

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.1
(1224 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
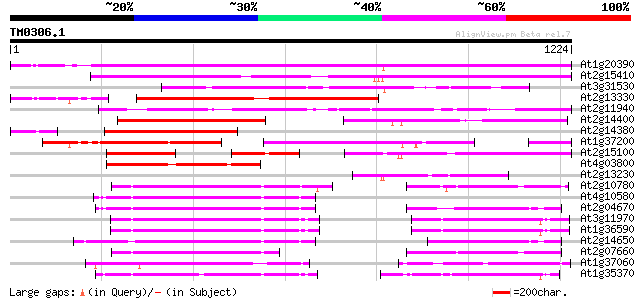
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 969 0.0
At2g15410 putative retroelement pol polyprotein 806 0.0
At3g31530 hypothetical protein 546 e-155
At2g13330 F14O4.9 449 e-126
At2g11940 putative retroelement gag/pol polyprotein 428 e-120
At2g14400 putative retroelement pol polyprotein 360 4e-99
At2g14380 putative retroelement pol polyprotein 339 5e-93
At1g37200 hypothetical protein 330 4e-90
At2g15100 putative retroelement pol polyprotein 315 8e-86
At4g03800 305 8e-83
At2g13230 pseudogene 243 6e-64
At2g10780 pseudogene 236 5e-62
At4g10580 putative reverse-transcriptase -like protein 231 1e-60
At2g04670 putative retroelement pol polyprotein 225 1e-58
At3g11970 hypothetical protein 224 2e-58
At1g36590 hypothetical protein 224 2e-58
At2g14650 putative retroelement pol polyprotein 219 6e-57
At2g07660 putative retroelement pol polyprotein 214 3e-55
At1g37060 Athila retroelment ORF 1, putative 213 5e-55
At1g35370 hypothetical protein 200 4e-51
>At1g20390 hypothetical protein
Length = 1791
Score = 969 bits (2506), Expect = 0.0
Identities = 531/1247 (42%), Positives = 753/1247 (59%), Gaps = 67/1247 (5%)
Query: 1 DQGSSADLIYGDAYEKLGLTEADLLPYDGALVGFSGERVFVRGYVELNTVFGEGINAESF 60
D GSS DLI+ D + +T+ + P L GF G+ V G ++L +F G+ A
Sbjct: 588 DTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTIKL-PIFVGGLIAW-- 644
Query: 61 AIKFLVVKCTSPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKGGVGILKA--DQVVAWRC 118
+KF+V+ + YNV++G P ++++ AI ST H VK+P + G+ L+A + R
Sbjct: 645 -VKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFP-THNGIFTLRAPKEAKTPSRS 702
Query: 119 YSESFKQYGHMGKRAVKEGHRVYGVDIDQEGVCLDPREGFLEHKMTPEEETKTVKVGERN 178
Y ES E R V+ID+ DP R
Sbjct: 703 YEES-------------ELCRTEMVNIDES----DP---------------------TRC 724
Query: 179 LKVGVNLTAIQEARLTQLLAENMDLFAWSAQDLPGIDPNFICHKLALNPGVKPIAQMKRK 238
+ VG ++ L LL N FAWS +D+ GIDP H+L ++P KP+ Q +RK
Sbjct: 725 VGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRK 784
Query: 239 MGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKD 298
+G E+A+AV E KL+ AG I EVKYP WLAN V+VKK NGKWR+C DYTDLNK CPKD
Sbjct: 785 LGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKD 844
Query: 299 SYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGL 358
SYPLP+ID+LV+ SG G+LS MDA+SGY+QI M+ D+EKT+F+T++ YCY+ M FGL
Sbjct: 845 SYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGL 904
Query: 359 KNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNP 418
KNAGATYQR ++++ Q+GR +E+Y+DDM+VKS + H L++ F + + M+LNP
Sbjct: 905 KNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNP 964
Query: 419 EKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFLPC 478
KC+FG+ SG+FLG+++T+RGIE NP + +AILE+ SP + +EVQ+L GRIAAL+RF+
Sbjct: 965 TKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISR 1024
Query: 479 SGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNA 538
S K PF+ L++ F W + E+AF+ LK+ LS PPIL +P G L ++I++SD+A
Sbjct: 1025 STDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHA 1084
Query: 539 VSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKT 598
VSSVL++E + E R I++ S +L AE RY IEKAALA++ SARKLRPYFQ I V T
Sbjct: 1085 VSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLT 1144
Query: 599 DFPLRQVLQKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQVLADFVNEM---SPEVKVS- 654
D PLR L P +GRM WAVELSE+ I F + +K+QVLADF+ E+ S E VS
Sbjct: 1145 DQPLRVALHSPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLADFLIELPLQSAERAVSG 1204
Query: 655 -EEAEWILSVDGSSYLKGSGAGVVLEGPGGVIIEQSLKFDFKASNNQAEYEAIIAGINLA 713
EW L VDGSS +GSG G+ L P ++EQS + F A+NN AEYE +IAG+ LA
Sbjct: 1205 NRGEEWSLYVDGSSSARGSGIGIRLVSPTAEVLEQSFRLRFVATNNVAEYEVLIAGLRLA 1264
Query: 714 IEMNVHCLVIKTDSQLVANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENT 773
M + + TDSQL+A Q+ G+Y+AK+ ++ YL Q + K ++ +++ +PR +N
Sbjct: 1265 AGMQITTIHAFTDSQLIAGQLSGEYEAKNEKMDAYLKIVQLMTKDFENFKLSKIPRGDNA 1324
Query: 774 RADVLCKLASTKKPGNNKSVIQETLKSPSINEDDVVMVTG----------AAPSDWMDRI 823
AD L LA T + + E++ PSI+ D V + A P+DW I
Sbjct: 1325 PADALAALALTSDSDLRRIIPVESIDKPSIDSTDAVEIVNTIRSSNAPDPADPTDWRVEI 1384
Query: 824 KMCLEADGADLALFSKDQVR-EASHYVLLGDQLYRRGVGVPLLRCVTRDEADRIMFEVHE 882
+ L ++ ++R +A+ Y L+ + L + +L C+ E + IM E HE
Sbjct: 1385 RDYLSDGTLPSDKWTARRLRIKAAKYTLMKEHLLKVSAFGAMLNCLHGTEINEIMKETHE 1444
Query: 883 GVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSA 942
G +H GGR+LA K+ + GFYWPT+ +DC + KCE+CQ +A P E+L + +
Sbjct: 1445 GAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTELLRAGVAP 1504
Query: 943 WPFAMWGVDILGPFTPAGAQIRFVLVAVDYFTKWIEAESMAKITAEKVKKFYWRKIICRF 1002
+PF W +DI+GP PA Q RF+LV DYFTKW+EAES A I A V+ F W+ IICR
Sbjct: 1505 YPFMRWAMDIVGPM-PASRQKRFILVMTDYFTKWVEAESYATIRANDVQNFVWKFIICRH 1563
Query: 1003 GVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANKVILNGIKKRLG 1062
G+P +++DNG+QF S +FC I + ++ +PQ NGQ EA NK IL+G+KKRL
Sbjct: 1564 GLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNKTILSGLKKRLD 1623
Query: 1063 DAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDAMVPVEIQDMTFRVAAYDENENH 1122
+ KG WADEL V+W+Y TTP+S T +TPF YG++AM P E+ + R + +N
Sbjct: 1624 EKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYSSLRRSMMVKNPEL 1683
Query: 1123 ENRLI--DLNLAEEVKTEVRLRQAAVKQRSERRYNTRVVPRHMQVGDLVLRRKAKGPDD- 1179
+R++ L+ EE++ R + + + YN +V RH VGDLVLR+ + +
Sbjct: 1684 NDRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGDLVLRKVFENTAEI 1743
Query: 1180 --SKLSPNWEGPYRILRDLGQGAYHLEELSGRRIPRAWNAQHLRYYY 1224
KL NWEG Y++ + + G Y L +SG +PR WN+ HL+ YY
Sbjct: 1744 NAGKLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTWNSMHLKRYY 1790
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 806 bits (2083), Expect = 0.0
Identities = 442/1115 (39%), Positives = 637/1115 (56%), Gaps = 123/1115 (11%)
Query: 177 RNLKVGVNLTAIQEARLTQLLAENMDLFAWSAQDLPGIDPNFICHKLALNPGVKPIAQMK 236
R + + ++L + + L L +N FAWS +D+PGID CH+L ++P KP+ Q +
Sbjct: 728 RCVGIRIDLPSELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKR 787
Query: 237 RKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCP 296
RK+G ++ + V E KL+DAG I EV+YP WL N V+VKK NGKWR+C D+TDLNK CP
Sbjct: 788 RKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACP 847
Query: 297 KDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPF 356
KDS+PLP+ID+LV+ +G +LS MDA+SGY+QI M+ D EKT F+T+Q YCY+ MPF
Sbjct: 848 KDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVMPF 907
Query: 357 GLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRL 416
GLKNAGATY RL++++F Q+ +ME+Y+DDM+VKS H L + F + ++NM+L
Sbjct: 908 GLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKL 967
Query: 417 NPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFL 476
NP KC+FG+ SG+FLG+++TRRGIE NP + AI+++ SP + +EVQ+LIGRIAAL+RF+
Sbjct: 968 NPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSPRNTREVQRLIGRIAALNRFI 1027
Query: 477 PCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISD 536
S K PF+Q LR N+ F+W ++CE+ G L ++I++S
Sbjct: 1028 SRSTDKCLPFYQLLRANKRFEWDEKCEE--------------------GETLYLYIAVST 1067
Query: 537 NAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKV 596
+AVS VL++E + E I++VS L GAELRY +EK A A++ISARKLRPYF+ + ++V
Sbjct: 1068 SAVSGVLVREDRGEQHPIFYVSKTLDGAELRYPTLEKLAFAVVISARKLRPYFKSYTVEV 1127
Query: 597 KTDFPLRQVLQKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQVLADFVNEMSPEVKVSEE 656
T+ PLR +L P +GR+ WAVELSE+ I ++ + K+ S + +
Sbjct: 1128 LTNQPLRTILHSPSQSGRLAKWAVELSEYDIAYKNRTCAKSHRAPTRPYRRSIPCRKVDL 1187
Query: 657 AEWILSVDGSSYLKGSGAGVVLEGPGGVIIEQSLKFDFKASNNQAEYEAIIAGINLAIEM 716
A F ASNN+AEYEA+IAG+ LA +
Sbjct: 1188 AR-----------------------------------FPASNNEAEYEALIAGLRLAHGI 1212
Query: 717 NVHCLVIKTDSQLVANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENTRAD 776
V + DSQLVA+Q G+Y+AK+ ++ YL +EL + ++ +PR +N AD
Sbjct: 1213 EVKKIQAYCDSQLVASQFSGNYEAKNERMDAYLKVVRELSYNFEVFELTKIPRSDNAPAD 1272
Query: 777 VLCKLASTKKPGNNKSV-----------IQETLKSPS--------INEDDVVMVT----- 812
L LAST P + + IQ T+ PS + +VMV
Sbjct: 1273 ALAVLASTSDPDLRRVIPVECIDVPSIKIQGTITKPSEYTVIDLTAEQLSIVMVILPPAT 1332
Query: 813 -------------------------------------GAAPSDWMDRIKMCLEADGADLA 835
A ++W++ I+ +
Sbjct: 1333 DRSEDAGPSGNLVCSSDPGQSSGCDRSDDSNRSADSDSPASTNWIEEIRSYIADRIVPKD 1392
Query: 836 LFSKDQVREAS-HYVLLGDQLYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSL 894
++ ++R S Y L + L R LL C+ +EA ++M E+HEG +H GGR+L
Sbjct: 1393 KWAARRLRARSAQYTLPHEHLLRWSATGVLLSCLDDEEAQQVMREIHEGAGGNHSGGRAL 1452
Query: 895 AAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILG 954
A K+ + G YWPT+ DC + +CEKCQ +A P EVL + +PF W +DI+
Sbjct: 1453 ALKIRKHGQYWPTMNADCEKFVARCEKCQGHAPFIHIPSEVLQTAMPPYPFMRWAMDIIR 1512
Query: 955 PFTPAGAQIRFVLVAVDYFTKWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGT 1014
PF PA Q +++LV DYFTKW EAES A+I +++V+ F W+ IIC G+P +V+DNGT
Sbjct: 1513 PF-PASRQKKYILVMTDYFTKWFEAESYARIQSKEVQNFVWKNIICHHGLPYEIVTDNGT 1571
Query: 1015 QFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLT 1074
QFTS FC + I + ++ +PQ NGQ EA NK IL+G+KKRL + KG WADEL
Sbjct: 1572 QFTSLQFEGFCAKWKIRLSKSTPRYPQCNGQAEATNKTILDGLKKRLDEKKGAWADELDG 1631
Query: 1075 VVWAYNTTPQSTTGETPFRLTYGVDAMVPVEIQDMTFR--VAAYDENENHENRLIDLNLA 1132
V+W+Y TTP+ +T TPF LTYG++A+ P E T R + D N + L +L+
Sbjct: 1632 VLWSYRTTPRRSTDRTPFSLTYGMEALAPCEAGLPTVRRSMLINDPTLNDQMLLDNLDTL 1691
Query: 1133 EEVKTEVRLRQAAVKQRSERRYNTRVVPRHMQVGDLVLRRKAKG---PDDSKLSPNWEGP 1189
EE + + LR +Q + + YN +V R GDLVLR+ + D KL +WEGP
Sbjct: 1692 EEQRDQALLRIQNYQQAAAKFYNKKVKNRLFAEGDLVLRKVFENTVEQDAGKLGAHWEGP 1751
Query: 1190 YRILRDLGQGAYHLEELSGRRIPRAWNAQHLRYYY 1224
Y I + + G Y L + G IPR+WN+ +L+ YY
Sbjct: 1752 YLISKVVKPGVYELLTMDGTPIPRSWNSANLKRYY 1786
>At3g31530 hypothetical protein
Length = 831
Score = 546 bits (1407), Expect = e-155
Identities = 314/820 (38%), Positives = 465/820 (56%), Gaps = 101/820 (12%)
Query: 332 MYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVK 391
M D+EKTAF T Q +CY+ MPFGLKNAGATY+R ++++F Q+G+ ME+Y+DDM+VK
Sbjct: 96 MNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKIFALQIGKTMEVYIDDMLVK 155
Query: 392 SEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAIL 451
S H L E F ++ +N++LNP KC FG++SG IE NP + +A+L
Sbjct: 156 SMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRSG-----------IEANPKQIEALL 204
Query: 452 EMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKE 511
M SP + +EVQ L GR+AAL+RF+ S K F+ LR+N+ F+WT CE+AFQ LK+
Sbjct: 205 GMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRRNKKFEWTTRCEEAFQELKK 264
Query: 512 LLSKPPILSRPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKI 571
L+ PPIL++P+ G P +++++SD AVS L++E + E ++I++VS AE RY ++
Sbjct: 265 YLATPPILAKPVIGEPQYLYVAVSDTAVSGELVREDRGEQKLIFYVSQTFTSAESRYPQM 324
Query: 572 EKAALALIISARKLRPYFQGFQIKVKTDFPLRQVLQKPDLAGRMVSWAVELSEFGIVFEK 631
EK ALA+++SA+KLRPYFQ I V PLR +L P +GR+ W +ELSE+ I ++
Sbjct: 325 EKLALAVVMSAQKLRPYFQSHSIIVMGSMPLRVILHSPSQSGRLAKWTIELSEYDIEYQN 384
Query: 632 KGQVKAQVLADFVNEMSPEVKVSE---EAEWILSVDGSSYLKGSGAGVVLEGPGGVIIEQ 688
K K+QVLADF+ E+ P + E + W+L VDGSS +GSG G+ L P G
Sbjct: 385 KTCAKSQVLADFIVEL-PTKEARENPLDTTWLLHVDGSSSKQGSGVGIRLTSPTG----- 438
Query: 689 SLKFDFKASNNQAEYEAIIAGINLAIEMNVHCLVIKTDSQLVANQIKGDYQAKDIQLAKY 748
+A+NN AEYEA++AG+NLA + + DSQL+ANQ G+Y +D ++ Y
Sbjct: 439 ------EATNNVAEYEALVAGLNLAWGLKIGKTRAFCDSQLIANQFNGEYTTQDKKMEAY 492
Query: 749 LTKTQELMKRMDSVQVNHVPREENTRADVLCKLASTKKPGNNKSVIQETLKSPSI---NE 805
L Q L K D ++ +PR ENT AD L LAST + + E ++ PSI E
Sbjct: 493 LIHVQNLAKNFDEFELTRIPRGENTSADALAALASTSDTSLKRVIPVEFIEKPSIELGKE 552
Query: 806 DDVVMVTGAAP--------SDWMDRIKMCLEADGADLALFSKDQVR-EASHYVLLGDQLY 856
+ V+ + +A S+WM+ I + + +++ +A+ +VL+ +LY
Sbjct: 553 EHVLPIQISADQDDPDDCNSEWMEPIISYISEGKLPSDKWKARKLKAQAARFVLVDTKLY 612
Query: 857 RRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYA 916
+ + PL+ CV + +IM E+H G SH PT+
Sbjct: 613 KWRLSGPLMTCVEAEAICKIMKEIHSG---SHA----------------PTI-------- 645
Query: 917 KKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLVAVDYFTKW 976
P E+LSS++S +PF W +DI+GP P+ Q + VLV DYF+KW
Sbjct: 646 -------------HQPAELLSSIASPYPFMRWSMDIIGPMHPS-KQKKLVLVLTDYFSKW 691
Query: 977 IEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFAS 1036
IEAES A I +V+ F + I+CR G+P +V+DNG+QF S + FC++ GI + ++
Sbjct: 692 IEAESYASIKDAQVENFVLKHILCRHGIPYEIVTDNGSQFISTRFQGFCDKWGIRLSKST 751
Query: 1037 VEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTY 1096
+PQ NGQ EAAN EL V+W + TTP+ T ETPF Y
Sbjct: 752 PRYPQGNGQAEAAN--------------------ELEGVLWLHRTTPRRATRETPFASIY 791
Query: 1097 GVDAMVPVEIQDMTFRVAAYDEN--ENHENRLIDLNLAEE 1134
G + ++P E+ + R + EN +N + L +L+L +E
Sbjct: 792 GTECVIPAEMIVPSLRRSLSPENDPDNTQTPLDELDLIDE 831
Score = 37.0 bits (84), Expect = 0.069
Identities = 30/118 (25%), Positives = 49/118 (41%), Gaps = 24/118 (20%)
Query: 62 IKFLVVKCTSPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKGGVGILKADQVVAWRCYSE 121
+ F + + YN +IG P LN+ A+ ST HL +K+P S D V
Sbjct: 19 VDFSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKFPTS---------DSV-------- 61
Query: 122 SFKQYGHMGKRAVKEGHRVYGVDIDQEGVCLDPREGFLEHKMTPEEETKTVKVGERNL 179
+++ GH AVK +++ F+ M PE++ KT E+ +
Sbjct: 62 -WEKLGHERAEAVKAENQLLETSYCHSWTL------FVVMMMNPEDQEKTAFYTEQGI 112
>At2g13330 F14O4.9
Length = 889
Score = 449 bits (1154), Expect = e-126
Identities = 235/530 (44%), Positives = 330/530 (61%), Gaps = 32/530 (6%)
Query: 277 KSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPD 336
+ NGKWR+C D+ DLNK CPKDS+PLP+ID+LV+ LS MDA+ GY+QI M
Sbjct: 289 EKNGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDG 348
Query: 337 EEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMG 396
+EKTAF+T++ Y Y+ MPFGLKNAG TYQRL++R+F Q+G+ +E+Y+DDM+VKS
Sbjct: 349 QEKTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEK 408
Query: 397 GHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSP 456
H L E F + K M+LNPEKCSF +QS +FL +++T RGIE NP + A +EM SP
Sbjct: 409 DHVPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSP 468
Query: 457 TSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKP 516
+EVQ+L GRI AL+ F+ S +K PF+Q LRK + F W +CEQAF+ LK L++P
Sbjct: 469 KMAREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTEP 528
Query: 517 PILSRPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAAL 576
PIL++P G PL ++ + + R +EK AL
Sbjct: 529 PILAKPEKGEPLYLYTN------------------------------RDKRCPAMEKLAL 558
Query: 577 ALIISARKLRPYFQGFQIKVKTDFPLRQVLQKPDLAGRMVSWAVELSEFGIVFEKKGQVK 636
+++S RKLR YFQ I V T P+R +L +GR+ WA+EL E+ I + + +K
Sbjct: 559 TVVMSVRKLRLYFQSHPIVVMTSQPIRTILHSLTQSGRLAKWAIELREYDIEYRTRTSLK 618
Query: 637 AQVLADFVNE--MSPEVKVSEEAEWILSVDGSSYLKGSGAGVVLEGPGGVIIEQSLKFDF 694
AQVLADFV + ++ + +W+L VDGSS +GSG G+ L P G +IEQSL+ F
Sbjct: 619 AQVLADFVIKLPLADLDGTNSNKKWLLHVDGSSNRQGSGVGIQLTSPTGEVIEQSLQLGF 678
Query: 695 KASNNQAEYEAIIAGINLAIEMNVHCLVIKTDSQLVANQIKGDYQAKDIQLAKYLTKTQE 754
ASNN++EYEA+IAGI LA E + + +DSQLV +Q G+Y+AKD ++ YL +
Sbjct: 679 NASNNESEYEALIAGIKLAQEKGIREIHAYSDSQLVTSQFHGEYEAKDERMEAYLELVKT 738
Query: 755 LMKRMDSVQVNHVPREENTRADVLCKLASTKKPGNNKSVIQETLKSPSIN 804
L ++ +S ++ +PR ENT AD L LAST P + + E ++ SI+
Sbjct: 739 LAQQFESFKLTRIPRGENTSADTLAALASTSDPFVKRIIPVEGIEHTSID 788
Score = 85.5 bits (210), Expect = 2e-16
Identities = 64/220 (29%), Positives = 106/220 (48%), Gaps = 21/220 (9%)
Query: 1 DQGSSADLIYGDAYEKLGLTEADLLPYDGALVGFSGERVFVRGYVELNTVFGEGINAESF 60
D GSS D+++ DA+++ G ++ L L GF+G+ F G ++L T+ G+ +
Sbjct: 82 DTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTGFAGDTTFSIGTIQLPTI-ARGVRQLT- 139
Query: 61 AIKFLVVKCTSPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKGGVGILKADQVVAWRCYS 120
FLVV +P+N ++GRP L+ + A+ ST H +K+P K G+ ++ Q + +CY
Sbjct: 140 --NFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQCIKFPSYK-GIAVVYGSQRSSRKCYM 196
Query: 121 ESFKQYGH------MGKRAVKEGHRVYGVDIDQEGVCLDPREGFLEHKMTPEEETKTVKV 174
S++ M + + E V +D Q G R+ + E + K
Sbjct: 197 GSYEDIKKADPVVLMIEDELAEMKTVRSLDPSQRGT----RKSLITQVCIDESDPK---- 248
Query: 175 GERNLKVGVNLTAIQEARLTQLLAENMDLFAWSAQDLPGI 214
R + +G +L L L EN D FAWS+ +L GI
Sbjct: 249 --RCVGIGHDLDLTVREDLITFLKENKDSFAWSSANLQGI 286
>At2g11940 putative retroelement gag/pol polyprotein
Length = 1212
Score = 428 bits (1101), Expect = e-120
Identities = 297/1040 (28%), Positives = 483/1040 (45%), Gaps = 200/1040 (19%)
Query: 193 LTQLLAENMDLFAWSAQDLPGIDPNFICHKLALNPGVKPIAQMKRKMGEEKAQAVKAETN 252
L Q L +N FAW+ +++ GI I H+L ++P KP+ Q ++K G ++A+AV +
Sbjct: 364 LIQFLEKNKSTFAWTIREMTGISTEVISHELNVDPTFKPVKQKRQKHGPDRAEAVNVKVV 423
Query: 253 KLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRA 312
+L+ A D +PLP+ID+ ++
Sbjct: 424 RLLKA----------------------------------------DCFPLPHIDRPIEST 443
Query: 313 SGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRV 372
+G MLS MDA+SGY+QI M D+EKT+F+T + Y Y+ MPFGLKN GATYQ L++ +
Sbjct: 444 TGQEMLSFMDAFSGYNQIMMNPDDQEKTSFITERGTYYYKVMPFGLKNVGATYQLLVNIM 503
Query: 373 FEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLG 432
F+ +G+ ME+Y+DDM+VK+ H + F + + M+LNP KC+F + S +FLG
Sbjct: 504 FKDLLGKTMEVYIDDMLVKTAFSASHLDHFQQCFDILNEFGMKLNPLKCTFAVPSREFLG 563
Query: 433 FMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRK 492
F I + DKC PF LRK
Sbjct: 564 F------ISRSTDKC------------------------------------LPFCTLLRK 581
Query: 493 -NRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNAVSSVLLQECKDEL 551
N+ F W + CE+AF+ LK LS+P +L++P G L ++IS+S+NA+S VL++ + +
Sbjct: 582 VNKGFVWDERCEEAFRQLKSYLSEPLVLAKPEFGERLFLYISVSENAISGVLVRVERSDE 641
Query: 552 RIIYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTDFPLRQVLQKPDL 611
R I++VS + E RY +EK ALA++ +ARKLRPYFQ I V T PLR +L P+
Sbjct: 642 RPIFYVSKSFSDVETRYPMMEKLALAVVTAARKLRPYFQSHPIVVLTSLPLRTILHSPNQ 701
Query: 612 AGRMVSWAVELSEFGIVFEKKGQVKAQVLADFVNEMSPEVKVSEEAEWILSVDGSSYLKG 671
+GR+ WA+ELSEF + F + +K+QVLADF+ E+ L+ S Y
Sbjct: 702 SGRLAKWAIELSEFNLEFHARTSLKSQVLADFLIELP------------LATTASDY--- 746
Query: 672 SGAGVVLEGPGGVIIEQSLKFDFKASNNQAE-YEAIIAGINLAIEMNVHCLVIKTDSQLV 730
SG D++A +N+ E Y ++ + E + +
Sbjct: 747 SG-------------------DYEAKDNRMEAYLDLLRELAEKFE-KFELIKVPRAENSA 786
Query: 731 ANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENTRADVLCKLASTKKPGNN 790
A+ + +I + + ++ + ++N V R + +V ++ ++
Sbjct: 787 ADALAALASTPEITVRRIISVETIAQPNIRIEEINFVTRAMRRQLEVQANNPELQQL-HD 845
Query: 791 KSVIQETLKSPSINEDDVVMVTGAAPSDWMDRIKMCLEADGADLALFSKDQVREASHYVL 850
+ I E + + + + +DW + I+ + + +++
Sbjct: 846 EDEIPEAVPLVADEDSTEYNLPHDWGADWREPIRNYIFNGVLPTDKWEARKLKATCARFC 905
Query: 851 LGDQ-LYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLK 909
L D LYRR + P C+ ++ ++ E+H+G C +H GGRSLA K+ + G++WPT+
Sbjct: 906 LADDILYRRIISAPDAICIFGEQTRTVIKEIHDGTCGNHSGGRSLAFKLKKYGYFWPTMV 965
Query: 910 NDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLVA 969
DC YA + + + S PF W +DI+GP + +RF+L+
Sbjct: 966 ADCEAYAHRSRRTFHFG-------------ISPLPFMKWSMDIVGPLHVSTRGVRFLLL- 1011
Query: 970 VDYFTKWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMG 1029
+W+EA + + IT +V+ F W++IICR G+P +V+DNG QF S + F E
Sbjct: 1012 -----EWVEAAAYSYITQVQVRHFIWKEIICRHGLPYEIVTDNGPQFISEQFKAFYAEWQ 1066
Query: 1030 IEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGE 1089
I + ++ +PQ G VEA
Sbjct: 1067 IRLNRSTPRYPQ--GNVEA----------------------------------------- 1083
Query: 1090 TPFRLTYGVDAMVPVEIQDMTFRVAAYDENENHENRLI--DLNLAEEVKTEVRLRQAAVK 1147
++P EI+ R +NE N+++ ++ +E + +R
Sbjct: 1084 -----------VIPAEIKVPRTRRIRNPKNEAANNKMMVDVIDTIDERRNRALVRMQNYH 1132
Query: 1148 QRSERRYNTRVVPRHMQVGDLVLRRKAKGPDD---SKLSPNWEGPYRILRDLGQGAYHLE 1204
+ + YN+ V R VG LVLRR + + KL +WEGPY+I + G L
Sbjct: 1133 NAAAQYYNSNVRSRSFDVGTLVLRRIHQNTAEKGAGKLGISWEGPYKITHIVRNGDC-LI 1191
Query: 1205 ELSGRRIPRAWNAQHLRYYY 1224
+ G+ + RAWNA HL+ +Y
Sbjct: 1192 NMEGKTVRRAWNAAHLKRFY 1211
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 360 bits (923), Expect = 4e-99
Identities = 167/322 (51%), Positives = 240/322 (73%)
Query: 236 KRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHC 295
+RK+G E+A+AV E +KL+ G IREV+YP WLAN V+VKK NGK R+C D+TDLNK C
Sbjct: 469 RRKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKAC 528
Query: 296 PKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMP 355
PKDS+PLP+ID+LV+ +G +LS MDA+SGY+QI M D+EKT F+T++ YCY+ MP
Sbjct: 529 PKDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMP 588
Query: 356 FGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMR 415
FGL+NAGATY RL++++F VG+ ME+Y+DDM++KS + H L E F + ++ M+
Sbjct: 589 FGLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMK 648
Query: 416 LNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRF 475
LNP KC+FG+ SG+FLG+++T+RGIE NP++ A L M SP + KEVQ+L GRIAAL+RF
Sbjct: 649 LNPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRF 708
Query: 476 LPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISIS 535
+ S K+ PF+Q L+ N+ F W ++CE+AF LK L+ PP+L +P L +++S+S
Sbjct: 709 ISRSTDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVS 768
Query: 536 DNAVSSVLLQECKDELRIIYFV 557
++AVS VL++E + E + IY++
Sbjct: 769 NHAVSGVLIREDRGEQKPIYYI 790
Score = 289 bits (740), Expect = 6e-78
Identities = 174/530 (32%), Positives = 258/530 (47%), Gaps = 84/530 (15%)
Query: 729 LVANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENTRADVLCKLASTKKPG 788
++ NQ GDY+AKD ++ YL ++L K ++ +PR +NT AD L LAST P
Sbjct: 790 IIVNQFNGDYEAKDSRMEAYLHVVKDLAKNFRKFELIRIPRGQNTTADALAALASTSDPE 849
Query: 789 NNKSVIQETLKSPSINEDDVVMVTGAAPSDWMDRIKMCLEADGADL-----ALFSKDQVR 843
N + E + SI E+ V + + DR +E A K V+
Sbjct: 850 VNSIIPVECISERSIKEEKEAFVVTRSRAAARDRGDTAVELPPVKRRKSKEATVPKPAVQ 909
Query: 844 EASHYVLLGD--------------------------------QLYRRGVGVPLLRCVTRD 871
E L+ + LYRRGV P L +
Sbjct: 910 ENIGIELIEEVISDAKIEAETEPWVDEDILAPNEHPEPEALNSLYRRGVSDPYLLSIFGP 969
Query: 872 EADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRA 931
+ +M EVHEG+C SH GR++A K+ + +YWPT+ DC+ YA++C++CQ++A L
Sbjct: 970 VVEIVMSEVHEGLCGSHSSGRAMAFKIKKMCYYWPTMITDCVKYAQRCKRCQLHAPLIHQ 1029
Query: 932 PPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLVAVDYFTKWIEAESMAKITAEKVK 991
P E+ SS+S+ +PF W +DI+GP + ++++LV DYF+KWIEAE++
Sbjct: 1030 PSELFSSISAPYPFMRWSMDIIGPLHRSTRGVQYLLVLTDYFSKWIEAEAIL-------- 1081
Query: 992 KFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANK 1051
W GI++ ++++ +PQ NGQ EAANK
Sbjct: 1082 -LNW---------------------------------GIKVSYSTLRYPQGNGQAEAANK 1107
Query: 1052 VILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDAMVPVEIQDMTF 1111
IL+ +KKRL KG W DEL V+WAY TTP+ +TGETPF L YG+ A+VP E+
Sbjct: 1108 TILSNLKKRLNHLKGGWYDELQPVLWAYRTTPRRSTGETPFSLVYGMKAVVPAELNVPGL 1167
Query: 1112 RVAAYDENENHENRLID--LNLAEEVKTEVRLRQAAVKQRSERRYNTRVVPRHMQVGDLV 1169
R NE + ++D L+ E + + ++ + + R YN++V R VGD V
Sbjct: 1168 RRTEAPLNEKENSAMLDDSLDTINERRDQALIQIQNYQHAAARYYNSKVKSRPFFVGDYV 1227
Query: 1170 LRR---KAKGPDDSKLSPNWEGPYRILRDLGQGAYHLEELSGRRIPRAWN 1216
L+R K KL NWEGPY + + G Y L++L R + R WN
Sbjct: 1228 LKRVFDNKKEEGAGKLGINWEGPYIVTEVVRNGVYRLKDLEDRPVQRPWN 1277
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 339 bits (870), Expect = 5e-93
Identities = 157/291 (53%), Positives = 218/291 (73%), Gaps = 1/291 (0%)
Query: 208 AQDLPGIDPNFICHKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPT 267
A D+ GIDP CH+L ++P K + Q +RK+G E+++AV E +KL+DAG I EVKYP
Sbjct: 474 AHDMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPE 533
Query: 268 WLANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGY 327
WLAN V+VKK N KWR+C D+TDLNK CPKDS+PLP+ID++V+ +G +LS MDA+SGY
Sbjct: 534 WLANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGY 593
Query: 328 HQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDD 387
+QI M+ D+EKT+F+ ++ YCY+ MPFGLKN GA YQRL++++F Q+G+ ME+Y+DD
Sbjct: 594 NQIPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDD 653
Query: 388 MVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKC 447
M+VKS H L F + K+NM+LNP KC FG+ SG+FLG+++T+RGIE NP +
Sbjct: 654 MLVKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQI 713
Query: 448 KAILEMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRK-NRVFQ 497
+AIL++QSP + KEVQ+L GRIA L+RF+ S K+ PF+Q LR N+ F+
Sbjct: 714 RAILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLRSANKTFE 764
Score = 53.5 bits (127), Expect = 7e-07
Identities = 30/103 (29%), Positives = 52/103 (50%), Gaps = 4/103 (3%)
Query: 1 DQGSSADLIYGDAYEKLGLTEADLLPYDGALVGFSGERVFVRGYVELNTVFGEGINAESF 60
D GSS ++++ D +K+ + + + P L GF G + G ++L G
Sbjct: 365 DTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTNGTIKLPIYLGGAATWH-- 422
Query: 61 AIKFLVVKCTSPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKG 103
KF+VV + YN+++G P ++ + AI S+ H +K P S G
Sbjct: 423 --KFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG 463
>At1g37200 hypothetical protein
Length = 1564
Score = 330 bits (845), Expect = 4e-90
Identities = 178/398 (44%), Positives = 252/398 (62%), Gaps = 23/398 (5%)
Query: 71 SPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKGGVGILKADQVVAWRCYSESFKQYGH-- 128
+P+N ++GRP L+ + A+ ST H +K+P KG + ++ Q + RCY S++
Sbjct: 530 APFNAILGRPWLHAMKAVPSTYHQCIKFPSEKG-IAVIYGSQRSSRRCYMGSYELIKKAE 588
Query: 129 ----MGKRAVKEGHRVYGVDIDQEGVCLDPREGFLEHKMTPEEETKTVKVGERNLKVGVN 184
M + + E V D Q G P++ + E + K + VG +
Sbjct: 589 LVVLMIEDKLAEMKTVRSSDPSQCG----PQKSSITQVYIDESDPKQC------VGVGQD 638
Query: 185 LT-AIQEARLTQLLAENMDLFAWSAQDLPGIDPNFICHKLALNPGVKPIAQMKRKMGEEK 243
L AI+E +T L EN D FAWS+ +L GI H+L ++P +PI Q +RK+G E+
Sbjct: 639 LDPAIREDFIT-FLKENKDSFAWSSANLQGISLEVTSHELNVDPTYRPIKQKRRKLGPER 697
Query: 244 AQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLP 303
A+AV+ E ++L+ G IREVKYP WLAN V+VK NGKWR+C D+ DLNK CPKDS+PLP
Sbjct: 698 AKAVQDEVDRLLKIGSIREVKYPDWLANPVVVKNKNGKWRVCIDFMDLNKACPKDSFPLP 757
Query: 304 NIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGA 363
+ID+LV +G +LS MDA+SGY+QI M D+EKTAF+T+ CY+ MPFGLKN GA
Sbjct: 758 HIDRLVKATAGHELLSFMDAFSGYNQILMRPDDQEKTAFITD----CYKVMPFGLKNTGA 813
Query: 364 TYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSF 423
TYQRL++R+F Q+G+ ME+Y+DDM+VKS H L E F + K M+LNPEKCSF
Sbjct: 814 TYQRLVNRMFADQLGKTMELYIDDMLVKSAHEKDHLPQLRECFKILNKFEMKLNPEKCSF 873
Query: 424 GIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKE 461
G+ SG+FLG+++T+RGIE NP + A ++M SP ++
Sbjct: 874 GVPSGEFLGYLVTQRGIEANPKQIAAFIDMPSPKMARD 911
Score = 264 bits (675), Expect = 2e-70
Identities = 174/509 (34%), Positives = 260/509 (50%), Gaps = 56/509 (11%)
Query: 554 IYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTDFPLRQVLQKPDLAG 613
IY+VS L AE Y+ +EK ALA+++SARKLR Y Q I V T P+R +L +
Sbjct: 934 IYYVSKTLIDAETHYRAMEKLALAVVMSARKLRQYLQSHPIVVMTSQPIRTILHSTTQSS 993
Query: 614 RMVSWAVELSEFGIVFEKKGQVKAQVLADFVNEM--SPEVKVSEEAEWILSVDGSSYLKG 671
R+ WA+ELSE+ I + + KAQVLADFV E+ + + +W+L VDGSS +G
Sbjct: 994 RLAKWAIELSEYDIEYRTRTSFKAQVLADFVIELPLADLDGTNSNKKWLLHVDGSSNRQG 1053
Query: 672 SGAGVVLEGPGGVIIEQSLKFDFKASNNQAEYEAIIAGINLAIEMNVHCLVIKTDSQLVA 731
SG G+ L P G +IEQS + F ASNN++EYEA+I GI LA M + + +DSQLV
Sbjct: 1054 SGEGIQLTSPTGEVIEQSFRLGFNASNNESEYEALIDGIKLAQGMRIRDIHAHSDSQLVT 1113
Query: 732 NQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENTRADVLCKLASTKKPGNNK 791
+Q G+Y+AKD ++ YL + L ++ +S ++ +PR ENT D L LAST P +
Sbjct: 1114 SQFHGEYEAKDERMEAYLELVKTLTQQFESFELTRIPRGENTSTDALAALASTLDPFVKR 1173
Query: 792 SVIQETLKSPSINEDDVVMVTGAAPSDWMD--RIKMCLEADGADLALFSKDQVREASHYV 849
+ E ++ PSI D V G S + R+ A LA ++ E +
Sbjct: 1174 IIPVEGIEHPSI--DLTVKHAGMETSATCNFTRVTRSTTAAARALAAAAEAGAAEEEAKL 1231
Query: 850 LLGDQL------------------YRRGVGVPLLRCVTRDEADRIMFEVHEG-------- 883
+QL RG+ P + +A + + EG
Sbjct: 1232 RTPEQLASYEPEPYNDWRIPIIDYIERGITPPDKWETRKLKAQSARYCIMEGRLMKRSVA 1291
Query: 884 ----VC-----------ASHVG--GRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYA 926
VC + H G G + + LR + DC+ ++ +C+KCQ +A
Sbjct: 1292 GPYMVCTYGQQTKDLMKSMHEGQCGSHYSGRTLRC-----IMLADCIAHSLRCDKCQRHA 1346
Query: 927 DLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRF--VLVAVDYFTKWIEAESMAK 984
PPE +SS+SS +PF W +D++ P +G + R +LV DY TKWIE ++ +
Sbjct: 1347 PTLHQPPEEMSSISSPYPFMKWSMDVVRPMEASGGKKRLKNLLVLTDYSTKWIEGKAFQQ 1406
Query: 985 ITAEKVKKFYWRKIICRFGVPATLVSDNG 1013
+T ++V+ F I+ R G+P +V+DNG
Sbjct: 1407 VTEKQVEVFLLENIVYRHGIPYEIVTDNG 1435
Score = 57.8 bits (138), Expect = 4e-08
Identities = 33/96 (34%), Positives = 52/96 (53%), Gaps = 4/96 (4%)
Query: 1133 EEVKTEVRLRQAAVKQRSERRYNTRVVPRHMQVGDLVLRR---KAKGPDDSKLSPNWEGP 1189
+E + ++ +Q + R YN+ + R +VG+LVLR+ + + KL NWEGP
Sbjct: 1468 DERRDHAMIQVQNYQQPAVRYYNSNIKIRRFEVGELVLRKVFSNTRELNAGKLGTNWEGP 1527
Query: 1190 YRILRDLGQGAYHLEEL-SGRRIPRAWNAQHLRYYY 1224
YRI + G Y L ++ +G R WNA HL+ Y+
Sbjct: 1528 YRITEVVRDGIYKLVKVFNGVPELRPWNAMHLKKYH 1563
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 315 bits (808), Expect = 8e-86
Identities = 188/561 (33%), Positives = 280/561 (49%), Gaps = 88/561 (15%)
Query: 731 ANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPREENTRADVLCKLASTKKPGNN 790
AN+ G+Y+AKD + YL +E+ R + ++ +PR EN+ A+ L LAST +
Sbjct: 789 ANRYSGEYEAKDACMEAYLNLVREVSGRFEQFELTRIPRAENSAANALAALASTFEVTLP 848
Query: 791 KSVIQETLKSPSINEDDVVMVTGAAPSDWMDRIKMCLEADGADLALF---SKDQVREASH 847
+ + ET+ PSI D++ VT A ++ L+A A+ L +++ +A H
Sbjct: 849 RVIPVETISQPSIRLDEISFVTTRA-------MRRRLDAQSAENGLHQLGDDEEISDAVH 901
Query: 848 ------------------------------------YVLLG------------------- 852
Y+L G
Sbjct: 902 PTEIVENQSLPDNHNAPLPDQPPHDWGADWREPIRDYILNGTLPAEKWAARKLKATCARF 961
Query: 853 ----DQLYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTL 908
D LYRR P C+ ++ +M EVH+G C +H GGRSLA KV + G+YWPTL
Sbjct: 962 CIANDILYRRIFSAPDAVCIFGEQTRTVMKEVHDGTCGNHTGGRSLAFKVRKYGYYWPTL 1021
Query: 909 KNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLV 968
DC YA+KCE+CQ +A L P E+L+++S+ +PF W +DI+GP
Sbjct: 1022 VADCEAYARKCEQCQKHAPLILQPAELLTTVSAPYPFMKWLMDIVGP------------- 1068
Query: 969 AVDYFTKWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEM 1028
+ T+ +EA + + IT +V F W+ IICR G+P +V+DNG+QF S FC E
Sbjct: 1069 -LHVSTRGVEAAAYSNITHVQVWNFIWKDIICRHGLPYEIVTDNGSQFISEQFEVFCEEW 1127
Query: 1029 GIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTG 1088
I + ++ +PQ NGQ EA NK I++ +KK+L KG W EL V+WA TTP+ T
Sbjct: 1128 QIRLSHSTPRYPQGNGQAEAMNKTIISNLKKKLNAYKGAWFGELQNVLWAVRTTPRRATD 1187
Query: 1089 ETPFRLTYGVDAMVPVEIQDMTFRVAAYDENENHENRLI--DLNLAEEVKTEVRLRQAAV 1146
ETPF L YG++A++P EI+ + R +NE N +I ++ +E + R
Sbjct: 1188 ETPFSLIYGMEAVIPAEIKVPSARRIRNPQNETENNEMIIDVIDTIDERRNRALARMQNY 1247
Query: 1147 KQRSERRYNTRVVPRHMQVGDLVLRRKAKGPDD---SKLSPNWEGPYRILRDLGQGAYHL 1203
R YN+ V R +VG LVLRR + + KL +WEGPY+I + G Y L
Sbjct: 1248 HNAGARYYNSNVRNRSFEVGTLVLRRVQQNKAEKGAGKLGISWEGPYKITHVVRNGVYRL 1307
Query: 1204 EELSGRRIPRAWNAQHLRYYY 1224
+ G+ + RAWN+ HL+ +Y
Sbjct: 1308 INMEGKTVRRAWNSMHLKRFY 1328
Score = 183 bits (465), Expect = 5e-46
Identities = 85/151 (56%), Positives = 111/151 (73%)
Query: 211 LPGIDPNFICHKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLA 270
+ GI I H+L ++P KP+ Q +RK+G ++AQAV E +L++ G IREVKYP WLA
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 271 NVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQI 330
N V+VKK NGKWR+C D+TDLNK C KD +PLP+ID+LV+ +G MLS MDA+SGY+QI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 331 RMYAPDEEKTAFMTNQANYCYQTMPFGLKNA 361
M D+EKT+F+T Y Y+ MPFGLKNA
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
Score = 111 bits (277), Expect = 3e-24
Identities = 59/149 (39%), Positives = 92/149 (61%), Gaps = 8/149 (5%)
Query: 485 PFFQCLRK-NRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNAVSSVL 543
PF K ++ F W + CE+AF+ LK LS+P +L++ G L ++I++S++AV+ V
Sbjct: 617 PFLHFTAKISKGFIWNESCEEAFKQLKRYLSEPSVLAKREFGEQLFLYIAVSESAVTGVQ 676
Query: 544 LQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTDFPLR 603
++ + + R I++V + RY +EK ALA++ +ARKLRPYFQ I V T PLR
Sbjct: 677 VRVERSDKRPIFYV-------KTRYPMMEKLALAVVTAARKLRPYFQSHPIVVLTSLPLR 729
Query: 604 QVLQKPDLAGRMVSWAVELSEFGIVFEKK 632
+L P +GR+ WA+ELSEF + F +
Sbjct: 730 TILHSPTQSGRLGKWAIELSEFDLEFRAR 758
Score = 32.7 bits (73), Expect = 1.3
Identities = 15/42 (35%), Positives = 24/42 (56%)
Query: 62 IKFLVVKCTSPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKG 103
++F V + YNV++G P L ++ + ST H VK+P G
Sbjct: 369 VEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVG 410
>At4g03800
Length = 637
Score = 305 bits (782), Expect = 8e-83
Identities = 151/336 (44%), Positives = 228/336 (66%), Gaps = 21/336 (6%)
Query: 211 LPGIDPNFICHKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLA 270
+P IDP+ ICH+L ++P KP+ Q +RK+G E+A+AV + +KL+ G IREV+YP W+A
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 271 NVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQI 330
V+VKK NGK R+C D+TDLNK CPKDS+PLP+ID+LV+ +G +L+ MDA+ GY+QI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 331 RMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVV 390
M D+EKT+F+T++ GATYQ L++++F + + ME+ +DD +V
Sbjct: 441 MMNPEDQEKTSFITDR---------------GATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 391 KSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAI 450
KS + H L E F + ++ M+LN KC+FG+ SG+FLG+++T+RGIE NP++ A
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINAF 545
Query: 451 LEMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLK 510
L+ S + KEVQ+L GRIA+ S K+ PF+Q L+ N F W ++CE+AF+ LK
Sbjct: 546 LKTPSLRNFKEVQRLTGRIASRST------DKSLPFYQILKGNNGFLWDEKCEEAFRQLK 599
Query: 511 ELLSKPPILSRPIPGTPLSVFISISDNAVSSVLLQE 546
L+ PP+LS+P L +++ +S++AVS VL++E
Sbjct: 600 AYLTTPPVLSKPEADEKLYLYVFVSNHAVSGVLVRE 635
Score = 42.4 bits (98), Expect = 0.002
Identities = 30/103 (29%), Positives = 48/103 (46%), Gaps = 18/103 (17%)
Query: 1 DQGSSADLIYGDAYEKLGLTEADLLPYDGALVGFSGERVFVRGYVELNTVFGEGINAESF 60
D GSS DLI+ L LV F+ E G ++L + G+ S
Sbjct: 220 DTGSSVDLIF--------------LGPPSPLVAFTSESAMSLGTIKLPVLAGK----MSK 261
Query: 61 AIKFLVVKCTSPYNVLIGRPSLNKLGAIISTRHLTVKYPLSKG 103
+ F+V + YN+++G P + ++ A+ ST H +K+P S G
Sbjct: 262 IVDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWLKFPTSNG 304
>At2g13230 pseudogene
Length = 353
Score = 243 bits (619), Expect = 6e-64
Identities = 136/358 (37%), Positives = 204/358 (55%), Gaps = 26/358 (7%)
Query: 748 YLTKTQELMKRMDSVQVNHVPREENTRADVLCKLASTKKPGNNKSVIQETLKSPSINEDD 807
YL Q+ K+ ++ +PR ENT AD L LAST + + E ++ PSI D
Sbjct: 4 YLALVQDHAKQFHEFKLTRIPRGENTSADALAALASTSDLNMRRVIPVEFIEKPSIELDK 63
Query: 808 VV------MVTG-----------AAPSDWMDRIKMCLEADGADLALFSKDQVR-EASHYV 849
V +V G S+W++ I+ + L + +++ EA+ +V
Sbjct: 64 VEHAFPIQVVAGYEDASDNSPSLGCDSEWIEPIRNFISKRKVRLDKWEARKLKAEAARFV 123
Query: 850 LLGDQLYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLK 909
L+ ++L++ + PL+ CV ++ A ++M EVH G C ++ GGR+LA K+ R G++WPT+
Sbjct: 124 LVEEKLFKWRLSGPLMTCVEKEAARKVMKEVHGGSCGNNSGGRALAIKIKRLGYFWPTMI 183
Query: 910 NDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLVA 969
DC EKCQ A P E+LSS++S +P W +DI+GP A Q + VLV
Sbjct: 184 KDC-------EKCQRNAPTIHLPAELLSSIASPYPLMRWSMDIIGPMH-ASKQKKLVLVL 235
Query: 970 VDYFTKWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMG 1029
DYF+K EAES A I +V+ F W+ I+CR GVP +V+DNG+QF S + FC++ G
Sbjct: 236 TDYFSKLREAESYASIKEAQVESFMWKHILCRHGVPYEIVTDNGSQFISTRFQGFCDKWG 295
Query: 1030 IEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTT 1087
I + ++ + Q NGQ +AANK IL+G+KKRL KG W DEL V+W++ TTP+ T
Sbjct: 296 IRLSKSTPRYLQGNGQADAANKTILDGLKKRLTAKKGSWVDELEGVLWSHRTTPRRAT 353
>At2g10780 pseudogene
Length = 1611
Score = 236 bits (603), Expect = 5e-62
Identities = 149/495 (30%), Positives = 251/495 (50%), Gaps = 20/495 (4%)
Query: 223 LALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKW 282
+ L PG PI++ +M + +K + +L+D GFIR P W A V+ VKK +G +
Sbjct: 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSF 709
Query: 283 RMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAF 342
R+C DY LNK K+ YPLP ID+L+D+ G S +D SGYHQI + D KTAF
Sbjct: 710 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 769
Query: 343 MTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDL 402
T ++ + MPFGL NA A + ++M+ VF + + I+++D++V S+ H L
Sbjct: 770 RTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHL 829
Query: 403 AEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEV 462
+R+H + KCSF +S FLG +I+ +G+ V+P+K ++I E P + E+
Sbjct: 830 RAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 889
Query: 463 QKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRP 522
+ +G RF+ S A P + K+ F W+DECE++F LK +L+ P+L P
Sbjct: 890 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLP 949
Query: 523 IPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISA 582
G P +V+ S + VL+Q+ +I + S L+ E Y + A++
Sbjct: 950 EEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKHEKNYPTHDLEMAAVVFFL 1005
Query: 583 RKLRPYFQGFQIKVKTDF-PLRQVLQKPDLAGRMVSWAVELSEFGI-VFEKKGQVKAQVL 640
+ R Y G ++++ TD L+ + +P+L R W ++++ + + G KA +
Sbjct: 1006 KIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVADYNLDIAYHPG--KANQV 1063
Query: 641 ADFVNEMSPEVKVSEEAEWILSVDGSSYLK---------GSGAGVVLEGPGGVIIEQSLK 691
AD ++ EV+ ++++ G+ ++ G GA + + + Q
Sbjct: 1064 ADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLGLGAADQADLLSRIRLAQERD 1123
Query: 692 FDFK--ASNNQAEYE 704
+ K A NN+ EY+
Sbjct: 1124 EEIKGWAQNNKTEYQ 1138
Score = 117 bits (292), Expect = 5e-26
Identities = 101/373 (27%), Positives = 167/373 (44%), Gaps = 45/373 (12%)
Query: 867 CVTRDEA--DRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQI 924
CV D A + I+ E H+ + H G + + L+ ++W +K D + KC CQ+
Sbjct: 1152 CVPNDRALKEEILREAHQSKFSIHPGSNKMY-RDLKRYYHWVGMKKDVARWVAKCPTCQL 1210
Query: 925 YADLHRAPPEVLSSMSSAWPFAMWGVD------ILGPFTPAGAQIRFVLVAVDYFTKWIE 978
H+ P +L ++ P W D + G T ++ V V VD TK
Sbjct: 1211 VKAEHQVPSGLLQNL----PIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTK--S 1264
Query: 979 AESMA---KITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFA 1035
A MA K AE + + Y +I+ G+P ++VSD T+FTS+ + F +G + +
Sbjct: 1265 AHFMAISDKDGAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLS 1324
Query: 1036 SVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLT 1095
+ HPQ++ Q E + + + ++ + D G W L V +AYN + Q++ G +P+
Sbjct: 1325 TAYHPQTDEQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEAL 1384
Query: 1096 YGVDAMVPVEIQDMTFRVAAYDENENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERR-- 1153
YG P+ + E RL + +E ++ + +K+ +R+
Sbjct: 1385 YGRACRTPLCWTPV------------GERRLFGPTIVDETTERMKFLKIKLKEAQDRQKS 1432
Query: 1154 -YNTRVVPRHMQVGDLV----LRRKAKGPDDS--KLSPNWEGPYRILRDLGQGAYHLEEL 1206
N R QVGDLV + K G S KLSP + GPY+++ +G AY L+
Sbjct: 1433 YANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLD-- 1490
Query: 1207 SGRRIPRAWNAQH 1219
+P NA H
Sbjct: 1491 ----LPPKLNAFH 1499
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 231 bits (590), Expect = 1e-60
Identities = 150/486 (30%), Positives = 246/486 (49%), Gaps = 15/486 (3%)
Query: 183 VNLTAIQEARLTQLLAENMDLFAWSAQDLPGIDPNFIC-HKLALNPGVKPIAQMKRKMGE 241
V A+ + R+ Q E D+F Q L G+ P+ + L PG P+++ +M
Sbjct: 449 VGQVAVSDIRVVQ---EFQDVF----QSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAP 501
Query: 242 EKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYP 301
+ +K + L+ GFIR P W A V+ VKK +G +R+C DY +LN+ K+ YP
Sbjct: 502 AEMAELKKQLKDLLGKGFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYP 560
Query: 302 LPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNA 361
LP ID+L+D+ G S +D SGYHQI + D KTAF T ++ + MPFGL NA
Sbjct: 561 LPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNA 620
Query: 362 GATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKC 421
A + RLM+ VF+ + + I++DD++V S+ + L ++R+ + KC
Sbjct: 621 PAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKC 680
Query: 422 SFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFLPCSGS 481
SF + FLG +++ G+ V+P+K +AI + PT+ E++ +G RF+ S
Sbjct: 681 SFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFAS 740
Query: 482 KATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNAVSS 541
A P + K+ F W+ ECE+ F SLKE+L+ P+L+ P G P V+ S +
Sbjct: 741 MAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGC 800
Query: 542 VLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTDF- 600
VL+Q K +I + S L E Y + A+I + + R Y G +++V TD
Sbjct: 801 VLMQHGK----VIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHK 856
Query: 601 PLRQVLQKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQVLADFVNEMSPEVKVSEEAEWI 660
L+ + +P+L R W ++++ + KA V+ D ++ + + E +
Sbjct: 857 SLKYIFTQPELNLRQRRWMELVADYDLEIAYH-PGKANVVVDALSRKRVGAALGQSVEVL 915
Query: 661 LSVDGS 666
+S G+
Sbjct: 916 VSEIGA 921
Score = 33.5 bits (75), Expect = 0.77
Identities = 23/72 (31%), Positives = 40/72 (54%), Gaps = 8/72 (11%)
Query: 1139 VRLRQAAVKQRSERRYNTRVVPRHMQVGDLVLRRKA--KGPD----DSKLSPNWEGPYRI 1192
+ +++A +QRS R + +VGD V + A +GP+ ++KLSP + GP++I
Sbjct: 1073 LNMKEAQDRQRSYADKRRREL--EFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFKI 1130
Query: 1193 LRDLGQGAYHLE 1204
+ + AY LE
Sbjct: 1131 VERVEPVAYRLE 1142
Score = 33.5 bits (75), Expect = 0.77
Identities = 36/152 (23%), Positives = 62/152 (40%), Gaps = 15/152 (9%)
Query: 899 LRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTP 958
L+ + W +K D + +C+ CQ L +A +VL M + P W D +
Sbjct: 965 LKRYYQWVGMKMDVANWVAECDVCQ----LVKAEHQVLGGMLQSLPIPEWKWDFITMDLV 1020
Query: 959 AGAQIR----FVLVAVDYFTKWIEAESMAKITAEKV--KKFYWRKIICRFGVPATL--VS 1010
G ++ + V VD TK ++ K V KKF +I+ GVP +
Sbjct: 1021 VGLRVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFV-SEIVKLHGVPLNMKEAQ 1079
Query: 1011 DNGTQFTSRIVRDFCNEMG--IEMRFASVEHP 1040
D + + R+ E+G + ++ A + P
Sbjct: 1080 DRQRSYADKRRRELEFEVGDRVYLKMAMLRGP 1111
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 225 bits (574), Expect = 1e-58
Identities = 148/482 (30%), Positives = 243/482 (49%), Gaps = 15/482 (3%)
Query: 187 AIQEARLTQLLAENMDLFAWSAQDLPGIDPNFIC-HKLALNPGVKPIAQMKRKMGEEKAQ 245
A+ + R+ Q E D+F Q L G+ P+ + L PG P+++ +M +
Sbjct: 479 AVSDIRVIQ---EFEDVF----QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMT 531
Query: 246 AVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNI 305
+K + L+ GFIR P W A V+ VKK +G +R+C DY LN K+ YPLP I
Sbjct: 532 ELKKQLEDLLGKGFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRI 590
Query: 306 DKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATY 365
D+L+D+ G S +D SGYHQI + D KTAF T ++ + MPF L NA A +
Sbjct: 591 DELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAF 650
Query: 366 QRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGI 425
RLM+ VF+ + + I++DD++V S+ H + L ++R+ + KCSF
Sbjct: 651 MRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQ 710
Query: 426 QSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATP 485
+ FLG +++ G+ V+P+K +AI + PT+ E++ + RF+ S A P
Sbjct: 711 REIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRFVKGFASMAQP 770
Query: 486 FFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNAVSSVLLQ 545
+ K+ F W+ ECE+ F SLKE+L+ P+L+ P G P V+ S + VL+Q
Sbjct: 771 MTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQ 830
Query: 546 ECKDELRIIYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTDF-PLRQ 604
K +I + S L+ E Y + +I + + R Y G +++V TD L+
Sbjct: 831 RGK----VIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKY 886
Query: 605 VLQKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQVLADFVNEMSPEVKVSEEAEWILSVD 664
+ +P+L R + W ++++ + KA V+AD ++ + E ++S
Sbjct: 887 IFNQPELNLRQMRWMELVADYDLEIAYH-PGKANVVADALSRKRVGAAPGQSVEALVSEI 945
Query: 665 GS 666
G+
Sbjct: 946 GA 947
Score = 122 bits (307), Expect = 1e-27
Identities = 97/350 (27%), Positives = 162/350 (45%), Gaps = 56/350 (16%)
Query: 867 CVTRDEADR--IMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQI 924
CV +DE R I+ E H + + H G + + L+ + W +K D + +C+ CQ+
Sbjct: 1008 CVPKDEELRREILSEAHASMFSIHPGATKMY-RDLKRYYQWVGMKRDVANWVAECDVCQL 1066
Query: 925 YADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVLVAVDYFTKWIEAESMAK 984
H+ P A+W V +D TK ++ K
Sbjct: 1067 VKAEHQVPD------------AIW-------------------VIMDRLTKSAHFLAIRK 1095
Query: 985 ITAEKV-KKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSN 1043
V K Y +I+ GVP ++VSD ++FT R F +MG +++ ++ HPQ++
Sbjct: 1096 TDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTD 1155
Query: 1044 GQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDAMVP 1103
GQ E + + + ++ + D G WAD L V +AYN + Q++ G PF YG P
Sbjct: 1156 GQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTP 1215
Query: 1104 V---EIQDMTFRVAAYDENENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERRYNTRVVP 1160
+ ++++ + A Y + R++ LN+ E + R R A K+R E +
Sbjct: 1216 LRWTQVEERSIYGADYVQETTERIRVLKLNMKE---AQARQRSYADKRRRELEF------ 1266
Query: 1161 RHMQVGDLVLRRKA--KGPD----DSKLSPNWEGPYRILRDLGQGAYHLE 1204
+VGD V + A +GP+ ++KLSP + GP+RI+ +G AY LE
Sbjct: 1267 ---EVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVERVGPVAYRLE 1313
>At3g11970 hypothetical protein
Length = 1499
Score = 224 bits (571), Expect = 2e-58
Identities = 145/456 (31%), Positives = 232/456 (50%), Gaps = 12/456 (2%)
Query: 221 HKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNG 280
HK+ L G P+ Q + + + L+ G ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 281 KWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKT 340
WR+C DY +LN KDS+P+P I+ L+D G + S +D +GYHQ+RM D +KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 341 AFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCL 400
AF T+ ++ Y MPFGL NA AT+Q LM+ +F+ + + + ++ DD++V S + H
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 401 DLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVK 460
L + F +R + + KC+F + ++LG I+ +GIE +P K KA+ E PT++K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 461 EVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILS 520
+++ +G RF+ G A P L K F+WT +QAF+ LK L + P+LS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 521 RPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALII 580
P+ V + +VL+QE + ++S L+G +L EK LA+I
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQEGHP----LAYISRQLKGKQLHLSIYEKELLAVIF 944
Query: 581 SARKLRPYFQGFQIKVKTD-FPLRQVLQKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQV 639
+ RK R Y +KTD L+ +L++ W +L EF + + Q K V
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQYR-QGKENV 1003
Query: 640 LADFVNEMSPEVKVSEEAEWILSVDGSSYLKGSGAG 675
+AD ++ V+ SE ++V LK AG
Sbjct: 1004 VADALS----RVEGSEVLHMAMTVVECDLLKDIQAG 1035
Score = 107 bits (266), Expect = 5e-23
Identities = 90/357 (25%), Positives = 157/357 (43%), Gaps = 24/357 (6%)
Query: 876 IMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEV 935
I+ +H H GR + + ++ FYW + D Y + C CQ A P +
Sbjct: 1084 ILLWLHGSGVGGH-SGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGL 1142
Query: 936 LSSMSSAWPFAMWG---VDILGPFTPAGAQIRFVLVAVDYFTKWIEAESMAK-ITAEKVK 991
L + P +W +D + +G + ++V VD +K +++ +A V
Sbjct: 1143 LQPLPI--PDTIWSEVSMDFIEGLPVSGGKT-VIMVVVDRLSKAAHFIALSHPYSALTVA 1199
Query: 992 KFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANK 1051
Y + G P ++VSD FTS R+F G+ ++ S HPQS+GQ E N+
Sbjct: 1200 HAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNR 1259
Query: 1052 VILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDAMVPVEIQDMTF 1111
+ ++ D LW+ L + YNT S++ TPF + YG V +
Sbjct: 1260 CLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGES 1319
Query: 1112 RVAAYDEN-ENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERRYN------TRVVPRHMQ 1164
+VA + + E+ L+ L ++ + R++Q A + R+ER + ++ P Q
Sbjct: 1320 KVAVVARSLQEREDMLLFLKF-HLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQ 1378
Query: 1165 VGDLVLRRKAKGPDDSKLSPNWEGPYRILRDLGQGAYHLEELSGRRIPRAWNAQHLR 1221
+V+R + KLSP + GPY+I+ G+ AY L S ++ ++ L+
Sbjct: 1379 --SVVMRA------NQKLSPKYFGPYKIIDRCGEVAYKLALPSYSQVHPVFHVSQLK 1427
>At1g36590 hypothetical protein
Length = 1499
Score = 224 bits (571), Expect = 2e-58
Identities = 145/456 (31%), Positives = 232/456 (50%), Gaps = 12/456 (2%)
Query: 221 HKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNG 280
HK+ L G P+ Q + + + L+ G ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 281 KWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKT 340
WR+C DY +LN KDS+P+P I+ L+D G + S +D +GYHQ+RM D +KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 341 AFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCL 400
AF T+ ++ Y MPFGL NA AT+Q LM+ +F+ + + + ++ DD++V S + H
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 401 DLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVK 460
L + F +R + + KC+F + ++LG I+ +GIE +P K KA+ E PT++K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 461 EVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILS 520
+++ +G RF+ G A P L K F+WT +QAF+ LK L + P+LS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 521 RPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALII 580
P+ V + +VL+QE + ++S L+G +L EK LA+I
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQEGHP----LAYISRQLKGKQLHLSIYEKELLAVIF 944
Query: 581 SARKLRPYFQGFQIKVKTD-FPLRQVLQKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQV 639
+ RK R Y +KTD L+ +L++ W +L EF + + Q K V
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQYR-QGKENV 1003
Query: 640 LADFVNEMSPEVKVSEEAEWILSVDGSSYLKGSGAG 675
+AD ++ V+ SE ++V LK AG
Sbjct: 1004 VADALS----RVEGSEVLHMAMTVVECDLLKDIQAG 1035
Score = 102 bits (254), Expect = 1e-21
Identities = 89/357 (24%), Positives = 157/357 (43%), Gaps = 24/357 (6%)
Query: 876 IMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEV 935
I+ +H H GR + + ++ FY + D Y + C CQ A P +
Sbjct: 1084 ILLWLHGSGVGGH-SGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGL 1142
Query: 936 LSSMSSAWPFAMWG---VDILGPFTPAGAQIRFVLVAVDYFTKWIEAESMAK-ITAEKVK 991
L + P +W +D + +G + ++V VD +K +++ +A V
Sbjct: 1143 LQPLPI--PDTIWSEVSMDFIEGLPVSGGKT-VIMVVVDRLSKAAHFIALSHPYSALTVA 1199
Query: 992 KFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANK 1051
+ Y + G P ++VSD FTS R+F G+ ++ S HPQS+GQ E N+
Sbjct: 1200 QAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNR 1259
Query: 1052 VILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDAMVPVEIQDMTF 1111
+ ++ D LW+ L + YNT S++ TPF + YG V +
Sbjct: 1260 CLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGES 1319
Query: 1112 RVAAYDEN-ENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERRYN------TRVVPRHMQ 1164
+VA + + E+ L+ L ++ + R++Q A + R+ER + ++ P Q
Sbjct: 1320 KVAVVARSLQEREDMLLFLKF-HLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQ 1378
Query: 1165 VGDLVLRRKAKGPDDSKLSPNWEGPYRILRDLGQGAYHLEELSGRRIPRAWNAQHLR 1221
+V+R + KLSP + GPY+I+ G+ AY L S ++ ++ L+
Sbjct: 1379 --SVVMRA------NQKLSPKYFGPYKIIDRCGEVAYKLALPSYSQVHPVFHVSQLK 1427
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 219 bits (559), Expect = 6e-57
Identities = 153/530 (28%), Positives = 256/530 (47%), Gaps = 24/530 (4%)
Query: 140 VYGVD-IDQEGVCLDPREGFLEHKMTPEEETKTVKVGERNLKVGVNLTAIQEARLTQLLA 198
+ G+D +D V LD G + + E V G R + ++A+Q ++ +
Sbjct: 388 ILGMDWLDHYRVHLDCHRGRVSFE---RPEGSLVYQGVRPTSGSLVISAVQAEKMIEKGC 444
Query: 199 ENMDLFAWSAQDLPGIDPNFIC-HKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDA 257
E F Q L G+ P+ + L PG P+++ +M + +K + L+ A
Sbjct: 445 EAYLEFEDVFQSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGA 504
Query: 258 GFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGM 317
V+ VKK +G +R+C DY LN K+ YPLP ID+L+D+ G
Sbjct: 505 P-------------VLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATC 551
Query: 318 LSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQV 377
S +D SGYH I + D KTAF T ++ + MPFGL NA A + RLM+ VF+ +
Sbjct: 552 FSKIDLTSGYHLIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVL 611
Query: 378 GRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITR 437
+ I++DD++V S+ + H + L ++R+ + KCSF + FLG +++
Sbjct: 612 DEFVIIFIDDILVYSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSA 671
Query: 438 RGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQ 497
G+ V+P+K +AI + +PT+ E++ +G RF+ S A P + K+ F
Sbjct: 672 EGVSVDPEKIEAIRDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFV 731
Query: 498 WTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYFV 557
W+ ECE+ F SLKE+L+ P+L+ P G P V+ S + VL+Q K +I +
Sbjct: 732 WSPECEEGFVSLKEMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGK----VIAYA 787
Query: 558 SHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTDF-PLRQVLQKPDLAGRMV 616
S L+ E Y + A+I + + R Y G ++V TD L+ + +P+L R
Sbjct: 788 SRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQR 847
Query: 617 SWAVELSEFGIVFEKKGQVKAQVLADFVNEMSPEVKVSEEAEWILSVDGS 666
W ++++ + KA V+AD ++ + E ++S G+
Sbjct: 848 QWMELVADYDLEIAYH-PGKANVVADALSHKRVGAAPGQSVEALVSEIGA 896
Score = 112 bits (280), Expect = 1e-24
Identities = 88/305 (28%), Positives = 145/305 (46%), Gaps = 24/305 (7%)
Query: 911 DCMGYAKKCEKCQIYADLHRAPPEVLSSMS-SAWPFAMWGVDILGPFTPAGAQIRFVLVA 969
D + +C+ CQ+ H+ P +L S+ W + +D + P + V
Sbjct: 942 DVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGL-PVSRTKDAIWVI 1000
Query: 970 VDYFTKWIEAESMAKITAEKV-KKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEM 1028
VD TK ++ K V K Y +I+ GVP ++VSD ++FTS R F EM
Sbjct: 1001 VDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEM 1060
Query: 1029 GIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTG 1088
G +++ ++ HPQ+ GQ E + + + ++ + D G WAD L V +AYN + ++ G
Sbjct: 1061 GTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASIG 1120
Query: 1089 ETPFRLTYGVDAMVPV---EIQDMTFRVAAYDENENHENRLIDLNLAEEVKTEVRLRQAA 1145
PF Y P+ ++ + + A Y + R++ LN+ E + R R A
Sbjct: 1121 MAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKE---AQDRQRSYA 1177
Query: 1146 VKQRSERRYNTRVVPRHMQVGDLVLRRKA--KGPD----DSKLSPNWEGPYRILRDLGQG 1199
K+R E + +VGD V + A +GP+ ++KLSP + GP+RI+ +G
Sbjct: 1178 DKRRRELEF---------EVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPV 1228
Query: 1200 AYHLE 1204
AY LE
Sbjct: 1229 AYRLE 1233
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 214 bits (544), Expect = 3e-55
Identities = 122/366 (33%), Positives = 192/366 (52%), Gaps = 5/366 (1%)
Query: 223 LALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKW 282
+ L PG PI++ +M + +K + +L+ GFIR P W A V+ VKK +G +
Sbjct: 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSF 204
Query: 283 RMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAF 342
R+C DY LNK K+ YPLP ID+L+D+ G S +D SGYHQI + D KTAF
Sbjct: 205 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 264
Query: 343 MTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDL 402
T ++ + MPFGL NA A + ++M+ VF + + I++DD++V S+ H L
Sbjct: 265 RTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHL 324
Query: 403 AEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEV 462
+R+H + K SF +S FLG +I+ +G+ V+P+K ++I E P + E+
Sbjct: 325 RAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 384
Query: 463 QKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRP 522
+ +G RF+ S A P + K+ F W+DECE++F LK +L P+L P
Sbjct: 385 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLP 444
Query: 523 IPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISA 582
G P +V+ S + VL+Q+ +I + S L+ E Y + A++ +
Sbjct: 445 EEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKHEKNYPTHDLEMAAVVFAL 500
Query: 583 RKLRPY 588
+ R Y
Sbjct: 501 KIWRSY 506
Score = 116 bits (291), Expect = 7e-26
Identities = 96/355 (27%), Positives = 164/355 (46%), Gaps = 33/355 (9%)
Query: 867 CVTRDEA--DRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQI 924
CV D A + I+ E H+ + H G + + L+ ++W ++ D + KC CQ+
Sbjct: 544 CVPNDRALKEEILREAHQSKFSIHPGSNKMY-RDLKRYYHWVGMRKDVARWVAKCPTCQL 602
Query: 925 YADLHRAPPEVLSSMS-SAWPFAMWGVDILGPFTPAGAQIRF--VLVAVDYFTKWIEAES 981
H+ P +L ++ S W + +D + P G + + V V VD TK A
Sbjct: 603 VKAEHQVPSGLLQNLPISEWKWDHITMDFVTRL-PTGIKSKHNAVWVVVDRLTK--SAHF 659
Query: 982 MA---KITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVE 1038
MA K AE + + Y +I+ G+P ++VSD T+FTS+ F +G + ++
Sbjct: 660 MAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLSTAY 719
Query: 1039 HPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGV 1098
HPQ++GQ E + + + ++ + D G W L + +AYN + Q++ G +P+ YG
Sbjct: 720 HPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALYGR 779
Query: 1099 DAMVPVEIQDMTFRVAAYDENENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERR---YN 1155
P+ + E RL + +E ++ + +K+ +R+ N
Sbjct: 780 ACRTPLCWTPV------------GERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYAN 827
Query: 1156 TRVVPRHMQVGDLV----LRRKAKGPDDS--KLSPNWEGPYRILRDLGQGAYHLE 1204
R QVGDLV + K G S KLSP + GPY+++ +G AY L+
Sbjct: 828 KRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLD 882
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 213 bits (542), Expect = 5e-55
Identities = 147/519 (28%), Positives = 232/519 (44%), Gaps = 60/519 (11%)
Query: 165 PEEETKTVKVGERNLKVGVN----------LTAIQEARLTQLLAENMDLFAWSAQDLPGI 214
P+ + K + G R + +G+N LTA Q L L + +S D+ GI
Sbjct: 823 PKVDLKPLPKGLRYVFLGLNSTYPVIVNDELTADQVNLLITELMKYRKAIGYSLDDIKGI 882
Query: 215 DPNFICHKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVM 274
P H++ L + +R++ + VK E KL+DAG I + TW++ V
Sbjct: 883 SPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHC 942
Query: 275 VKKSNGKW------------------RMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFG 316
V K G RMC +Y LN K+ +PLP ID +++R +
Sbjct: 943 VPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHP 1002
Query: 317 MLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQ 376
+D+YSG+ QI ++ D+ KT F + Y+ MPFGL NA AT+QR M +F
Sbjct: 1003 YYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDL 1062
Query: 377 VGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMIT 436
+ +E+++DD V L+L + N+ LN EKC F ++ G LG I+
Sbjct: 1063 IEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKIS 1122
Query: 437 RRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVF 496
GIEV+ K ++++Q P +VK+++ +G F+ A P + L K F
Sbjct: 1123 EEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEF 1182
Query: 497 QWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYF 556
+ DEC AF+ +KE L PI+ P P +
Sbjct: 1183 AFDDECLTAFKLIKEALITAPIVQAPNWDFPFEII------------------------- 1217
Query: 557 VSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTDF-PLRQVLQKPDLAGRM 615
+ A++RY EK LA++ + K R Y G ++ + TD LR + K D R+
Sbjct: 1218 ---TMDDAQVRYATTEKELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRL 1274
Query: 616 VSWAVELSEFGI-VFEKKGQVKAQVLADFVNEMSPEVKV 653
+ W + L EF + + +KKG +AD ++ M E +V
Sbjct: 1275 LRWILLLQEFDMEIVDKKGIENG--VADHLSRMRIEDEV 1311
Score = 166 bits (420), Expect = 8e-41
Identities = 107/380 (28%), Positives = 179/380 (46%), Gaps = 44/380 (11%)
Query: 848 YVLLGDQLYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPT 907
Y L D++YR CV+ DE + I+ H H +K+L+AGF+WP+
Sbjct: 1375 YTLCKDKIYRT--------CVSEDEIEGILLHCHGFAYGGHFATFKTMSKILQAGFWWPS 1426
Query: 908 LKNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTPAGAQIRFVL 967
+ D + KC+ + F +WG+D +GPF P+ +++L
Sbjct: 1427 MFKDAQEFISKCDSFE--------------------NFDVWGIDFMGPF-PSSYGNKYIL 1465
Query: 968 VAVDYFTKWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNE 1027
VA+DY +KW+EA + A V K + I RFGVP ++SD G F ++ + +
Sbjct: 1466 VAIDYVSKWVEAIASHTNDARVVLKLFKTIIFPRFGVPRIVISDGGKHFINKGFENLLKK 1525
Query: 1028 MGIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTT 1087
G++ + ++GQVE +N+ I ++K +G + W+ +L +WAY T ++
Sbjct: 1526 HGVKHK--------TSGQVEISNREIKAILEKTVGSTRKDWSAKLNDTLWAYRTAFKTPI 1577
Query: 1088 GETPFRLTYGVDAMVPVEIQDMTF---RVAAYDENENHENRLIDLNLAEEVKTEVRLRQA 1144
G TPF L YG +PVE++ ++ +D E RLI LN +++ E
Sbjct: 1578 GTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLNDLNKIRLEAYESSK 1637
Query: 1145 AVKQRSERRYNTRVVPRHMQVGDLVLRRKAK-GPDDSKLSPNWEGPYRILRDLGQGAYHL 1203
K+R++ ++ ++V R +VGD VL ++ KL W GP+ + GA
Sbjct: 1638 IYKERTKSFHDKKIVSRDFKVGDQVLLFNSRLRLFPGKLKSRWSGPFSVTAVRPYGAI-- 1695
Query: 1204 EELSGRRIPRAWNAQHLRYY 1223
L+G+ N Q L+ Y
Sbjct: 1696 -TLAGKNGDFTVNGQRLKKY 1714
>At1g35370 hypothetical protein
Length = 1447
Score = 200 bits (509), Expect = 4e-51
Identities = 148/484 (30%), Positives = 237/484 (48%), Gaps = 22/484 (4%)
Query: 188 IQEARLTQLLAENMDLFAWSAQDLPGIDPNFICHKLALNPGVKPIAQMKRKMGEEKAQAV 247
++E+ + ++ E D+FA DLP HK+ L G P+ Q + + +
Sbjct: 539 VEESVVQNIVEEFPDVFA-EPTDLPPFREKHD-HKIKLLEGANPVNQRPYRYVVHQKDEI 596
Query: 248 KAETNKLIDAGFIREVKYPTWLANVVMVKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDK 307
+I +G I+ P + + VV+VKK +G WR+C DYT+LN KD + +P I+
Sbjct: 597 DKIVQDMIKSGTIQVSSSP-FASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIED 655
Query: 308 LVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQR 367
L+D G + S +D +GYHQ+RM D +KTAF T+ ++ Y M FGL NA AT+Q
Sbjct: 656 LMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQS 715
Query: 368 LMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQS 427
LM+ VF + + + ++ DD+++ S + H L F +R H + F S
Sbjct: 716 LMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHKL--------FAKGS 767
Query: 428 GKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGRIAALSRFLPCSGSKATPFF 487
+ LG I+ R IE +P K +A+ E +PT+VK+V+ +G RF+ G A P
Sbjct: 768 KEHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIAGP-L 826
Query: 488 QCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPGTPLSVFISISDNAVSSVLLQEC 547
L K F W+ E + AF +LK +L P+L+ P+ V + +VL+Q+
Sbjct: 827 HALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQKG 886
Query: 548 KDELRIIYFVSHALQGAELRYQKIEKAALALIISARKLRPYFQGFQIKVKTD-FPLRQVL 606
+ ++S L+G +L EK LA I + RK R Y +KTD L+ +L
Sbjct: 887 HP----LAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLL 942
Query: 607 QKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQVLADFVNEMSPEVKVSEEAEWILSVDGS 666
++ W +L EF + + Q K ++AD ++ V+ SE LS+
Sbjct: 943 EQRLNTPVQQQWLPKLLEFDYEIQYR-QGKENLVADALS----RVEGSEVLHMALSIVEC 997
Query: 667 SYLK 670
+LK
Sbjct: 998 DFLK 1001
Score = 108 bits (269), Expect = 2e-23
Identities = 101/405 (24%), Positives = 181/405 (43%), Gaps = 35/405 (8%)
Query: 810 MVTGAAPSDWMDRIKMCLEADGA--DLALFSKDQVREASHYVLLGDQLYRRGVGVPLLRC 867
M D++ I++ E+DG D+ + HY D L R+ +
Sbjct: 990 MALSIVECDFLKEIQVAYESDGVLKDIISALQQHPDAKKHYSWSQDILRRKS------KI 1043
Query: 868 VTRDEADRIMFEVHEGVCASHVGGRS---LAAKVLRAGFYWPTLKNDCMGYAKKCEKCQI 924
V ++ + I ++ + + S +GGRS + + +++ FYW + D + + C CQ
Sbjct: 1044 VVPNDVE-ITNKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQ 1102
Query: 925 YADLHRAPPEVLSSMSSAWPFAMW---GVDILGPFTPAGAQIRFVLVAVDYFTKWIEAES 981
+ A P +L + P +W +D + +G + ++V VD +K +
Sbjct: 1103 CKSDNAAYPGLLQPLPI--PDKIWCDVSMDFIEGLPNSGGK-SVIMVVVDRLSKAAHFVA 1159
Query: 982 MAK-ITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHP 1040
+A +A V + + + G P ++VSD FTS ++F G+E+R +S HP
Sbjct: 1160 LAHPYSALTVAQAFLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHP 1219
Query: 1041 QSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDA 1100
QS+GQ E N+ + N ++ LW L + YNT S++ TPF L YG
Sbjct: 1220 QSDGQTEVVNRCLENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAP 1279
Query: 1101 MVPVEIQDMTFRVAAYDEN-ENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERRYN---- 1155
+ + +VA + + EN L+ L ++ + R++Q A + R+ER ++
Sbjct: 1280 PIHLPYLPGKSKVAVVARSLQERENMLLFLKF-HLMRAQHRMKQFADQHRTERTFDIGDF 1338
Query: 1156 --TRVVPRHMQVGDLVLRRKAKGPDDSKLSPNWEGPYRILRDLGQ 1198
++ P Q +VLR + KLSP + GPY+I+ G+
Sbjct: 1339 VYVKLQPYRQQ--SVVLR------VNQKLSPKYFGPYKIIEKCGE 1375
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,920,563
Number of Sequences: 26719
Number of extensions: 1153940
Number of successful extensions: 3142
Number of sequences better than 10.0: 142
Number of HSP's better than 10.0 without gapping: 102
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 2780
Number of HSP's gapped (non-prelim): 264
length of query: 1224
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1113
effective length of database: 8,352,787
effective search space: 9296651931
effective search space used: 9296651931
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0306.1


