

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
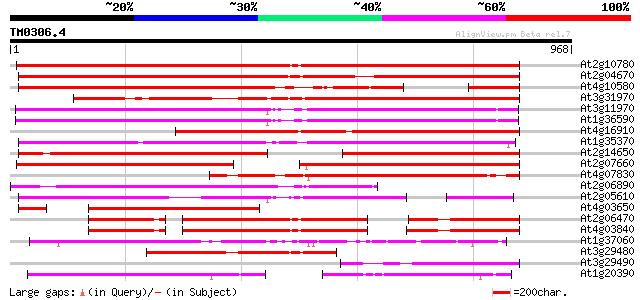
Query= TM0306.4
(968 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 969 0.0
At2g04670 putative retroelement pol polyprotein 920 0.0
At4g10580 putative reverse-transcriptase -like protein 701 0.0
At3g31970 hypothetical protein 662 0.0
At3g11970 hypothetical protein 643 0.0
At1g36590 hypothetical protein 639 0.0
At4g16910 retrotransposon like protein 594 e-170
At1g35370 hypothetical protein 577 e-165
At2g14650 putative retroelement pol polyprotein 554 e-157
At2g07660 putative retroelement pol polyprotein 488 e-138
At4g07830 putative reverse transcriptase 462 e-130
At2g06890 putative retroelement integrase 455 e-128
At2g05610 putative retroelement pol polyprotein 425 e-119
At4g03650 putative reverse transcriptase 401 e-111
At2g06470 putative retroelement pol polyprotein 311 9e-85
At4g03840 putative transposon protein 307 2e-83
At1g37060 Athila retroelment ORF 1, putative 281 2e-75
At3g29480 hypothetical protein 276 4e-74
At3g29490 hypothetical protein 255 1e-67
At1g20390 hypothetical protein 215 8e-56
>At2g10780 pseudogene
Length = 1611
Score = 969 bits (2504), Expect = 0.0
Identities = 481/870 (55%), Positives = 608/870 (69%), Gaps = 7/870 (0%)
Query: 12 VPRLPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGA 71
V +PP R F I++ G PIS APYRMAPAE+ +LK QLE+L KGFIRPS SPWGA
Sbjct: 638 VSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGA 697
Query: 72 PVLLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQI 131
PVL VKKKDG RLC+DYR LNK TVKN+YPLPRID+LMDQL G FSKIDL S YHQI
Sbjct: 698 PVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQI 757
Query: 132 RVKTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILI 191
++ D+ KTAFRTRY H+E++VMPFG+TNAPA FM MN +F +FLD FV++FI+DIL+
Sbjct: 758 PIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILV 817
Query: 192 YS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETV 251
YS + H+EHL VL LRE +L+A SKC FW V FLGHVIS +G+ VDP K+ ++
Sbjct: 818 YSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSI 877
Query: 252 LAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLK 311
W +P+ TEIRSF+GLAGYYRRF+ FA + PLT+LT KD F W++ C+ SF LK
Sbjct: 878 KEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELK 937
Query: 312 ERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLE 371
LT +PVL+LP+ EPY VY DAS GLGCVLMQ V+AYASRQL+ HEKNYPTHDLE
Sbjct: 938 AMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLE 997
Query: 372 LTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHP 431
+ A+VF LKIWR YLYG I++DHKSLKY+F Q ELN+RQRRWME + DY + YHP
Sbjct: 998 MAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVADYNLDIAYHP 1057
Query: 432 GKANVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYE-GMLKFGMIRIASGLMEEI 490
GKAN VADALSR+ +++L+ + + E L G A L+ I
Sbjct: 1058 GKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLGLGAADQAD-LLSRI 1116
Query: 491 RERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKW 550
R Q +D E+ Q E++ + + RVCVP + L+ I+ E H+SK+
Sbjct: 1117 RLAQERD----EEIKGWAQNNKTEYQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSKF 1172
Query: 551 RIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKW 610
IHPG KMY+DLK + W GMKK V +VA C TCQ K EHQ P+G+LQ+L +PEWKW
Sbjct: 1173 SIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPIPEWKW 1232
Query: 611 DSISMDFVVALPR-TQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHG 669
D I+MDFV LP + +H+++WV+VDRLTKSAHF+ E +AE Y+ EIVRLHG
Sbjct: 1233 DHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIVRLHG 1292
Query: 670 VSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLD 729
+ SIVSD D +FTS FW A ALGT++ LS+AYHPQ D Q+ERTIQ+LED+LRACVLD
Sbjct: 1293 IPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQTLEDMLRACVLD 1352
Query: 730 NRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQT 789
G+W+ L+EF YNN+F ASIGM+PYEALYGR CRTPLCW GE L GP ++ +T
Sbjct: 1353 WGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDET 1412
Query: 790 TEKVKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKF 849
TE++K ++ K++ +Q RQKSYA++RR+ LEF+ GD V+L+ G GR +KL+P++
Sbjct: 1413 TERMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRY 1472
Query: 850 IGPYQITRRVGPVAYQIALPPFLPIFTMYF 879
+GPY++ RVG VAY++ LPP L F F
Sbjct: 1473 VGPYKVIERVGAVAYKLDLPPKLNAFHNVF 1502
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 920 bits (2377), Expect = 0.0
Identities = 462/865 (53%), Positives = 586/865 (67%), Gaps = 38/865 (4%)
Query: 15 LPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPVL 74
LPP R F I++ G P+S APYRMAPAE+ ELK QLEDL KGFIRPS SPWGAPVL
Sbjct: 499 LPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVL 558
Query: 75 LVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVK 134
VKKKDG RLC+DYR LN TVKN+YPLPRID+L+DQLRG FSKIDL S YHQI +
Sbjct: 559 FVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIA 618
Query: 135 TEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS* 194
D+ KTAFRTRYGH+E++VMPF +TNAPA FM MN +F EFLD FV++FIDDIL+YS
Sbjct: 619 EADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSK 678
Query: 195 DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAW 254
+EHE HL +V+ LRE+KL+A SKC FW E+ FLGH++S EG+ VDP K+E + W
Sbjct: 679 SPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDW 738
Query: 255 EQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERL 314
+P TEIRSF+ L GYYRRF++GFA + P+T+LT KD PF W+ C+ F +LKE L
Sbjct: 739 PRPTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEML 798
Query: 315 TTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTA 374
T++PVL LP+ +PY VY DAS GLGCVLMQ KV+AYASRQL+ HE NYPTHDLE+
Sbjct: 799 TSTPVLALPEHGQPYMVYTDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAV 858
Query: 375 IVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGKA 434
++F+LKIWR YLYG +F+DHKSLKY+F+Q ELN+RQ RWME + DY+ + YHPGKA
Sbjct: 859 VIFALKIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADYDLEIAYHPGKA 918
Query: 435 NVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIASGLMEEIRERQ 494
NVVADALSRK + L+ + + L V E + + R + L+ R Q
Sbjct: 919 NVVADALSRKRVGAAPGQSVEALVSEIGALRLCVVAREPLGLEAVDR--ADLLTRARLAQ 976
Query: 495 LQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKWRIHP 554
+D L+ + + E++ + + RVCVP + ELRR I+ E H S + IHP
Sbjct: 977 EKDEGLI----AASKAEGSEYQFAANGTIFVYGRVCVPKDEELRREILSEAHASMFSIHP 1032
Query: 555 GMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSIS 614
G TKMY+DLK + W GMK+ V +VA C CQ K EHQ P
Sbjct: 1033 GATKMYRDLKRYYQWVGMKRDVANWVAECDVCQLVKAEHQVP------------------ 1074
Query: 615 MDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSI 674
D+IWVI+DRLTKSAHFL R T LA+ YV+EIV+LHGV SI
Sbjct: 1075 --------------DAIWVIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSI 1120
Query: 675 VSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSW 734
VSD D KFT FW A +GTK+++S+AYHPQ DGQ+ERTIQ+LED+LR CVLD G W
Sbjct: 1121 VSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHW 1180
Query: 735 DDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVK 794
D L+EF YNN++ ASIGMAP+EALYGR C TPL W Q E + G + +Q+TTE+++
Sbjct: 1181 ADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPLRWTQVEERSIYGADYVQETTERIR 1240
Query: 795 QIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQ 854
++ M+ +Q+RQ+SYAD+RRR LEFE GD V+L++ G R+I KL+P+++GP++
Sbjct: 1241 VLKLNMKEAQARQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFR 1300
Query: 855 ITRRVGPVAYQIALPPFLPIFTMYF 879
I RVGPVAY++ LP + F F
Sbjct: 1301 IVERVGPVAYRLELPDVMRAFHKVF 1325
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 701 bits (1810), Expect = 0.0
Identities = 359/665 (53%), Positives = 445/665 (65%), Gaps = 57/665 (8%)
Query: 15 LPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPVL 74
LPP + F I++ G P+S APYRMAPAE+ ELK QL+DL KGFIRPS SPWGAPVL
Sbjct: 473 LPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVL 532
Query: 75 LVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVK 134
VKKKDG RLC+DYR+LN+ TVKNRYPLPRID+L+DQLRG FSKIDL S YHQI +
Sbjct: 533 FVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIA 592
Query: 135 TEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS* 194
D+ KTAFRTRYGH+E++VMPFG+TNAPAVFM MN +F EFLD FV++FIDDIL+YS
Sbjct: 593 EADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSK 652
Query: 195 DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAW 254
+E E HL +V+ LRE+KL+A SKC FW E+ FLGH++S EG+ VDP K+E + W
Sbjct: 653 SPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW 712
Query: 255 EQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERL 314
+P TEIRSF+G AGYYRRF++GFA + P+T+LT KD PF W++ C+ F +LKE L
Sbjct: 713 PRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEML 772
Query: 315 TTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTA 374
T++PVL LP+ +PY VY DAS GLGCVLMQH KV+AYASRQL HE NYPTHDLE+ A
Sbjct: 773 TSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHGKVIAYASRQLMKHEGNYPTHDLEMAA 832
Query: 375 IVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGKA 434
++F+LKIWR YLYG +F+DHKSLKY+F Q ELN+RQRRWME + DY+ + YHPGKA
Sbjct: 833 VIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAYHPGKA 892
Query: 435 NVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIASGLMEEIRERQ 494
NVV DALSRK V + + +E+L + + G +R+ + E +
Sbjct: 893 NVVVDALSRK-RVGAALGQSVEVL---------------VSEIGALRLCAVAREPLG--- 933
Query: 495 LQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKWRIHP 554
L DR + T L + DEG ++
Sbjct: 934 ----------------------------LEAVDRADLLTRVRLAQK-KDEGLRA------ 958
Query: 555 GMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSIS 614
TKMY+DLK + W GMK V +VA C CQ K EHQ GMLQSL +PEWKWD I+
Sbjct: 959 --TKMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFIT 1016
Query: 615 MDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSI 674
MD VV L R + D+IWVIVDRLTKSAHFL R T LA+ +V+EIV+LHGV ++
Sbjct: 1017 MDLVVGL-RVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNM 1075
Query: 675 VSDSD 679
D
Sbjct: 1076 KEAQD 1080
Score = 84.3 bits (207), Expect = 3e-16
Identities = 39/88 (44%), Positives = 58/88 (65%)
Query: 792 KVKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIG 851
K+ + M+ +Q RQ+SYAD+RRR LEFE GD V+L++ G R+I KL+P+++G
Sbjct: 1067 KLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMG 1126
Query: 852 PYQITRRVGPVAYQIALPPFLPIFTMYF 879
P++I RV PVAY++ LP + F F
Sbjct: 1127 PFKIVERVEPVAYRLELPDVMRAFHKVF 1154
>At3g31970 hypothetical protein
Length = 1329
Score = 662 bits (1707), Expect = 0.0
Identities = 358/771 (46%), Positives = 468/771 (60%), Gaps = 80/771 (10%)
Query: 110 MDQLRGDAVFSKIDLKSSYHQIRVKTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDY 169
M + G S I + + I + ++ KTAFRTRYGH+E++VMPFG+TN P FM
Sbjct: 552 MPESVGQVAVSDIRVVHEFEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRL 611
Query: 170 MNRIFHEFLDRFVVVFIDDILIYS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEV 229
MN +F EFLD FV++FIDDIL+YS +EHE + + S E EV
Sbjct: 612 MNSVFQEFLDEFVIIFIDDILVYSKSPEEHE--------------VQSEESDGEAAGAEV 657
Query: 230 NFLGHVISKEGIVVDPAKVETVLAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQ 289
G+ VD K+E + W +P TEIRSF+GLAGYY+RF++ FA + P+T+
Sbjct: 658 ----------GVSVDLEKIEAIRDWPRPTNATEIRSFLGLAGYYKRFVKVFASMAQPMTK 707
Query: 290 LTRKDQPFAWTEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWK 349
LT KD PF W+ C F +LKE LT++PVL LP+ EPY VY DAS GL CVLMQ K
Sbjct: 708 LTGKDVPFVWSPECDEGFMSLKEMLTSTPVLALPEHGEPYMVYTDASRVGLDCVLMQRGK 767
Query: 350 VVAYASRQLKTHEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKEL 409
V F+DHKSLKY+F Q EL
Sbjct: 768 V-------------------------------------------FTDHKSLKYIFTQPEL 784
Query: 410 NMRQRRWMEFIKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELE-LLEQFRDMSLGV 468
N+RQRRWME + DY+ + YH GKANVVADALSRK ++ +E L+ + + L V
Sbjct: 785 NLRQRRWMELVADYDLEIAYHSGKANVVADALSRK-----RVGGSVEALVSEIGALRLCV 839
Query: 469 TPYEGMLKFGMIRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDR 528
E + + R + L+ +R Q +D L+ + E++ + + R
Sbjct: 840 MAQEPLGLEAVDR--ADLLTRVRLAQEKDEGLIAAS----KADGSEYQFAANGTILVHGR 893
Query: 529 VCVPTNPELRRLIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQK 588
VCVP + ELRR I+ E H S + IHP TKMY+DLK + W GMK+ V +V C CQ
Sbjct: 894 VCVPKDEELRREILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQL 953
Query: 589 AKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGR 648
K EHQ P G+LQSL + EWKWD I+MDFVV LP ++ + D+IWVIVDRLTKSAHFL R
Sbjct: 954 VKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLPVSRTK-DAIWVIVDRLTKSAHFLAIR 1012
Query: 649 TTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQI 708
T LA+ YV+EIV LHGV SIVSD D KFTS FW A +GTK+++S+AYHPQ
Sbjct: 1013 KTDGAVLLAKKYVSEIVELHGVPVSIVSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQT 1072
Query: 709 DGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRT 768
DGQ+ERTIQ+LED+LR CVLD G W D L+EF YNN++ ASI MAP+EALYGR CRT
Sbjct: 1073 DGQSERTIQTLEDMLRMCVLDRGGHWADHLSLVEFAYNNSYQASIRMAPFEALYGRPCRT 1132
Query: 769 PLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFL 828
PLCW Q GE + G + + +TTE+++ ++ M+ +Q RQ+SYAD+RRR LEFE GD V+L
Sbjct: 1133 PLCWTQVGERSIYGADYVLETTERIRVLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYL 1192
Query: 829 RVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFLPIFTMYF 879
++ G R+I KLTP+++GP++I RVGPVAY++ LP + F F
Sbjct: 1193 KMAMLRGPNRSISETKLTPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVF 1243
>At3g11970 hypothetical protein
Length = 1499
Score = 643 bits (1659), Expect = 0.0
Identities = 360/880 (40%), Positives = 518/880 (57%), Gaps = 48/880 (5%)
Query: 11 EVPRLPPVRDVE-FVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPW 69
E LPP R+ I ++ G P++ PYR + + E+ +EDL G ++ S SP+
Sbjct: 578 EPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPY 637
Query: 70 GAPVLLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYH 129
+PV+LVKKKDG RLCVDYR+LN TVK+ +P+P I+DLMD+L G +FSKIDL++ YH
Sbjct: 638 ASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYH 697
Query: 130 QIRVKTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDI 189
Q+R+ +DI KTAF+T GH+EYLVMPFG+TNAPA F MN IF FL +FV+VF DDI
Sbjct: 698 QVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDI 757
Query: 190 LIYS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVE 249
L+YS +EH +HL QV V+R KL+A SKC F + +V +LGH IS +GI DPAK++
Sbjct: 758 LVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIK 817
Query: 250 TVLAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQT 309
V W QP T+ ++R F+GLAGYYRRF+ F I GPL LT+ D F WT + +F+
Sbjct: 818 AVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTD-AFEWTAVAQQAFED 876
Query: 310 LKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHD 369
LK L +PVL LP ++ + V DA QG+G VLMQ +AY SRQLK + + ++
Sbjct: 877 LKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYE 936
Query: 370 LELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQY 429
EL A++F+++ WRHYL F I +D +SLKYL +Q+ Q++W+ + ++++ +QY
Sbjct: 937 KELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY 996
Query: 430 HPGKANVVADALSR-----KMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIAS 484
GK NVVADALSR +H+++ +V E +LL +D+ G
Sbjct: 997 RQGKENVVADALSRVEGSEVLHMAMTVV-ECDLL---KDIQAGYA--------------- 1037
Query: 485 GLMEEIRERQLQDVFLLEKRNLVIQGKDPEFK---IGTDNILRCKDRVCVPTNPELRRLI 541
+ QLQD+ +R DP+ K + NILR K ++ VP N ++ I
Sbjct: 1038 ------NDSQLQDIITALQR-------DPDSKKYFSWSQNILRRKSKIVVPANDNIKNTI 1084
Query: 542 MDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQ 601
+ H S H G +Q +K F+W GM K + Y+ +C TCQ+ K + G+LQ
Sbjct: 1085 LLWLHGSGVGGHSGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQ 1144
Query: 602 SLDVPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYV 661
L +P+ W +SMDF+ LP + + I V+VDRL+K+AHF+ Y+ +A Y+
Sbjct: 1145 PLPIPDTIWSEVSMDFIEGLPVSGGK-TVIMVVVDRLSKAAHFIALSHPYSALTVAHAYL 1203
Query: 662 AEIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLED 721
+ +LHG TSIVSD D FTS FW G L+L+SAYHPQ DGQTE + LE
Sbjct: 1204 DNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLET 1263
Query: 722 LLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLI 781
LR D W L E+ YN N+H+S M P+E +YG+ L + + +
Sbjct: 1264 YLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAV 1323
Query: 782 GPELLQQTTEKVKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRV---TQTTGVGR 838
LQ+ + + ++ + +Q R K +ADQ R EFE GD+V++++ Q + V R
Sbjct: 1324 VARSLQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMR 1383
Query: 839 AIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFLPIFTMY 878
A ++KL+PK+ GPY+I R G VAY++ALP + + ++
Sbjct: 1384 A--NQKLSPKYFGPYKIIDRCGEVAYKLALPSYSQVHPVF 1421
>At1g36590 hypothetical protein
Length = 1499
Score = 639 bits (1647), Expect = 0.0
Identities = 359/880 (40%), Positives = 518/880 (58%), Gaps = 48/880 (5%)
Query: 11 EVPRLPPVRDVE-FVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPW 69
E LPP R+ I ++ G P++ PYR + + E+ +EDL G ++ S SP+
Sbjct: 578 EPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPY 637
Query: 70 GAPVLLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYH 129
+PV+LVKKKDG RLCVDYR+LN TVK+ +P+P I+DLMD+L G +FSKIDL++ YH
Sbjct: 638 ASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYH 697
Query: 130 QIRVKTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDI 189
Q+R+ +DI KTAF+T GH+EYLVMPFG+TNAPA F MN IF FL +FV+VF DDI
Sbjct: 698 QVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDI 757
Query: 190 LIYS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVE 249
L+YS +EH +HL QV V+R KL+A SKC F + +V +LGH IS +GI DPAK++
Sbjct: 758 LVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIK 817
Query: 250 TVLAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQT 309
V W QP T+ ++R F+GLAGYYRRF+ F I GPL LT+ D F WT + +F+
Sbjct: 818 AVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTD-AFEWTAVAQQAFED 876
Query: 310 LKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHD 369
LK L +PVL LP ++ + V DA QG+G VLMQ +AY SRQLK + + ++
Sbjct: 877 LKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYE 936
Query: 370 LELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQY 429
EL A++F+++ WRHYL F I +D +SLKYL +Q+ Q++W+ + ++++ +QY
Sbjct: 937 KELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY 996
Query: 430 HPGKANVVADALSR-----KMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIAS 484
GK NVVADALSR +H+++ +V E +LL +D+ G
Sbjct: 997 RQGKENVVADALSRVEGSEVLHMAMTVV-ECDLL---KDIQAGYA--------------- 1037
Query: 485 GLMEEIRERQLQDVFLLEKRNLVIQGKDPEFK---IGTDNILRCKDRVCVPTNPELRRLI 541
+ QLQD+ +R DP+ K + NILR K ++ VP N ++ I
Sbjct: 1038 ------NDSQLQDIITALQR-------DPDSKKYFSWSQNILRRKSKIVVPANDNIKNTI 1084
Query: 542 MDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQ 601
+ H S H G +Q +K F+ GM K + Y+ +C TCQ+ K + G+LQ
Sbjct: 1085 LLWLHGSGVGGHSGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQ 1144
Query: 602 SLDVPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYV 661
L +P+ W +SMDF+ LP + + I V+VDRL+K+AHF+ Y+ +A+ Y+
Sbjct: 1145 PLPIPDTIWSEVSMDFIEGLPVSGGK-TVIMVVVDRLSKAAHFIALSHPYSALTVAQAYL 1203
Query: 662 AEIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLED 721
+ +LHG TSIVSD D FTS FW G L+L+SAYHPQ DGQTE + LE
Sbjct: 1204 DNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLET 1263
Query: 722 LLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLI 781
LR D W L E+ YN N+H+S M P+E +YG+ L + + +
Sbjct: 1264 YLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAV 1323
Query: 782 GPELLQQTTEKVKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRV---TQTTGVGR 838
LQ+ + + ++ + +Q R K +ADQ R EFE GD+V++++ Q + V R
Sbjct: 1324 VARSLQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMR 1383
Query: 839 AIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFLPIFTMY 878
A ++KL+PK+ GPY+I R G VAY++ALP + + ++
Sbjct: 1384 A--NQKLSPKYFGPYKIIDRCGEVAYKLALPSYSQVHPVF 1421
>At4g16910 retrotransposon like protein
Length = 687
Score = 594 bits (1531), Expect = e-170
Identities = 303/595 (50%), Positives = 392/595 (64%), Gaps = 13/595 (2%)
Query: 286 PLTQLTRKDQPFAWTEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLM 345
P+TQLT KD F W+E C+ SF LK LT +PVL+LP+ EPY VY DAS GLGCVL+
Sbjct: 4 PMTQLTGKDTAFNWSEECERSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLI 63
Query: 346 QHWKVVAYASRQLKTHEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFD 405
Q V+AYASRQL+ HEKNYPT+DLE+ A+VF+LKIWR YLYG IF+DHKSLKY+F
Sbjct: 64 QKGSVIAYASRQLRKHEKNYPTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKYIFT 123
Query: 406 QKELNMRQRRWMEFIKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELELLEQFRDMS 465
Q ELN+RQRRW++ + DY+ + YHPGKAN V DALSR V ++ L+ +
Sbjct: 124 QPELNLRQRRWIKLVADYDLNIAYHPGKANQVVDALSRLRTVVEAERNQVNLVNMMGTLH 183
Query: 466 LGVTPYEGMLKFGMIRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRC 525
L E + L+ IR Q +D E+ Q E++ + +
Sbjct: 184 LNALSKEVEPLGLRAANQADLLSRIRSAQERD----EEIKGWAQNNKTEYQSSNNGTIVV 239
Query: 526 KDRVCVPTNPELRRLIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLT 585
RVC P + L+ I+ E H+SK+ IHPG KMY+DLK + W GMKK V +VA
Sbjct: 240 NGRVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVA---- 295
Query: 586 CQKAKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRTQK-RHDSIWVIVDRLTKSAHF 644
K EHQ P+GMLQ+L +PEWKWD I MDFV LP K +H+++WV+VDRLTKSAHF
Sbjct: 296 ----KEEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHF 351
Query: 645 LPGRTTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAY 704
+ E +AE Y+ EIVRLHG+ SIVSD D +FTS FW LGT++ LS+AY
Sbjct: 352 MAISDKDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNLSTAY 411
Query: 705 HPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGR 764
HPQ DGQ+ERTIQ+LED+LRACVLD G+W+ L+EF YNN+F ASIGM+PYEALYGR
Sbjct: 412 HPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGR 471
Query: 765 KCRTPLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQSRQKSYADQRRRTLEFEEGD 824
RTPLCW GE L GP ++ +TT+K+K ++ K++ +Q RQKSYA++RR+ LEF+ GD
Sbjct: 472 AGRTPLCWTPVGERRLFGPAVVDETTKKMKFLKIKLKEAQDRQKSYANKRRKELEFQVGD 531
Query: 825 HVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFLPIFTMYF 879
V+L+ G GR +KL P+++GPY++ RVG VAY++ LPP L F F
Sbjct: 532 LVYLKAMTYKGAGRFTSRKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNVF 586
>At1g35370 hypothetical protein
Length = 1447
Score = 577 bits (1488), Expect = e-165
Identities = 333/869 (38%), Positives = 493/869 (56%), Gaps = 47/869 (5%)
Query: 15 LPPVRDV-EFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPV 73
LPP R+ + I ++ G P++ PYR + E+ ++D+ G I+ S SP+ +PV
Sbjct: 561 LPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPV 620
Query: 74 LLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRV 133
+LVKKKDG RLCVDY +LN TVK+R+ +P I+DLMD+L G VFSKIDL++ YHQ+R+
Sbjct: 621 VLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRM 680
Query: 134 KTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS 193
+DI KTAF+T GH+EYLVM FG+TNAPA F MN +F +FL +FV+VF DDILIYS
Sbjct: 681 DPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYS 740
Query: 194 *DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLA 253
+EH+EHL V V+R KL+A SK LGH IS I DPAK++ V
Sbjct: 741 SSIEEHKEHLRLVFEVMRLHKLFAKGSK--------EHLGHFISAREIETDPAKIQAVKE 792
Query: 254 WEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKER 313
W P TV ++R F+G AGYYRRF+ F I GPL LT+ D F W+ +++F TLK
Sbjct: 793 WPTPTTVKQVRGFLGFAGYYRRFVRNFGVIAGPLHALTKTD-GFCWSLEAQSAFDTLKAV 851
Query: 314 LTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELT 373
L +PVL LP ++ + V DA QG+ VLMQ +AY SRQLK + + ++ EL
Sbjct: 852 LCNAPVLALPVFDKQFMVETDACGQGIRAVLMQKGHPLAYISRQLKGKQLHLSIYEKELL 911
Query: 374 AIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGK 433
A +F+++ WRHYL F I +D +SLKYL +Q+ Q++W+ + ++++ +QY GK
Sbjct: 912 AFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPKLLEFDYEIQYRQGK 971
Query: 434 ANVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIASGLMEEIRER 493
N+VADALSR V+ E+L M+L + + + + + + G++++I
Sbjct: 972 ENLVADALSR--------VEGSEVLH----MALSIVECDFLKEIQVAYESDGVLKDIISA 1019
Query: 494 QLQDVFLLEKRNLVIQGKDPEFK---IGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKW 550
Q P+ K + +ILR K ++ VP + E+ ++ H S
Sbjct: 1020 LQQ---------------HPDAKKHYSWSQDILRRKSKIVVPNDVEITNKLLQWLHCSGM 1064
Query: 551 RIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKW 610
G +Q +K F+W GM K + ++ +C TCQ+ K ++ G+LQ L +P+ W
Sbjct: 1065 GGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIW 1124
Query: 611 DSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGV 670
+SMDF+ LP + + I V+VDRL+K+AHF+ Y+ +A+ ++ + + HG
Sbjct: 1125 CDVSMDFIEGLPNSGGK-SVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHGC 1183
Query: 671 STSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDN 730
TSIVSD D FTS FW G +LR+SSAYHPQ DGQTE + LE+ LR
Sbjct: 1184 PTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHAR 1243
Query: 731 RGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTT 790
W+ PL E+ YN N+H+S M P+E +YG+ L + + + LQ+
Sbjct: 1244 PHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQERE 1303
Query: 791 EKVKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIK-SRKLTPKF 849
+ ++ + +Q R K +ADQ R F+ GD V++++ ++ ++KL+PK+
Sbjct: 1304 NMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKY 1363
Query: 850 IGPYQITRR-----VGPVAYQIALPPFLP 873
GPY+I + VG V LP LP
Sbjct: 1364 FGPYKIIEKCGEVMVGNVTTSTQLPSVLP 1392
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 554 bits (1427), Expect = e-157
Identities = 265/430 (61%), Positives = 320/430 (73%), Gaps = 12/430 (2%)
Query: 15 LPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPVL 74
LPP R F I++ G P+S APYRMAPAE+ ELK QLEDL GAPVL
Sbjct: 460 LPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLL------------GAPVL 507
Query: 75 LVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVK 134
VKKKDG RLC+DYR LN TVKN+YPLPRID+L+DQLRG FSKIDL S YH I +
Sbjct: 508 FVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIA 567
Query: 135 TEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS* 194
D+ KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F E LD FV++FIDDIL+YS
Sbjct: 568 EADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSK 627
Query: 195 DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAW 254
+EHE HL +V+ LRE+KL+A SKC FW E+ FLGH++S EG+ VDP K+E + W
Sbjct: 628 SLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW 687
Query: 255 EQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERL 314
P TEIRSF+GLAGYYRRF++GFA + P+T+LT KD PF W+ C+ F +LKE L
Sbjct: 688 HTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEML 747
Query: 315 TTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTA 374
T++PVL LP+ EPY VY DAS GLGCVLMQ KV+AYASRQL+ HE NYPTHDLE+ A
Sbjct: 748 TSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAA 807
Query: 375 IVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGKA 434
++F+LKIWR YLYG +F+DHKSLKY+F Q ELN+RQR+WME + DY+ + YHPGKA
Sbjct: 808 VIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLEIAYHPGKA 867
Query: 435 NVVADALSRK 444
NVVADALS K
Sbjct: 868 NVVADALSHK 877
Score = 348 bits (893), Expect = 8e-96
Identities = 171/306 (55%), Positives = 221/306 (71%), Gaps = 1/306 (0%)
Query: 574 KQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHDSIWV 633
+ V +VA C CQ K EHQ P GMLQSL +PEWKWD I++DFVV LP ++ + D+IWV
Sbjct: 941 EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTK-DAIWV 999
Query: 634 IVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALHDA 693
IVDRLTKSAHFL R T LA+ YV+EIV+LHGV SIVSD D KFTS FW A
Sbjct: 1000 IVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAE 1059
Query: 694 LGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASI 753
+GTK+++S+AYHPQ GQ+ERTIQ+LED+LR CVLD G W D L+EF YNN++ ASI
Sbjct: 1060 MGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASI 1119
Query: 754 GMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQSRQKSYADQ 813
GMAP+EALY R CRTPLC Q GE + G + +Q+TTE+++ ++ M+ +Q RQ+SYAD+
Sbjct: 1120 GMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADK 1179
Query: 814 RRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFLP 873
RRR LEFE GD V+L++ G R+I KL+P+++GP++I RVGPVAY++ LP +
Sbjct: 1180 RRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPVAYRLELPDVMR 1239
Query: 874 IFTMYF 879
F F
Sbjct: 1240 AFHKVF 1245
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 488 bits (1256), Expect = e-138
Identities = 236/375 (62%), Positives = 283/375 (74%)
Query: 12 VPRLPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGA 71
V +PP R F I++ G PIS APYRMAPAE+ ELK QLE+L AKGFIRPS SPWGA
Sbjct: 133 VSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGA 192
Query: 72 PVLLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQI 131
PVL VKKKDG RLC+DYR LNK TVKN+YPLPRID+LMDQL G FSKIDL S YHQI
Sbjct: 193 PVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQI 252
Query: 132 RVKTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILI 191
++ D+ KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F +FLD FV++FIDDIL+
Sbjct: 253 PIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILV 312
Query: 192 YS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETV 251
+S + H+EHL VL LRE +L+A SK FW V FLGHVIS +G+ VDP K+ ++
Sbjct: 313 HSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSI 372
Query: 252 LAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLK 311
W +P+ TEIRSF+GLAGYYRRF+ FA + PLT+LT KD F W++ C+ SF LK
Sbjct: 373 KEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELK 432
Query: 312 ERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLE 371
L +PVL+LP+ EPY VY DAS GLGCVLMQ V+AYASRQL+ HEKNYPTHDLE
Sbjct: 433 AMLINAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLE 492
Query: 372 LTAIVFSLKIWRHYL 386
+ A+VF+LKIWR YL
Sbjct: 493 MAAVVFALKIWRSYL 507
Score = 413 bits (1062), Expect = e-115
Identities = 200/385 (51%), Positives = 269/385 (68%), Gaps = 5/385 (1%)
Query: 500 LLEKRNLVIQG----KDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKWRIHPG 555
L ++R+ I+G E++ + + RVCVP + L+ I+ E H+SK+ IHPG
Sbjct: 510 LAQERDEEIKGWTLNNKTEYQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPG 569
Query: 556 MTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSISM 615
KMY+DLK + W GM+K V +VA C TCQ K EHQ P+G+LQ+L + EWKWD I+M
Sbjct: 570 SNKMYRDLKRYYHWVGMRKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHITM 629
Query: 616 DFVVALPRTQK-RHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSI 674
DFV LP K +H+++WV+VDRLTKSAHF+ E +AE Y+ EI+RLHG+ SI
Sbjct: 630 DFVTRLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSI 689
Query: 675 VSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSW 734
VSD D +FTS FW A ALGT++ LS+AYHPQ DGQ+ERTIQ+LED+LRACVLD G+W
Sbjct: 690 VSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNW 749
Query: 735 DDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVK 794
+ LIEF YNN+F ASIGM+PYEALYGR CRTPLCW GE L GP ++ +TTE++K
Sbjct: 750 EKYLRLIEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMK 809
Query: 795 QIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQ 854
++ K++ +Q RQKSYA++RR+ LEF+ GD V+L+ G GR +KL+P+++GPY+
Sbjct: 810 FLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYK 869
Query: 855 ITRRVGPVAYQIALPPFLPIFTMYF 879
+ RVG VAY++ LPP L +F F
Sbjct: 870 VIERVGAVAYKLDLPPKLNVFHNVF 894
>At4g07830 putative reverse transcriptase
Length = 611
Score = 462 bits (1188), Expect = e-130
Identities = 256/546 (46%), Positives = 334/546 (60%), Gaps = 68/546 (12%)
Query: 345 MQHWKVVAYASRQLKTHEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLF 404
MQ KV+AY SRQLK HE NYPTHDLE+ A+ F+DHKSLKY+F
Sbjct: 1 MQRGKVIAYGSRQLKKHEGNYPTHDLEMAAV------------------FTDHKSLKYIF 42
Query: 405 DQKELNMRQRRWMEFIKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELELLEQFRDM 464
Q ELN+R RRWM+ + DY+ + YHPGKANVV DALSRK V + +E L
Sbjct: 43 TQLELNLRLRRWMKLVADYDLEIAYHPGKANVVTDALSRK-RVGAAPGQSVETL------ 95
Query: 465 SLGVTPYEGMLKFGMIRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDP----------- 513
+++ G +R+ + E + + LL + L Q KD
Sbjct: 96 ---------VIEIGALRLCAVAREPLGLEAVDQTDLLSRVQLA-QEKDEGLIAASKAEGF 145
Query: 514 EFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMK 573
E++ + + RVCVP + ELR+ I+ E H S + IHPG TKMY+DLK + W GMK
Sbjct: 146 EYQFAANGTILVHGRVCVPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMK 205
Query: 574 KQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHDSIWV 633
+ V +V C CQ K+EHQ +LQSL +PEWKWD I+MDFVV LP ++ + D+IWV
Sbjct: 206 RDVGNWVEECDVCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRTK-DAIWV 264
Query: 634 IVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALHDA 693
IVDRLTKSAHFL R T LA+ YV+EIV+LHGV SIVSD D KFTS FW A
Sbjct: 265 IVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAE 324
Query: 694 LGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASI 753
+GTK+++S+AYHPQ DGQ+ERTIQ+LED+LR CVLD RG W D L+EF YNN++ ASI
Sbjct: 325 MGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASI 384
Query: 754 GMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQSRQKSYADQ 813
GMAP+E LYGR CRT LCW Q GE + G + +Q+ TE+++ ++ M+ +Q+RQ+SYAD+
Sbjct: 385 GMAPFEVLYGRPCRT-LCWTQVGERSIYGADYVQEITERIRVLKLNMKEAQNRQRSYADK 443
Query: 814 RRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALPPFLP 873
RR+ LEFE GD V+Q V R+ +RVGPVA+++ L +
Sbjct: 444 RRKELEFEVGD----SVSQDGHVARS----------------EQRVGPVAFRLELSDVMR 483
Query: 874 IFTMYF 879
F F
Sbjct: 484 AFHKVF 489
>At2g06890 putative retroelement integrase
Length = 1215
Score = 455 bits (1171), Expect = e-128
Identities = 261/635 (41%), Positives = 377/635 (59%), Gaps = 61/635 (9%)
Query: 1 IRDFC*CCSEEVPR-LPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAK 59
++D+ E+ P+ LPP+R +E ID V G + YR P E EL+ Q
Sbjct: 412 LQDYKDVFPEDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELQRQ------- 464
Query: 60 GFIRPSVSPWGAPVLLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVF 119
KDG R+C D R +N TVK +P+PR+DD++D+L G ++F
Sbjct: 465 -------------------KDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIF 505
Query: 120 SKIDLKSSYHQIRVKTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLD 179
SKIDLKS YHQIR+ D KTAF+T++G YE+LVMPFG+T+AP+ FM MN + F+
Sbjct: 506 SKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIG 565
Query: 180 RFVVVFIDDILIYS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKE 239
FV+V+ DDIL+YS +EH EHL VL+VLR+++LYAN KC F + + FLG V+S +
Sbjct: 566 IFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSAD 625
Query: 240 GIVVDPAKVETVLAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAW 299
G+ VD KV+ + W PKTV E+RSF GLAG+YRRF + F+ IV PLT++ +KD F W
Sbjct: 626 GVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKW 685
Query: 300 TEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLK 359
+A + +FQ+LK++LT +PVLIL + + +E+ CDAS G+G VLMQ K++A+ S +L
Sbjct: 686 EKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQDQKLIAFFSEKLG 745
Query: 360 THEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEF 419
NYPT+D EL A+V +L+ W+HYL+ F I +DH+SLK+L Q++LN R RW+EF
Sbjct: 746 GATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLNKRHARWVEF 805
Query: 420 IKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGM 479
I+ + + ++Y GK NVVADALS++ + + +L EQ +++ YE
Sbjct: 806 IETFAYVIKYKKGKDNVVADALSQRYTLLSTLNVKLMGFEQIKEV------YE------- 852
Query: 480 IRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRR 539
+ Q+V+ ++ G+ D L ++R+CVP N LR
Sbjct: 853 -----------TDHDFQEVYKACEK--FASGR----YFRQDKFLFYENRLCVP-NCSLRD 894
Query: 540 LIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGM 599
L + E H H G+ K + + +F WP MK V C TC++AK + Q P G+
Sbjct: 895 LFVREAHGGGLMGHFGIAKTLEVMTEHFRWPHMKCDVKRICGRCNTCKQAKSKIQ-PNGL 953
Query: 600 LQSLDVPEWKWDSISMDFVVALPRTQKRHDSIWVI 634
L +P+ W+ ISMDFV+ LPRT K DSI+V+
Sbjct: 954 YTPLPIPKHPWNDISMDFVMGLPRTGK--DSIFVV 986
Score = 40.8 bits (94), Expect = 0.004
Identities = 28/86 (32%), Positives = 46/86 (52%), Gaps = 6/86 (6%)
Query: 787 QQTTEKVKQIREKMRAS-QSRQKSYADQR---RRTLEFEEGDHVFLRVTQTTGVGRAIKS 842
++ E V+QI E R + + + K YA Q RR FE GD V++ + + + +
Sbjct: 1018 KKKAELVQQIHENARRNIEEKTKLYAKQANKGRREQIFEVGDMVWIHLRKERFPAQ--RK 1075
Query: 843 RKLTPKFIGPYQITRRVGPVAYQIAL 868
KL P+ GP++I +R+ AYQ+ L
Sbjct: 1076 SKLMPRIDGPFKIIKRINDNAYQLDL 1101
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 425 bits (1093), Expect = e-119
Identities = 249/677 (36%), Positives = 368/677 (53%), Gaps = 88/677 (12%)
Query: 15 LPPVRDV-EFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPV 73
LPP R + I+++ P++ PYR + E+ ++D+ A G I+ S SP+ +PV
Sbjct: 24 LPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLASGTIQASSSPYASPV 83
Query: 74 LLVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRV 133
+LVKKKDG RLCVDYR+LN TVK+R+P+P I+DLMD+L G V+SKIDL++ YHQ+R+
Sbjct: 84 VLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVYSKIDLRAGYHQVRM 143
Query: 134 KTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS 193
DI KTAF+T GHYEYLVMPFG+TNAPA F MN F FL +FV+VF DDILIYS
Sbjct: 144 DPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFLRKFVLVFFDDILIYS 203
Query: 194 *DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLA 253
+EH++HL V V+R L+A SKC F + V +LGH IS EGI DPAK++ V
Sbjct: 204 TSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISGEGIATDPAKIKAVQD 263
Query: 254 WEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKER 313
W P + ++ F+GL GYYRRF
Sbjct: 264 WPVPVNLKQLCGFLGLTGYYRRF------------------------------------- 286
Query: 314 LTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELT 373
+ V DA G+G VLMQ +A+ SRQLK + + ++ EL
Sbjct: 287 ---------------FVVEMDACGHGIGAVLMQEGHPLAFISRQLKGKQLHLSIYEKELL 331
Query: 374 AIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGK 433
A++F ++ WRHYL F I +D +SLKYL +Q+ Q++W+ + ++++ +QY GK
Sbjct: 332 AVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQYKQGK 391
Query: 434 ANVVADALSR-----KMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIASGLME 488
N+VADALSR +H++L +V E +LL++ I +G
Sbjct: 392 ENLVADALSRVEGSEVLHMALSVV-ECDLLKE---------------------IQAGY-- 427
Query: 489 EIRERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKS 548
+ + +Q + + L Q + + +LR K+++ VP N ++ I+ H S
Sbjct: 428 -VTDGDIQGIITI----LQQQADSKKHYTWSQGVLRRKNKIVVPNNSGIKDTILRWLHCS 482
Query: 549 KWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEW 608
H G +Q +K F+W M K + ++ +C TCQ+ K ++ G+LQ L +P+
Sbjct: 483 GMGGHSGKEVTHQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDR 542
Query: 609 KWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLH 668
W +SMDF+ LP + + I V+VDRL+K+AHF+ Y+ +A+ Y+ + +LH
Sbjct: 543 IWSDVSMDFIDGLPLSNGK-TVIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVFKLH 601
Query: 669 GVSTSIVSDSDPKFTSH 685
G +SIV S H
Sbjct: 602 GCPSSIVVSSRRVLVQH 618
Score = 67.8 bits (164), Expect = 3e-11
Identities = 37/116 (31%), Positives = 62/116 (52%), Gaps = 1/116 (0%)
Query: 755 MAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQSRQKSYADQR 814
M PYEA+YG+ L + + + +Q+ + ++ + +Q R K ADQ
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 815 RRTLEFEEGDHVFLRVTQTTGVGRAIKS-RKLTPKFIGPYQITRRVGPVAYQIALP 869
EFE GD+VF+++ ++S +KL+PK+ GPY++ R G VAY++ LP
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLP 740
>At4g03650 putative reverse transcriptase
Length = 839
Score = 401 bits (1030), Expect = e-111
Identities = 182/294 (61%), Positives = 227/294 (76%)
Query: 137 DIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DT 196
D+ KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F EFLD FV++FIDDIL+YS
Sbjct: 520 DVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSP 579
Query: 197 KEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAWEQ 256
+EHE HL +V+ LRE+KL+A SKC FW E+ FLGH++S EG+ VDP K+E + W +
Sbjct: 580 EEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPR 639
Query: 257 PKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLTT 316
P TEIRSF+GLAGYYRRFI+GFA + P+T+LT KD PF W+ C+ F +LKE LT+
Sbjct: 640 PTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTS 699
Query: 317 SPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTAIV 376
+PVL LP+ EPY VY DAS GLGC LMQ KV+AYASRQL+ HE NYPTHDLE+ A++
Sbjct: 700 TPVLALPEHGEPYMVYTDASGVGLGCALMQRGKVIAYASRQLRKHEGNYPTHDLEMAAVI 759
Query: 377 FSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYH 430
F+LKIWR YLYG +F+DHKSLKY+F Q ELN+RQRRWME + DY+ + YH
Sbjct: 760 FALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAYH 813
Score = 52.8 bits (125), Expect = 9e-07
Identities = 26/49 (53%), Positives = 33/49 (67%)
Query: 15 LPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIR 63
LPP R F I++ P+S APYRMAPA++ ELK QLEDL A+ +R
Sbjct: 474 LPPSRSDPFTIELEPKTAPLSKAPYRMAPAKMAELKKQLEDLLAEADVR 522
Score = 40.0 bits (92), Expect = 0.006
Identities = 19/43 (44%), Positives = 28/43 (64%), Gaps = 5/43 (11%)
Query: 674 IVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTI 716
+V+D D + H +GTK+++S+ YHPQ DGQ+ERTI
Sbjct: 802 LVADYDLEIAYHL-----AEMGTKVQMSTTYHPQTDGQSERTI 839
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 311 bits (798), Expect = 9e-85
Identities = 161/320 (50%), Positives = 205/320 (63%), Gaps = 6/320 (1%)
Query: 299 WTEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQL 358
W +C+ SF LK LT +PVL+LP+ EPY VY DAS GLGCVLMQ V+AYASRQL
Sbjct: 375 WQRSCEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQL 434
Query: 359 KTHEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWME 418
HEKNYPTHDLE+ A+VF+LKIWR YLYG I++DHKSLKY+F Q ELN+RQRRWME
Sbjct: 435 WKHEKNYPTHDLEMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYIFIQPELNLRQRRWME 494
Query: 419 FIKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYE-GMLKF 477
+ DY+ + YHPGKAN VADALSR+ +++L+ + + E L
Sbjct: 495 LVADYDLDIAYHPGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVESLGL 554
Query: 478 GMIRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPEL 537
G A L+ IR Q +D E + + K E++ + + RVCVP + L
Sbjct: 555 GAADQAD-LLSRIRLAQERDE---EIKGWALNNK-TEYQTSNNGTIVVNGRVCVPNDRAL 609
Query: 538 RRLIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPA 597
+ I+ E H+SK+ IHPG KMY+DLK + W GMKK V +VA C TCQ K EHQ P+
Sbjct: 610 KEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPS 669
Query: 598 GMLQSLDVPEWKWDSISMDF 617
G+LQ+L +PEWKWD I+MDF
Sbjct: 670 GLLQNLPIPEWKWDHITMDF 689
Score = 182 bits (462), Expect = 8e-46
Identities = 91/191 (47%), Positives = 124/191 (64%), Gaps = 19/191 (9%)
Query: 689 ALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNN 748
A ALGT++ LS+AYHPQ DGQ+ERTIQ+LED+LRACVLD G+W+ L
Sbjct: 690 AFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLTL-------- 741
Query: 749 FHASIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQSRQK 808
ALYGR CRTPLCW GE L GP ++ +TTE++K ++ K++ + RQK
Sbjct: 742 -----------ALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQK 790
Query: 809 SYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIAL 868
SYA++RR+ LEF+ GD V+L+ G GR +KL+P+++GPY++ RVG VAY++ L
Sbjct: 791 SYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDL 850
Query: 869 PPFLPIFTMYF 879
PP L F F
Sbjct: 851 PPKLNAFHNVF 861
Score = 126 bits (317), Expect = 5e-29
Identities = 69/137 (50%), Positives = 90/137 (65%), Gaps = 11/137 (8%)
Query: 137 DIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DT 196
D+ KTAFRTRYGH+E++VMPFG+TNAPA FM MN +F +FLD FV++FIDDIL+YS
Sbjct: 287 DVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVYSKSW 346
Query: 197 KEHEEHLCQVLHVLREKKLYANPSKCEFWLE--EVNF--LGHVISKEGIVVDPAKVETVL 252
+ H+EHL VL LRE +L+A SKC FW E +F L +++ ++V P + E
Sbjct: 347 EAHQEHLRAVLERLREHELFAKLSKCSFWQRSCEKSFLELKAMLTNAPVLVLPEEGE--- 403
Query: 253 AWEQPKTVTEIRSFVGL 269
P TV S VGL
Sbjct: 404 ----PYTVYTDASIVGL 416
>At4g03840 putative transposon protein
Length = 973
Score = 307 bits (787), Expect = 2e-83
Identities = 158/320 (49%), Positives = 205/320 (63%), Gaps = 6/320 (1%)
Query: 299 WTEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQL 358
W +C+ +F LK LT +PVL+LP+ EPY VY DAS GL CVLMQ V+AYASRQL
Sbjct: 375 WQRSCEKNFLELKAMLTNAPVLVLPEEGEPYTVYTDASVVGLECVLMQKGSVIAYASRQL 434
Query: 359 KTHEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWME 418
+ HEKNYPTHDLE+ A+VF+LKIWR YLYG I++DHKSLKY+F Q ELN+RQRRWME
Sbjct: 435 RKHEKNYPTHDLEMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWME 494
Query: 419 FIKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYE-GMLKF 477
+ DY+ + YH GKAN VADALSR+ +++L+ + + E L
Sbjct: 495 LVADYDLDIAYHAGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLGL 554
Query: 478 GMIRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPEL 537
G A+ L+ IR Q +D E + + K E++ + + RVCVP N L
Sbjct: 555 GAADQAN-LLSRIRLAQERDE---EIKGWALNNK-TEYQTSNNGTIVVNGRVCVPNNRAL 609
Query: 538 RRLIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPA 597
+ I+ E H+SK+ IHPG K+Y+DLK + W GMKK V +VA C TCQ K EHQ P+
Sbjct: 610 KEEILREAHQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPS 669
Query: 598 GMLQSLDVPEWKWDSISMDF 617
G+LQ+L +PEWKWD I+MDF
Sbjct: 670 GLLQNLPIPEWKWDHITMDF 689
Score = 184 bits (467), Expect = 2e-46
Identities = 92/194 (47%), Positives = 125/194 (64%), Gaps = 19/194 (9%)
Query: 686 FWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTY 745
FW A ALGT++ LS+AYHPQ DGQ+ERTIQ+LED+LRAC LD G+W+ L
Sbjct: 690 FWKAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACALDWGGNWEKYLRL----- 744
Query: 746 NNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQS 805
ALYGR CRTPLCW GE L GP ++ +TTE++K ++ K++ +Q
Sbjct: 745 --------------ALYGRACRTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQD 790
Query: 806 RQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQ 865
RQKSYA++RR+ LEF+ D V+L+ G GR +KL+P+++GPY++ RVG VAY+
Sbjct: 791 RQKSYANKRRKELEFQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYK 850
Query: 866 IALPPFLPIFTMYF 879
+ LPP L F F
Sbjct: 851 LDLPPKLNAFHNVF 864
Score = 124 bits (311), Expect = 3e-28
Identities = 68/137 (49%), Positives = 88/137 (63%), Gaps = 11/137 (8%)
Query: 137 DIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DT 196
D+ KTAF TRYGH+E++VMPFG+TNAP FM MN +F +FLD FV++FIDDIL+YS
Sbjct: 287 DVRKTAFLTRYGHFEFVVMPFGLTNAPTAFMKMMNGVFRDFLDEFVIIFIDDILVYSKSW 346
Query: 197 KEHEEHLCQVLHVLREKKLYANPSKCEFWLE--EVNF--LGHVISKEGIVVDPAKVETVL 252
+ H+EHL VL LRE +L+A SKC FW E NF L +++ ++V P + E
Sbjct: 347 EAHQEHLRAVLEQLREHELFAKLSKCSFWQRSCEKNFLELKAMLTNAPVLVLPEEGE--- 403
Query: 253 AWEQPKTVTEIRSFVGL 269
P TV S VGL
Sbjct: 404 ----PYTVYTDASVVGL 416
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 281 bits (718), Expect = 2e-75
Identities = 240/878 (27%), Positives = 399/878 (45%), Gaps = 146/878 (16%)
Query: 35 SIAPYRMAPAELVEL-KSQLEDLSAKGFIRP-SVSPWGAPVLLVKKKDGKS--------- 83
SI P R L E+ K ++ L G I P S S W +PV V KK G +
Sbjct: 900 SIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDEL 959
Query: 84 ---------RLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVK 134
R+C++YR+LN A+ K +PLP ID ++++L + +D S + QI +
Sbjct: 960 IPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIH 1019
Query: 135 TEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS* 194
D KT F YG + Y MPFG+ NAPA F M IF + ++ V VF+DD +Y
Sbjct: 1020 PNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGS 1079
Query: 195 DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAW 254
+LC+VL E L N KC F + E LG IS+EGI VD AK++ ++
Sbjct: 1080 SFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQL 1139
Query: 255 EQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERL 314
+ PKTV +IRSF+G AG+YR FI+ F+K+ PLT+L K+ FA+ + C T+F+ +KE L
Sbjct: 1140 QPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEAL 1199
Query: 315 TTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTA 374
T+P++ P + P+E+ + M +V Y T + EL A
Sbjct: 1200 ITAPIVQAPNWDFPFEI-----------ITMDDAQV-------------RYATTEKELLA 1235
Query: 375 IVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGKA 434
+VF+ + +R YL G TI++DH +L++++ +K+ R RW+ +++++ + G
Sbjct: 1236 VVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGIE 1295
Query: 435 NVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIASGLMEEIRERQ 494
N VAD LSR +++ E+ + + P E ++ ++++ E++
Sbjct: 1296 NGVADHLSR-----MRIEDEVLIDDSM--------PEEQLM----------AIQQLNEKK 1332
Query: 495 LQDVFLLEKRNLVIQGKDP------EFKIGTDNI-----------LRCKD---RVCVPTN 534
L + + N ++ G++P E K +I CKD R CV +
Sbjct: 1333 LP--WYADHVNYLVSGEEPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKIYRTCV-SE 1389
Query: 535 PELRRLIMDEGHKSKWRIHPGMTK-MYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEH 593
E+ +++ H + H K M + L+ FWWP M K E+++ C
Sbjct: 1390 DEIEGILL-HCHGFAYGGHFATFKTMSKILQAGFWWPSMFKDAQEFISKC---------- 1438
Query: 594 QRPAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNV 653
++ DV W +DF+ P + + I V +D ++K + T +
Sbjct: 1439 ----DSFENFDV--W-----GIDFMGPFP-SSYGNKYILVAIDYVSKWVEAIASHTN-DA 1485
Query: 654 EKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTE 713
+ +++ I GV ++SD F + + L G K + S GQ E
Sbjct: 1486 RVVLKLFKTIIFPRFGVPRIVISDGGKHFINKGFENLLKKHGVKHKTS--------GQVE 1537
Query: 714 RTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWY 773
+ + ++ +L V R W + Y F IG P+ LYG+ C P+
Sbjct: 1538 ISNREIKAILEKTVGSTRKDWSAKLNDTLWAYRTAFKTPIGTTPFNLLYGKSCHLPV--- 1594
Query: 774 QDGESLLIGPELLQ---QTTEKVKQIR------------EKMRASQSRQKSYADQRRRTL 818
+ + +LL +T E+ + I+ E + + R KS+ D++ +
Sbjct: 1595 ELEYKAMWAVKLLNFDIKTAEEKRLIQLNDLNKIRLEAYESSKIYKERTKSFHDKKIVSR 1654
Query: 819 EFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQIT 856
+F+ GD V L ++ +KSR + GP+ +T
Sbjct: 1655 DFKVGDQVLLFNSRLRLFPGKLKSR-----WSGPFSVT 1687
>At3g29480 hypothetical protein
Length = 718
Score = 276 bits (706), Expect = 4e-74
Identities = 151/329 (45%), Positives = 205/329 (61%), Gaps = 26/329 (7%)
Query: 237 SKEGIVVDPAKVETVLAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQP 296
++ G+ VD K+E + W +P TEIRSF+GLAGYYRRF++GFA + PLT+LT K+ P
Sbjct: 412 AEAGVSVDLEKIEAIKDWPRPTNATEIRSFLGLAGYYRRFVKGFASMAPPLTKLTGKNVP 471
Query: 297 FAWTEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASR 356
F W+ C+ F +LKE LT++PVL LP+ E Y VY DAS GLGCVLMQ KV+AYASR
Sbjct: 472 FVWSPECEEGFVSLKEILTSTPVLALPEHGESYMVYTDASRVGLGCVLMQRGKVIAYASR 531
Query: 357 QLKTHEKNYPTHDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRW 416
QL+ HE NYPTHDLE++A +F+DHKSLKY+ Q ELN+RQRRW
Sbjct: 532 QLRKHEGNYPTHDLEMSA------------------VFTDHKSLKYIITQPELNLRQRRW 573
Query: 417 MEFIKDYEFTLQYHPGKANVVADALSRKMHVSLKMVKELE-LLEQFRDMSLGVTPYEGML 475
ME + DY+ + YH GKA+VVADALSRK V + + +E L+ + R + L E +
Sbjct: 574 MELVADYDLEIAYHHGKASVVADALSRK-RVGVAPGQSVEALVSEIRALRLCAVAREPLG 632
Query: 476 KFGMIRIASGLMEEIRERQLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNP 535
+ R+ L+ +R Q +D L+ + + +++ + + RVCV +
Sbjct: 633 LEAVDRV--DLLSRVRLAQEKDEGLI----AASKAEGSKYQFAANGTILVHGRVCVLNDE 686
Query: 536 ELRRLIMDEGHKSKWRIHPGMTKMYQDLK 564
ELRR I+ E H S + IHPG TKMY+DLK
Sbjct: 687 ELRREILSEAHASMFSIHPGATKMYRDLK 715
>At3g29490 hypothetical protein
Length = 438
Score = 255 bits (651), Expect = 1e-67
Identities = 136/308 (44%), Positives = 177/308 (57%), Gaps = 53/308 (17%)
Query: 572 MKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHDSI 631
MK+ V +VA C CQ K EHQ P GMLQSL +PEW
Sbjct: 1 MKRDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWNT--------------------- 39
Query: 632 WVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALH 691
FL R T LA+ YV+EIV+LHGV
Sbjct: 40 ------------FLAIRKTDGAAVLAKKYVSEIVKLHGVPAE------------------ 69
Query: 692 DALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHA 751
+GTK+++S+ YHPQ DGQ ERTIQ+LED+LR CVLD G W D L+EF YNN++ A
Sbjct: 70 --MGTKVQMSTPYHPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQA 127
Query: 752 SIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEKVKQIREKMRASQSRQKSYA 811
IGMAP+EALYGR CRTPLCW Q GE + G + +Q+TTE+++ ++ M+ +Q RQ SYA
Sbjct: 128 GIGMAPFEALYGRPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYA 187
Query: 812 DQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIALPPF 871
D+RRR LEFE GD V+L++ G R+I KL+ +++GP++I RVGPVAY + LP
Sbjct: 188 DKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDV 247
Query: 872 LPIFTMYF 879
+ F F
Sbjct: 248 MRAFHKVF 255
>At1g20390 hypothetical protein
Length = 1791
Score = 215 bits (548), Expect = 8e-56
Identities = 129/417 (30%), Positives = 209/417 (49%), Gaps = 7/417 (1%)
Query: 31 FGPISIAPYRMAPAELVELKSQLEDLSAKG-FIRPSVSPWGAPVLLVKKKDGKSRLCVDY 89
F P+ ++ P + ++E L G I W A ++VKKK+GK R+CVDY
Sbjct: 775 FKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDY 834
Query: 90 RQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVKTEDIPKTAFRTRYGH 149
LNKA K+ YPLP ID L++ G+ + S +D S Y+QI + +D KT+F T G
Sbjct: 835 TDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGT 894
Query: 150 YEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DTKEHEEHLCQVLHV 209
Y Y VM FG+ NA A + ++N++ + + R V V+IDD+L+ S ++H EHL + V
Sbjct: 895 YCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDV 954
Query: 210 LREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAWEQPKTVTEIRSFVGL 269
L + NP+KC F + FLG+V++K GI +P ++ +L P+ E++ G
Sbjct: 955 LNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGR 1014
Query: 270 AGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLTTSPVLILPQPEEPY 329
RFI P L ++ F W + + +F+ LK+ L+T P+L+ P+ E
Sbjct: 1015 IAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETL 1074
Query: 330 EVYCDASHQGLGCVLMQ----HWKVVAYASRQLKTHEKNYPTHDLELTAIVFSLKIWRHY 385
+Y S + VL++ + + Y S+ L E YP + A+V S + R Y
Sbjct: 1075 YLYIAVSDHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPY 1134
Query: 386 LYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPG-KANVVADAL 441
T + +D + L+ + R +W + +Y+ + P K+ V+AD L
Sbjct: 1135 FQSHTIAVLTD-QPLRVALHSPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLADFL 1190
Score = 80.9 bits (198), Expect = 3e-15
Identities = 79/340 (23%), Positives = 149/340 (43%), Gaps = 22/340 (6%)
Query: 541 IMDEGHKSKWRIHPGMTKMYQDLK-LNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGM 599
IM E H+ H G + LK L F+WP M + A C CQ+ +P +
Sbjct: 1438 IMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTEL 1497
Query: 600 LQS--LDVPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLA 657
L++ P +W +MD V +P ++++ I V+ D TK T +
Sbjct: 1498 LRAGVAPYPFMRW---AMDIVGPMPASRQKR-FILVMTDYFTKWVE-AESYATIRANDVQ 1552
Query: 658 EIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQ 717
I+ HG+ I++D+ +F S + + +L S+ +PQ +GQ E T +
Sbjct: 1553 NFVWKFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNK 1612
Query: 718 SLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTP--LCWYQD 775
++ L+ + + +G+W D + ++Y ++ P+ YG + P + +
Sbjct: 1613 TILSGLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYSSL 1672
Query: 776 GESLLI-GPELL-QQTTEKVKQIREKMRASQSRQKSYA-------DQRRRTLEFEEGDHV 826
S+++ PEL + +++ + E A+ R ++Y +Q+ F+ GD V
Sbjct: 1673 RRSMMVKNPELNDRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGDLV 1732
Query: 827 FLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQI 866
+V + T I + KL + G YQ+++ V P Y++
Sbjct: 1733 LRKVFENTA---EINAGKLGANWEGSYQVSKIVRPGDYEL 1769
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.141 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,305,920
Number of Sequences: 26719
Number of extensions: 898441
Number of successful extensions: 2897
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 2678
Number of HSP's gapped (non-prelim): 136
length of query: 968
length of database: 11,318,596
effective HSP length: 109
effective length of query: 859
effective length of database: 8,406,225
effective search space: 7220947275
effective search space used: 7220947275
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0306.4


