

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.5
(165 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
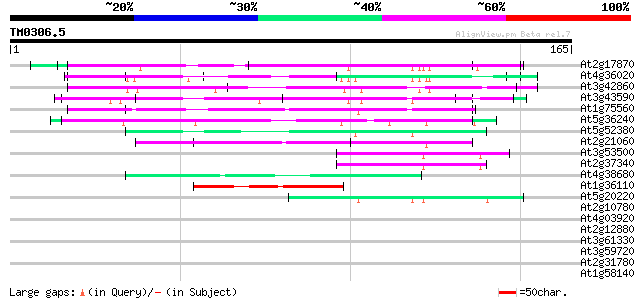
Score E
Sequences producing significant alignments: (bits) Value
At2g17870 putative glycine-rich, zinc-finger DNA-binding protein 88 2e-18
At4g36020 glycine-rich protein 73 6e-14
At3g42860 unknown protein (At3g42860) 68 2e-12
At3g43590 putative protein 63 8e-11
At1g75560 DNA-binding protein 63 8e-11
At5g36240 putative protein 62 1e-10
At5g52380 unknown protein 54 3e-08
At2g21060 glycine-rich protein (AtGRP2) 51 3e-07
At3g53500 splicing factor - like protein 44 3e-05
At2g37340 unknown protein 44 4e-05
At4g38680 glycine-rich protein 2 (GRP2) 44 5e-05
At1g36110 hypothetical protein 44 5e-05
At5g20220 zinc finger protein 42 1e-04
At2g10780 pseudogene 39 0.001
At4g03920 putative protein 38 0.003
At2g12880 pseudogene 37 0.004
At3g61330 copia-type polyprotein 37 0.006
At3g59720 copia-type reverse transcriptase-like protein 37 0.006
At2g31780 similar to Ariadne protein from Drosophila 37 0.006
At1g58140 hypothetical protein 37 0.006
>At2g17870 putative glycine-rich, zinc-finger DNA-binding protein
Length = 301
Score = 88.2 bits (217), Expect = 2e-18
Identities = 53/162 (32%), Positives = 67/162 (40%), Gaps = 38/162 (23%)
Query: 18 CFCCGKFGQFANNCSVIGPR-----------CFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
C+ CG G FA +C G C+ C GHLA +C+ GG ++ G
Sbjct: 132 CYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCR---GGSGGNRYGGG 188
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD--------GGPLCFNCGEIGHYASKCKI 118
GG G+ G CY C VGH+ C GG C+ CG +GH A C
Sbjct: 189 ---GGRGSGGDG----CYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTS 241
Query: 119 K--------GRLCFNCNQPGHFSRDC-KAPKGESSGNTGKGK 151
K GR C+ C GH +RDC + G S G G K
Sbjct: 242 KIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNK 283
Score = 79.7 bits (195), Expect = 6e-16
Identities = 51/162 (31%), Positives = 65/162 (39%), Gaps = 38/162 (23%)
Query: 15 DVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGN 74
+VT G + N+ G CF C GH+A +C GG S GG G
Sbjct: 73 EVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKS---------FGGGGG 123
Query: 75 KTSGKIVTCYRCKKVGHYTNKCPD-----------GGPLCFNCGEIGHYASKCK------ 117
+ SG CY C VGH+ C GG C++CGE+GH A C+
Sbjct: 124 RRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGGN 183
Query: 118 ---------IKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKG 150
G C+ C GHF+RDC+ G GN G G
Sbjct: 184 RYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGG---GNVGGG 222
Score = 57.8 bits (138), Expect = 3e-09
Identities = 40/148 (27%), Positives = 49/148 (33%), Gaps = 55/148 (37%)
Query: 7 GG*PIGDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
GG G C+ CG G FA +C + G
Sbjct: 187 GGGGRGSGGDGCYMCGGVGHFARDC------------------------------RQNGG 216
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD--------GGPLCFNCGEIGHYASKCKI 118
GG G+ TCY C VGH C GG C+ CG GH A C
Sbjct: 217 GNVGGGGS-------TCYTCGGVGHIAKVCTSKIPSGGGGGGRACYECGGTGHLARDCDR 269
Query: 119 KG----------RLCFNCNQPGHFSRDC 136
+G CF C + GHF+R+C
Sbjct: 270 RGSGSSGGGGGSNKCFICGKEGHFAREC 297
Score = 51.2 bits (121), Expect = 2e-07
Identities = 32/96 (33%), Positives = 39/96 (40%), Gaps = 16/96 (16%)
Query: 71 GTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIK----------G 120
G+ KT VT + N G CFNCGE+GH A C G
Sbjct: 64 GSDGKTKAIEVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGG 123
Query: 121 RL------CFNCNQPGHFSRDCKAPKGESSGNTGKG 150
R C+ C GHF+RDC+ G +SG G G
Sbjct: 124 RRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGG 159
Score = 35.8 bits (81), Expect = 0.011
Identities = 15/50 (30%), Positives = 23/50 (46%)
Query: 107 GEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAE 156
G + + + G CFNC + GH ++DC G S G G++ E
Sbjct: 80 GSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGE 129
Score = 32.3 bits (72), Expect = 0.12
Identities = 15/47 (31%), Positives = 21/47 (43%), Gaps = 10/47 (21%)
Query: 18 CFCCGKFGQFANNCSVIGP----------RCFKCNREGHLAVNCKTL 54
C+ CG G A +C G +CF C +EGH A C ++
Sbjct: 254 CYECGGTGHLARDCDRRGSGSSGGGGGSNKCFICGKEGHFARECTSV 300
>At4g36020 glycine-rich protein
Length = 299
Score = 73.2 bits (178), Expect = 6e-14
Identities = 46/158 (29%), Positives = 61/158 (38%), Gaps = 42/158 (26%)
Query: 18 CFCCGKFGQFANNCSVIG------------PRCFKCNREGHLAVNC--KTLQGGPSDSKM 63
C+ CG G FA +C+ G C+ C GH+A +C K++ G +
Sbjct: 134 CYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQKSVGNGDQRGAV 193
Query: 64 KGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD------------GGPLCFNCGEIGH 111
KG G G CY C VGH+ C G C++CG +GH
Sbjct: 194 KG----GNDG---------CYTCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCGGVGH 240
Query: 112 YASKCKIK---GRLCFNCNQPGHFSRDCKAPKGESSGN 146
A C K R C+ C GH +RDC GN
Sbjct: 241 IARDCATKRQPSRGCYQCGGSGHLARDCDQRGSGGGGN 278
Score = 60.8 bits (146), Expect = 3e-10
Identities = 37/142 (26%), Positives = 51/142 (35%), Gaps = 39/142 (27%)
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
G C+ C GH++ +C G GG G + S CY C GH+
Sbjct: 99 GSGCYNCGELGHISKDC-------------GIGGGGGGGERRSRGGEGCYNCGDTGHFAR 145
Query: 95 KCPDGG------------PLCFNCGEIGHYASKCK------------IKGRL--CFNCNQ 128
C G C+ CG++GH A C +KG C+ C
Sbjct: 146 DCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQKSVGNGDQRGAVKGGNDGCYTCGD 205
Query: 129 PGHFSRDCKAPKGESSGNTGKG 150
GHF+RDC + +G G
Sbjct: 206 VGHFARDCTQKVAAGNVRSGGG 227
Score = 56.6 bits (135), Expect = 6e-09
Identities = 33/122 (27%), Positives = 47/122 (38%), Gaps = 30/122 (24%)
Query: 58 PSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC------------PDGGPLCFN 105
P +K + + G G + G CY C ++GH + C GG C+N
Sbjct: 77 PGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGGGERRSRGGEGCYN 136
Query: 106 CGEIGHYASKCKIKGR------------LCFNCNQPGHFSRDCKAPKGESSGNTGKGKQL 153
CG+ GH+A C G C+ C GH +RDC + + G G Q
Sbjct: 137 CGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDC------TQKSVGNGDQR 190
Query: 154 AA 155
A
Sbjct: 191 GA 192
Score = 47.8 bits (112), Expect = 3e-06
Identities = 25/83 (30%), Positives = 36/83 (43%), Gaps = 21/83 (25%)
Query: 17 TCFCCGKFGQFANNCSV---IGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTG 73
TC+ CG G A +C+ C++C GHLA +C +GG G
Sbjct: 231 TCYSCGGVGHIARDCATKRQPSRGCYQCGGSGHLARDC-------------DQRGSGGGG 277
Query: 74 NKTSGKIVTCYRCKKVGHYTNKC 96
N + CY+C K GH+ +C
Sbjct: 278 NDNA-----CYKCGKEGHFAREC 295
Score = 35.4 bits (80), Expect = 0.014
Identities = 15/44 (34%), Positives = 21/44 (47%), Gaps = 7/44 (15%)
Query: 18 CFCCGKFGQFANNCSVIGP-------RCFKCNREGHLAVNCKTL 54
C+ CG G A +C G C+KC +EGH A C ++
Sbjct: 255 CYQCGGSGHLARDCDQRGSGGGGNDNACYKCGKEGHFARECSSV 298
Score = 29.3 bits (64), Expect = 1.0
Identities = 11/23 (47%), Positives = 13/23 (55%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVI 34
G D C+ CGK G FA CS +
Sbjct: 276 GGNDNACYKCGKEGHFARECSSV 298
>At3g42860 unknown protein (At3g42860)
Length = 372
Score = 68.2 bits (165), Expect = 2e-12
Identities = 43/146 (29%), Positives = 66/146 (44%), Gaps = 31/146 (21%)
Query: 18 CFCCGKFGQFANNCSV---IGP--------RCFKCNREGHLAVNCKTLQGGPS--DSKMK 64
C+ CGK G +A +C+V GP CFKC + GH + +C G P +MK
Sbjct: 239 CYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPGQMK 298
Query: 65 GNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP-DGGPLCFNCGEIGHYASKCKIKGRLC 123
+ ++G CY+C K GH++ C F G+ +S C
Sbjct: 299 SSSSSG-----------ECYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSSTGD-----C 342
Query: 124 FNCNQPGHFSRDCKAPKGESSGNTGK 149
+ C + GH+SRDC +P +++ GK
Sbjct: 343 YKCGKAGHWSRDCTSP-AQTTNTPGK 367
Score = 56.2 bits (134), Expect = 8e-09
Identities = 33/118 (27%), Positives = 51/118 (42%), Gaps = 29/118 (24%)
Query: 65 GNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP---DGGPL--------CFNCGEIGHYA 113
G+ T G N +G CY+C K GH+ C D GP+ CF CG+ GH++
Sbjct: 223 GDSDTRGYQNAKTG--TPCYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWS 280
Query: 114 SKCKIK--------GRL--------CFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAA 155
C + G++ C+ C + GH+SRDC +G+ K ++
Sbjct: 281 RDCTAQSGNPKYEPGQMKSSSSSGECYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSS 338
>At3g43590 putative protein
Length = 551
Score = 62.8 bits (151), Expect = 8e-11
Identities = 35/127 (27%), Positives = 52/127 (40%), Gaps = 18/127 (14%)
Query: 14 KDVTCFCCGKFGQFA----NNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDAT 69
KDV C+ C FG N C++C + GH + C G + ND+
Sbjct: 259 KDVQCYICKSFGHLCCVEPGNSLSWAVSCYRCGQLGHSGLAC-----GRHYEESNENDSA 313
Query: 70 GGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIKGRLCFNCNQP 129
S + CYRC + GH+ +CP+ + + G LC+ CN
Sbjct: 314 TPERLFNSREASECYRCGEEGHFARECPNSSSISTSHGRESQ---------TLCYRCNGS 364
Query: 130 GHFSRDC 136
GHF+R+C
Sbjct: 365 GHFAREC 371
Score = 58.9 bits (141), Expect = 1e-09
Identities = 35/110 (31%), Positives = 46/110 (41%), Gaps = 22/110 (20%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGT---GNKTSGKIVTCYRCKKVGHYTN 94
C+ C +GH + NC P+ +K + G+ G K K CY CKK GH
Sbjct: 168 CYSCGEQGHTSFNC------PTPTKRRKPCFICGSLEHGAKQCSKGHDCYICKKTGHRAK 221
Query: 95 KCPD------GGPLCFNCGEIGHYASKCK-------IKGRLCFNCNQPGH 131
CPD G +C CG+ GH CK +K C+ C GH
Sbjct: 222 DCPDKYKNGSKGAVCLRCGDFGHDMILCKYEYSKEDLKDVQCYICKSFGH 271
Score = 50.8 bits (120), Expect = 3e-07
Identities = 26/71 (36%), Positives = 37/71 (51%), Gaps = 6/71 (8%)
Query: 81 VTCYRCKKVGHYTNKCPDGGPL---CFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCK 137
V+CY C + GH + CP CF CG + H A +C KG C+ C + GH ++DC
Sbjct: 166 VSCYSCGEQGHTSFNCPTPTKRRKPCFICGSLEHGAKQCS-KGHDCYICKKTGHRAKDC- 223
Query: 138 APKGESSGNTG 148
P +G+ G
Sbjct: 224 -PDKYKNGSKG 233
Score = 40.8 bits (94), Expect = 3e-04
Identities = 36/157 (22%), Positives = 56/157 (34%), Gaps = 44/157 (28%)
Query: 16 VTCFCCGKFGQFANNC--------------------SVIGPRCFKCNREGHLAVNCKTLQ 55
V+C+ CG+ G C S C++C EGH A C
Sbjct: 285 VSCYRCGQLGHSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFAREC---- 340
Query: 56 GGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASK 115
P+ S + + G ++T CYRC GH+ +CP+ + E + K
Sbjct: 341 --PNSSSIS---TSHGRESQT-----LCYRCNGSGHFARECPNSSQVSKRDRETSTTSHK 390
Query: 116 CKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQ 152
+ K + + H ES+G T K K+
Sbjct: 391 SRKKNKENSEHDSTPH----------ESNGKTKKKKK 417
>At1g75560 DNA-binding protein
Length = 257
Score = 62.8 bits (151), Expect = 8e-11
Identities = 38/126 (30%), Positives = 55/126 (43%), Gaps = 15/126 (11%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C C + G FA +CS + C C GH+A C ++S+ G + S
Sbjct: 57 CNNCKRPGHFARDCSNVSV-CNNCGLPGHIAAECT------AESRCWNCREPGHVASNCS 109
Query: 78 GKIVTCYRCKKVGHYTNKCPDGGP------LCFNCGEIGHYASKCKIKGRLCFNCNQPGH 131
+ + C+ C K GH C + LC NC + GH A+ C + C NC GH
Sbjct: 110 NEGI-CHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQGHLAADCT-NDKACKNCRTSGH 167
Query: 132 FSRDCK 137
+RDC+
Sbjct: 168 IARDCR 173
Score = 51.6 bits (122), Expect = 2e-07
Identities = 30/102 (29%), Positives = 39/102 (37%), Gaps = 28/102 (27%)
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
G C C R GH A +C + C C GH
Sbjct: 54 GNLCNNCKRPGHFARDCSNVS--------------------------VCNNCGLPGHIAA 87
Query: 95 KCPDGGPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDC 136
+C C+NC E GH AS C +G +C +C + GH +RDC
Sbjct: 88 ECT-AESRCWNCREPGHVASNCSNEG-ICHSCGKSGHRARDC 127
Score = 36.2 bits (82), Expect = 0.008
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 2/38 (5%)
Query: 119 KGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAE 156
+G LC NC +PGHF+RDC N G +AAE
Sbjct: 53 QGNLCNNCKRPGHFARDCS--NVSVCNNCGLPGHIAAE 88
Score = 35.0 bits (79), Expect = 0.018
Identities = 24/91 (26%), Positives = 35/91 (38%), Gaps = 9/91 (9%)
Query: 15 DVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDA---TGG 71
D C C G A +C P C C+ GH+A +C SD + D GG
Sbjct: 156 DKACKNCRTSGHIARDCRN-DPVCNICSISGHVARHCPKGDSNYSDRGSRVRDGGMQRGG 214
Query: 72 TGNKTSGK-----IVTCYRCKKVGHYTNKCP 97
+ + ++ C+ C GH +CP
Sbjct: 215 LSRMSRDREGVSAMIICHNCGGRGHRAYECP 245
>At5g36240 putative protein
Length = 254
Score = 62.0 bits (149), Expect = 1e-10
Identities = 41/143 (28%), Positives = 62/143 (42%), Gaps = 31/143 (21%)
Query: 16 VTCFCCGKFGQFANNCS-----VIGPRCFKCNREGHLAVNCKT-----LQGGPSDSKMKG 65
V+C+ CG+ G C + P CF C REGH C S+ + +G
Sbjct: 77 VSCYRCGQLGHTGLACGRHYDDSVSPSCFICGREGHFEHQCHNSFSVCFPEDSSEDECQG 136
Query: 66 NDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIG--------HYASKCK 117
D++ + + R ++ GH+ ++CPD +CF EI + +SK
Sbjct: 137 PDSSSVRFQENT-------REEEEGHFEHQCPDSSSVCFQ--EISREEGFISLNSSSKST 187
Query: 118 IKGR----LCFNCNQPGHFSRDC 136
KGR LC+ C GH +RDC
Sbjct: 188 SKGRETRRLCYECKGKGHIARDC 210
Score = 48.9 bits (115), Expect = 1e-06
Identities = 39/144 (27%), Positives = 52/144 (36%), Gaps = 38/144 (26%)
Query: 13 DKDVTCFCCGKFGQFANNCS-------VIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKG 65
D+ C CG FG C + +C+ CN GHL ++ G
Sbjct: 23 DEAEVCLRCGGFGHDMTLCKYEYSHEDLKNIKCYVCNSLGHLCC----IEPG-------- 70
Query: 66 NDATGGTGNKTSGKIVTCYRCKKVGHYTNKC-----PDGGPLCFNCGEIGHYASKCKIKG 120
T V+CYRC ++GH C P CF CG GH+ +C
Sbjct: 71 ---------HTQSWTVSCYRCGQLGHTGLACGRHYDDSVSPSCFICGREGHFEHQCHNSF 121
Query: 121 RLCFNCNQPGHFSRD-CKAPKGES 143
+CF P S D C+ P S
Sbjct: 122 SVCF----PEDSSEDECQGPDSSS 141
>At5g52380 unknown protein
Length = 268
Score = 54.3 bits (129), Expect = 3e-08
Identities = 34/119 (28%), Positives = 47/119 (38%), Gaps = 33/119 (27%)
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
G CF C+ + H+A C P S+ + N C +C++ GH
Sbjct: 73 GEGCFICHSKTHIAKLC------PEKSEWERNKI--------------CLQCRRRGHSLK 112
Query: 95 KCPDGG------PLCFNCGEIGHYASKCK-------IKGRLCFNCNQPGHFSRDCKAPK 140
CP+ LC+NCG+ GH S C K CF C GH S++C K
Sbjct: 113 NCPEKNNESSEKKLCYNCGDTGHSLSHCPYPMEDGGTKFASCFICKGQGHISKNCPENK 171
Score = 32.0 bits (71), Expect = 0.16
Identities = 16/50 (32%), Positives = 21/50 (42%), Gaps = 5/50 (10%)
Query: 100 GPLCFNCGEIGHYASKCKIKG-----RLCFNCNQPGHFSRDCKAPKGESS 144
G CF C H A C K ++C C + GH ++C ESS
Sbjct: 73 GEGCFICHSKTHIAKLCPEKSEWERNKICLQCRRRGHSLKNCPEKNNESS 122
Score = 31.2 bits (69), Expect = 0.26
Identities = 22/88 (25%), Positives = 28/88 (31%), Gaps = 25/88 (28%)
Query: 18 CFCCGKFGQFANNC-------SVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATG 70
C+ CG G ++C CF C +GH++ NC G G
Sbjct: 127 CYNCGDTGHSLSHCPYPMEDGGTKFASCFICKGQGHISKNCP--------ENKHGIYPMG 178
Query: 71 GTGNKTSGKIVTCYRCKKVGHYTNKCPD 98
G C C V H CPD
Sbjct: 179 GC----------CKVCGSVAHLVKDCPD 196
Score = 27.3 bits (59), Expect = 3.8
Identities = 20/67 (29%), Positives = 24/67 (34%), Gaps = 9/67 (13%)
Query: 12 GDKDVTCFCCGKFGQFANNCS-------VIGPRCFKCNREGHLAVNC--KTLQGGPSDSK 62
G K +CF C G + NC +G C C HL +C K Q K
Sbjct: 148 GTKFASCFICKGQGHISKNCPENKHGIYPMGGCCKVCGSVAHLVKDCPDKFNQESAQPKK 207
Query: 63 MKGNDAT 69
DAT
Sbjct: 208 TSRFDAT 214
>At2g21060 glycine-rich protein (AtGRP2)
Length = 201
Score = 50.8 bits (120), Expect = 3e-07
Identities = 24/63 (38%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
CFKC GH+A C GG S G +GG G G + +CY C + GH+ C
Sbjct: 138 CFKCGEPGHMARECSQGGGGYSGGGGGGRYGSGGGGGGGGGGL-SCYSCGESGHFARDCT 196
Query: 98 DGG 100
GG
Sbjct: 197 SGG 199
Score = 46.6 bits (109), Expect = 6e-06
Identities = 28/86 (32%), Positives = 39/86 (44%), Gaps = 6/86 (6%)
Query: 55 QGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYAS 114
+GG S G +GG G G +C++C + GH +C GG G G Y S
Sbjct: 112 RGGGSYGGGYGGRGSGGRGG--GGGDNSCFKCGEPGHMARECSQGGGGYSGGGGGGRYGS 169
Query: 115 KCKIKGR----LCFNCNQPGHFSRDC 136
G C++C + GHF+RDC
Sbjct: 170 GGGGGGGGGGLSCYSCGESGHFARDC 195
Score = 35.8 bits (81), Expect = 0.011
Identities = 13/29 (44%), Positives = 17/29 (57%)
Query: 123 CFNCNQPGHFSRDCKAPKGESSGNTGKGK 151
CF C +PGH +R+C G SG G G+
Sbjct: 138 CFKCGEPGHMARECSQGGGGYSGGGGGGR 166
Score = 28.5 bits (62), Expect = 1.7
Identities = 12/31 (38%), Positives = 18/31 (57%)
Query: 7 GG*PIGDKDVTCFCCGKFGQFANNCSVIGPR 37
GG G ++C+ CG+ G FA +C+ G R
Sbjct: 171 GGGGGGGGGLSCYSCGESGHFARDCTSGGAR 201
>At3g53500 splicing factor - like protein
Length = 243
Score = 44.3 bits (103), Expect = 3e-05
Identities = 21/56 (37%), Positives = 27/56 (47%), Gaps = 5/56 (8%)
Query: 97 PDGGPLCFNCGEIGHYASKCKIKG--RLCFNCNQPGHFSRDCK---APKGESSGNT 147
P G CFNCG GH+A C C+ C + GH R+CK +PK G +
Sbjct: 54 PPGSGRCFNCGVDGHWARDCTAGDWKNKCYRCGERGHIERNCKNSPSPKKARQGGS 109
Score = 37.4 bits (85), Expect = 0.004
Identities = 23/75 (30%), Positives = 33/75 (43%), Gaps = 15/75 (20%)
Query: 2 TTAATGG*PIGDKDV----------TCFCCGKFGQFANNCSVIG--PRCFKCNREGHLAV 49
T A+ G P G +D CF CG G +A +C+ +C++C GH+
Sbjct: 34 TVEASRGAPRGSRDNGSRGPPPGSGRCFNCGVDGHWARDCTAGDWKNKCYRCGERGHIER 93
Query: 50 NCKTLQGGPSDSKMK 64
NCK PS K +
Sbjct: 94 NCK---NSPSPKKAR 105
Score = 30.0 bits (66), Expect = 0.59
Identities = 30/122 (24%), Positives = 43/122 (34%), Gaps = 26/122 (21%)
Query: 37 RCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
RCF C +GH A +C G NK CYRC + GH C
Sbjct: 59 RCFNCGVDGHWARDC----------------TAGDWKNK-------CYRCGERGHIERNC 95
Query: 97 PDGGPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAE 156
+ P + G Y S+ +K R P SR + S + ++ + E
Sbjct: 96 KN-SPSPKKARQGGSY-SRSPVKSRSPRRRRSPSR-SRSYSRGRSYSRSRSPVRREKSVE 152
Query: 157 ER 158
+R
Sbjct: 153 DR 154
>At2g37340 unknown protein
Length = 249
Score = 43.9 bits (102), Expect = 4e-05
Identities = 20/47 (42%), Positives = 25/47 (52%), Gaps = 3/47 (6%)
Query: 97 PDGGPLCFNCGEIGHYASKCKIKG--RLCFNCNQPGHFSRDCK-APK 140
P G CFNCG GH+A C C+ C + GH R+CK +PK
Sbjct: 54 PPGAGRCFNCGVDGHWARDCTAGDWKNKCYRCGERGHIERNCKNSPK 100
Score = 35.8 bits (81), Expect = 0.011
Identities = 14/37 (37%), Positives = 21/37 (55%), Gaps = 2/37 (5%)
Query: 18 CFCCGKFGQFANNCSVIG--PRCFKCNREGHLAVNCK 52
CF CG G +A +C+ +C++C GH+ NCK
Sbjct: 60 CFNCGVDGHWARDCTAGDWKNKCYRCGERGHIERNCK 96
Score = 34.3 bits (77), Expect = 0.031
Identities = 14/37 (37%), Positives = 17/37 (45%), Gaps = 2/37 (5%)
Query: 83 CYRCKKVGHYTNKCPDGG--PLCFNCGEIGHYASKCK 117
C+ C GH+ C G C+ CGE GH CK
Sbjct: 60 CFNCGVDGHWARDCTAGDWKNKCYRCGERGHIERNCK 96
Score = 29.6 bits (65), Expect = 0.77
Identities = 18/60 (30%), Positives = 21/60 (35%), Gaps = 23/60 (38%)
Query: 37 RCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
RCF C +GH A +C G NK CYRC + GH C
Sbjct: 59 RCFNCGVDGHWARDC----------------TAGDWKNK-------CYRCGERGHIERNC 95
>At4g38680 glycine-rich protein 2 (GRP2)
Length = 203
Score = 43.5 bits (101), Expect = 5e-05
Identities = 28/87 (32%), Positives = 34/87 (38%), Gaps = 11/87 (12%)
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
G C+KC GH+A +C GG G GG G G G
Sbjct: 128 GSDCYKCGEPGHMARDCSEGGGGYGGGG-GGYGGGGGYGGGGGG----------YGGGGR 176
Query: 95 KCPDGGPLCFNCGEIGHYASKCKIKGR 121
GG C++CGE GH+A C GR
Sbjct: 177 GGGGGGGSCYSCGESGHFARDCTSGGR 203
Score = 35.0 bits (79), Expect = 0.018
Identities = 28/94 (29%), Positives = 34/94 (35%), Gaps = 13/94 (13%)
Query: 7 GG*PIGDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
GG G D C+ CG+ G A +CS G G GG G
Sbjct: 122 GGGGRGGSD--CYKCGEPGHMARDCSEGG---------GGYGGGGGGYGGGGGYGGGGGG 170
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGG 100
GG G G +CY C + GH+ C GG
Sbjct: 171 YGGGGRGGGGGGG--SCYSCGESGHFARDCTSGG 202
Score = 32.7 bits (73), Expect = 0.091
Identities = 12/29 (41%), Positives = 15/29 (51%)
Query: 120 GRLCFNCNQPGHFSRDCKAPKGESSGNTG 148
G C+ C +PGH +RDC G G G
Sbjct: 128 GSDCYKCGEPGHMARDCSEGGGGYGGGGG 156
>At1g36110 hypothetical protein
Length = 745
Score = 43.5 bits (101), Expect = 5e-05
Identities = 20/44 (45%), Positives = 28/44 (63%), Gaps = 5/44 (11%)
Query: 55 QGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD 98
QGG + + +G G G++ + K VTCYRC K+GHY + CPD
Sbjct: 225 QGGGFNGRGRGR----GRGSRDTSK-VTCYRCDKLGHYASNCPD 263
Score = 30.8 bits (68), Expect = 0.35
Identities = 20/54 (37%), Positives = 27/54 (49%), Gaps = 5/54 (9%)
Query: 16 VTCFCCGKFGQFANNC-SVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDA 68
VTC+ C K G +A+NC + P F+ +G N L G S + M GN A
Sbjct: 246 VTCYRCDKLGHYASNCPDSVKPSVFETELDGD---NVWYLDNGAS-NHMTGNRA 295
>At5g20220 zinc finger protein
Length = 270
Score = 42.0 bits (97), Expect = 1e-04
Identities = 30/93 (32%), Positives = 36/93 (38%), Gaps = 24/93 (25%)
Query: 83 CYRCKKVGHYTNKCPDGGP------LCFNCGEIGHYASKCK------IKG-----RLCFN 125
C C + GH + CP+ G C CG GH C KG C
Sbjct: 143 CKNCGQEGHRRHYCPELGTNADRKFRCRGCGGKGHNRRTCPKSKSIVTKGISTRYHKCGI 202
Query: 126 CNQPGHFSRDCKAP-------KGESSGNTGKGK 151
C + GH SR C+ P GE+SG G GK
Sbjct: 203 CGERGHNSRTCRKPTGVNPSCSGENSGEDGVGK 235
Score = 32.7 bits (73), Expect = 0.091
Identities = 28/97 (28%), Positives = 33/97 (33%), Gaps = 18/97 (18%)
Query: 13 DKDVTCFCCGKFGQFANNC-----------SVIGPRCFKCNREGHLAVNCKTLQG-GPSD 60
D+ C CG G C S +C C GH + C+ G PS
Sbjct: 164 DRKFRCRGCGGKGHNRRTCPKSKSIVTKGISTRYHKCGICGERGHNSRTCRKPTGVNPSC 223
Query: 61 SKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
S N G G T C CKK+GH CP
Sbjct: 224 SGE--NSGEDGVGKIT----YACGFCKKMGHNVRTCP 254
>At2g10780 pseudogene
Length = 1611
Score = 38.9 bits (89), Expect = 0.001
Identities = 24/90 (26%), Positives = 36/90 (39%), Gaps = 16/90 (17%)
Query: 58 PSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKC- 116
P+D K K + + +G+ VTC + +++ C C CG H C
Sbjct: 359 PADKKRKFDKVDNTKSDAKTGECVTCGK-----NHSGTCWKAIGACGRCGSKDHAIQSCP 413
Query: 117 -------KIKG---RLCFNCNQPGHFSRDC 136
K+ G R CF C + GH R+C
Sbjct: 414 KMEPGQSKVLGEETRTCFYCGKTGHLKREC 443
Score = 37.0 bits (84), Expect = 0.005
Identities = 21/80 (26%), Positives = 31/80 (38%), Gaps = 16/80 (20%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C CGK + C C +C + H +C ++ P SK+ G +
Sbjct: 381 CVTCGK--NHSGTCWKAIGACGRCGSKDHAIQSCPKME--PGQSKVLGEETR-------- 428
Query: 78 GKIVTCYRCKKVGHYTNKCP 97
TC+ C K GH +CP
Sbjct: 429 ----TCFYCGKTGHLKRECP 444
Score = 26.6 bits (57), Expect = 6.5
Identities = 9/21 (42%), Positives = 12/21 (56%)
Query: 11 IGDKDVTCFCCGKFGQFANNC 31
+G++ TCF CGK G C
Sbjct: 423 LGEETRTCFYCGKTGHLKREC 443
>At4g03920 putative protein
Length = 463
Score = 37.7 bits (86), Expect = 0.003
Identities = 16/45 (35%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 101 PLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSG 145
P+C +C E+GH KCK+ C CN H + C P+ + G
Sbjct: 286 PVCAHCKEVGHSLRKCKVAPITCKACNSTLHLADAC--PRDQPQG 328
Score = 30.8 bits (68), Expect = 0.35
Identities = 13/34 (38%), Positives = 14/34 (40%)
Query: 83 CYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKC 116
C CK+VGH KC C C H A C
Sbjct: 288 CAHCKEVGHSLRKCKVAPITCKACNSTLHLADAC 321
>At2g12880 pseudogene
Length = 119
Score = 37.4 bits (85), Expect = 0.004
Identities = 23/99 (23%), Positives = 37/99 (37%), Gaps = 26/99 (26%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
C+KC + GH A +C + + + +TCY C + GH +N CP
Sbjct: 36 CYKCGKLGHFARSCHVV-------------------TQPTTAYITCYFCSEEGHRSNGCP 76
Query: 98 DGGP-------LCFNCGEIGHYASKCKIKGRLCFNCNQP 129
+ C+ CG H + + R F +P
Sbjct: 77 NKRTDQVNPKGHCYWCGNQDHRFNLIIWRSRCLFRLMKP 115
Score = 36.6 bits (83), Expect = 0.006
Identities = 13/40 (32%), Positives = 22/40 (54%), Gaps = 6/40 (15%)
Query: 103 CFNCGEIGHYASKCKIKGR------LCFNCNQPGHFSRDC 136
C+ CG++GH+A C + + C+ C++ GH S C
Sbjct: 36 CYKCGKLGHFARSCHVVTQPTTAYITCYFCSEEGHRSNGC 75
Score = 36.6 bits (83), Expect = 0.006
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 6/40 (15%)
Query: 18 CFCCGKFGQFANNCSVIGP------RCFKCNREGHLAVNC 51
C+ CGK G FA +C V+ C+ C+ EGH + C
Sbjct: 36 CYKCGKLGHFARSCHVVTQPTTAYITCYFCSEEGHRSNGC 75
Score = 26.9 bits (58), Expect = 5.0
Identities = 8/16 (50%), Positives = 11/16 (68%)
Query: 121 RLCFNCNQPGHFSRDC 136
R C+ C + GHF+R C
Sbjct: 34 RACYKCGKLGHFARSC 49
>At3g61330 copia-type polyprotein
Length = 1352
Score = 36.6 bits (83), Expect = 0.006
Identities = 13/28 (46%), Positives = 18/28 (63%)
Query: 90 GHYTNKCPDGGPLCFNCGEIGHYASKCK 117
GH ++ C+NCG+ GHYAS+CK
Sbjct: 268 GHPKSRYDKSSVKCYNCGKFGHYASECK 295
Score = 33.1 bits (74), Expect = 0.070
Identities = 12/33 (36%), Positives = 19/33 (57%)
Query: 110 GHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGE 142
GH S+ C+NC + GH++ +CKAP +
Sbjct: 268 GHPKSRYDKSSVKCYNCGKFGHYASECKAPSNK 300
Score = 32.0 bits (71), Expect = 0.16
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Query: 53 TLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
T Q G + S+ +G +K+S V CY C K GHY ++C
Sbjct: 254 TNQRGENSSRGRGKGHPKSRYDKSS---VKCYNCGKFGHYASEC 294
Score = 28.5 bits (62), Expect = 1.7
Identities = 10/25 (40%), Positives = 15/25 (60%)
Query: 16 VTCFCCGKFGQFANNCSVIGPRCFK 40
V C+ CGKFG +A+ C + F+
Sbjct: 279 VKCYNCGKFGHYASECKAPSNKKFE 303
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 36.6 bits (83), Expect = 0.006
Identities = 13/28 (46%), Positives = 18/28 (63%)
Query: 90 GHYTNKCPDGGPLCFNCGEIGHYASKCK 117
GH ++ C+NCG+ GHYAS+CK
Sbjct: 268 GHPKSRYDKSSVKCYNCGKFGHYASECK 295
Score = 34.7 bits (78), Expect = 0.024
Identities = 18/54 (33%), Positives = 27/54 (49%), Gaps = 4/54 (7%)
Query: 110 GHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAEERIHTKE 163
GH S+ C+NC + GH++ +CKAP S K K EE+I ++
Sbjct: 268 GHPKSRYDKSSVKCYNCGKFGHYASECKAP----SNKKFKEKANYVEEKIQEED 317
Score = 32.0 bits (71), Expect = 0.16
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Query: 53 TLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
T Q G + S+ +G +K+S V CY C K GHY ++C
Sbjct: 254 TNQRGENSSRGRGKGHPKSRYDKSS---VKCYNCGKFGHYASEC 294
Score = 30.0 bits (66), Expect = 0.59
Identities = 11/25 (44%), Positives = 15/25 (60%)
Query: 16 VTCFCCGKFGQFANNCSVIGPRCFK 40
V C+ CGKFG +A+ C + FK
Sbjct: 279 VKCYNCGKFGHYASECKAPSNKKFK 303
Score = 26.9 bits (58), Expect = 5.0
Identities = 11/28 (39%), Positives = 15/28 (53%), Gaps = 4/28 (14%)
Query: 37 RCFKCNREGHLAVNCKTLQGGPSDSKMK 64
+C+ C + GH A CK PS+ K K
Sbjct: 280 KCYNCGKFGHYASECK----APSNKKFK 303
>At2g31780 similar to Ariadne protein from Drosophila
Length = 542
Score = 36.6 bits (83), Expect = 0.006
Identities = 34/144 (23%), Positives = 54/144 (36%), Gaps = 31/144 (21%)
Query: 15 DVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGN 74
DV+C C KF C+ C + H V+C+T+ S +K D + N
Sbjct: 248 DVSCLCSYKF-------------CWNCCEDAHSPVDCETV----SKWLLKNKDESENM-N 289
Query: 75 KTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSR 134
K C +CK+ P+ N G H + + C+ C QP +
Sbjct: 290 WILAKTKPCPKCKR------------PIEKNTG-CNHMSCSAPCRHYFCWACLQPLSDHK 336
Query: 135 DCKAPKGESSGNTGKGKQLAAEER 158
C A K ++ T + + A +R
Sbjct: 337 ACNAFKADNEDETKRKRAKDAIDR 360
>At1g58140 hypothetical protein
Length = 1320
Score = 36.6 bits (83), Expect = 0.006
Identities = 13/28 (46%), Positives = 18/28 (63%)
Query: 90 GHYTNKCPDGGPLCFNCGEIGHYASKCK 117
GH ++ C+NCG+ GHYAS+CK
Sbjct: 268 GHPKSRYDKSSVKCYNCGKFGHYASECK 295
Score = 33.1 bits (74), Expect = 0.070
Identities = 12/33 (36%), Positives = 19/33 (57%)
Query: 110 GHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGE 142
GH S+ C+NC + GH++ +CKAP +
Sbjct: 268 GHPKSRYDKSSVKCYNCGKFGHYASECKAPSNK 300
Score = 32.0 bits (71), Expect = 0.16
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Query: 53 TLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKC 96
T Q G + S+ +G +K+S V CY C K GHY ++C
Sbjct: 254 TNQRGENSSRGRGKGHPKSRYDKSS---VKCYNCGKFGHYASEC 294
Score = 28.5 bits (62), Expect = 1.7
Identities = 10/25 (40%), Positives = 15/25 (60%)
Query: 16 VTCFCCGKFGQFANNCSVIGPRCFK 40
V C+ CGKFG +A+ C + F+
Sbjct: 279 VKCYNCGKFGHYASECKAPSNKKFE 303
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.140 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,559,897
Number of Sequences: 26719
Number of extensions: 215438
Number of successful extensions: 1199
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 597
Number of HSP's gapped (non-prelim): 470
length of query: 165
length of database: 11,318,596
effective HSP length: 92
effective length of query: 73
effective length of database: 8,860,448
effective search space: 646812704
effective search space used: 646812704
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0306.5


