Query: 288 NTFAEESRQLESSFETLKDLRDNCKNDLDSTDNTH 322
N E+ +E+ E ++ + K +L++ N H
Sbjct: 144 NFEIEKFEVVEAGIEAVQRKEEELKKELENVKNQH 178
Score = 31.2 bits (69), Expect = 0.80
Identities = 21/66 (31%), Positives = 35/66 (52%), Gaps = 5/66 (7%)
Query: 233 RLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAE 292
+ ALKK Q DA+++VQ+L E K ++ + +SS E + M S + +
Sbjct: 413 KTQALKKEQ-DATSSVQRLLEEKKKILSELESSKEEEEKSKKAMESLASAL----HEVSS 467
Query: 293 ESRQLE 298
ESR+L+
Sbjct: 468 ESRELK 473
>At3g54010 pasticcino 1-A (PAS1-A)
Length = 635
Score = 31.2 bits (69), Expect = 0.80
Identities = 21/81 (25%), Positives = 39/81 (47%), Gaps = 13/81 (16%)
Query: 238 KKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQL 297
K S+ DA+ A+ KL + +Q++ S++ K Q+K LFD E ++
Sbjct: 514 KSSEADATAALLKLKQ------KEQEAESKARK-------QFKGLFDKRPGEITEVGSEI 560
Query: 298 ESSFETLKDLRDNCKNDLDST 318
+T++++ + ND D T
Sbjct: 561 REESKTIEEVDETKDNDDDET 581
>At1g11660 heat-shock protein like
Length = 773
Score = 30.8 bits (68), Expect = 1.0
Identities = 17/62 (27%), Positives = 34/62 (54%), Gaps = 2/62 (3%)
Query: 249 QKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETLKDLR 308
+ L E ++ + D SE++ +E ++ K L D +EN F + ++++S + LK +
Sbjct: 624 RNLQETEEWLYEDGDDESENAYIEK--LNDVKKLIDPIENRFKDGEERVQASKDLLKTIA 681
Query: 309 DN 310
DN
Sbjct: 682 DN 683
>At1g79150 hypothetical protein
Length = 495
Score = 30.4 bits (67), Expect = 1.4
Identities = 23/98 (23%), Positives = 49/98 (49%), Gaps = 8/98 (8%)
Query: 221 IWSDLHYQDVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKL-ELALMHQW 279
+ D H + R KKS+++A + ++ D ++ +++ + +++ L E+
Sbjct: 127 VLEDEHVLNKSQRREKAKKSKREAKKHEK---DLPDEILQEEEETPQAAVLAEVKEELSA 183
Query: 280 KSLFDFMENTFAEESRQL----ESSFETLKDLRDNCKN 313
+ F+ +N AE L E++ +TLKD+ D CK+
Sbjct: 184 EESFENKKNKIAELGMLLLSDPEANIKTLKDMLDICKD 221
>At1g63350 disease resistance protein, putative
Length = 535
Score = 30.4 bits (67), Expect = 1.4
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 12/89 (13%)
Query: 185 LERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDA 244
L R ++ A A+L + C C FCSK+ + + Y G+ LK +
Sbjct: 73 LNRVETIESRVNDLLNARNAELQRLCLCGFCSKS--LTTSYRY----GKSVFLKLRE--- 123
Query: 245 SNAVQKLSEIKDSVMPDQQSSSESSKLEL 273
V+KL V+ DQ S+SE + +L
Sbjct: 124 ---VEKLERRVFEVISDQASTSEVEEQQL 149
>At5g24810 unknown protein
Length = 1009
Score = 29.6 bits (65), Expect = 2.3
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 6/92 (6%)
Query: 235 SALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEES 294
S+L + + V K + +KD+ + +++ + HQ + L+D E F S
Sbjct: 804 SSLSQPPLGSHTHVPKFTSLKDTTKKKKGKEMAATEKGKSKDHQERKLYD--EKQFMSAS 861
Query: 295 RQLESSFETLKDLRDN----CKNDLDSTDNTH 322
ES+ E+L L D K +++S D+ H
Sbjct: 862 SSRESNTESLARLVDTNSSAGKTEINSDDHQH 893
>At1g13640 unknown protein
Length = 622
Score = 29.6 bits (65), Expect = 2.3
Identities = 14/45 (31%), Positives = 25/45 (55%)
Query: 245 SNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENT 289
SN L E+K+S+ +++ E +KL L+L S+ + + NT
Sbjct: 498 SNGRSSLGELKESIAEEEEDDEEEAKLTLSLSKLSTSMKNNLSNT 542
>At5g23150 transcription factor-like protein (gb|AAD31171.1)
Length = 1392
Score = 29.3 bits (64), Expect = 3.0
Identities = 21/69 (30%), Positives = 34/69 (48%), Gaps = 5/69 (7%)
Query: 253 EIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETLKDLRDNCK 312
EI PD Q+ + +K +L Q K++ FA+ Q+ ++FE L++ + N
Sbjct: 62 EIAFVAPPDIQAFTSEAKSKLLARCQGKTV-----KYFAQAVEQICTAFEGLQNHKSNAL 116
Query: 313 NDLDSTDNT 321
D DS D T
Sbjct: 117 GDEDSLDAT 125
>At4g21230 receptor kinase - like protein
Length = 597
Score = 29.3 bits (64), Expect = 3.0
Identities = 10/20 (50%), Positives = 16/20 (80%)
Query: 305 KDLRDNCKNDLDSTDNTHFD 324
K L++N +N+ +STD+ HFD
Sbjct: 303 KTLKENAENEFESTDSLHFD 322
>At2g34730 putative myosin heavy chain
Length = 829
Score = 29.3 bits (64), Expect = 3.0
Identities = 22/100 (22%), Positives = 44/100 (44%), Gaps = 11/100 (11%)
Query: 217 KAAHIWSDLHYQDVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALM 276
+A + S + VKG + + +++ A + L E+KD ++ SE+ + L
Sbjct: 634 EALSVVSARELEKVKGYETKISSLREELELARESLKEMKDEKRKTEEKLSETKAEKETLK 693
Query: 277 HQWKSL-----------FDFMENTFAEESRQLESSFETLK 305
Q SL FD +E AE++++ S + ++
Sbjct: 694 KQLVSLDLVVPPQLIKGFDILEGLIAEKTQKTNSRLKNMQ 733
>At5g61200 putative protein
Length = 389
Score = 28.1 bits (61), Expect = 6.8
Identities = 23/67 (34%), Positives = 35/67 (51%), Gaps = 5/67 (7%)
Query: 247 AVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSF-ETLK 305
+V+KL E S+ + Q ES KL++ + Q +LFD + F ES Q E +K
Sbjct: 165 SVEKLEESVSSLTLESQCEIESIKLDIVALEQ--ALFDAQK--FQGESIQENDKLREIVK 220
Query: 306 DLRDNCK 312
+LR N +
Sbjct: 221 ELRLNSR 227
>At4g31570 putative protein
Length = 2712
Score = 28.1 bits (61), Expect = 6.8
Identities = 19/94 (20%), Positives = 44/94 (46%)
Query: 227 YQDVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFM 286
Y+++K + L + + ++++QK+ ++ + S+E + LE+ + + D
Sbjct: 1142 YEELKQSFNTLFEKNEFTASSMQKVYADLTKLITESCGSAEMTSLEVENVAVFDPFRDGS 1201
Query: 287 ENTFAEESRQLESSFETLKDLRDNCKNDLDSTDN 320
E R++ S L+ + D ++DL S N
Sbjct: 1202 FENLLEAVRKILSERLELQSVIDKLQSDLSSKSN 1235
>At3g57060 putative protein
Length = 1439
Score = 28.1 bits (61), Expect = 6.8
Identities = 18/68 (26%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Query: 47 KKVQSKLGVQAQQGRSNKDKVVVSQQK-GQNPCLELPFCGDDSLGNVSSQESNLQLPSGG 105
++ ++K A + ++ ++ + +Q+ G N L L DD+L + ++ + ++ SGG
Sbjct: 857 RRQKTKKDKPAAESQNTEENLEATQENNGINAELGLA-ASDDALLDTLAERAEREIVSGG 915
Query: 106 LEEKNLLG 113
EKNL+G
Sbjct: 916 SVEKNLIG 923
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,457,466
Number of Sequences: 26719
Number of extensions: 320620
Number of successful extensions: 825
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 799
Number of HSP's gapped (non-prelim): 35
length of query: 326
length of database: 11,318,596
effective HSP length: 100
effective length of query: 226
effective length of database: 8,646,696
effective search space: 1954153296
effective search space used: 1954153296
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0307.15


