

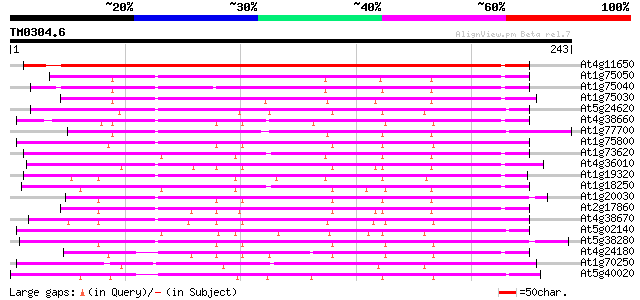
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0304.6
(243 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g11650 osmotin precursor like protein 361 e-100
At1g75050 thaumatin-like protein 206 9e-54
At1g75040 thaumatin-like protein 200 5e-52
At1g75030 thaumatin-like protein 186 1e-47
At5g24620 thaumatin-like protein 181 3e-46
At4g38660 thaumatin-like protein 177 3e-45
At1g77700 thaumatin-like protein 177 3e-45
At1g75800 thaumatin, putative 176 1e-44
At1g73620 thaumatin like protein 175 2e-44
At4g36010 thaumatin-like predicted GPI-anchored protein 174 5e-44
At1g19320 pathogenesis-related protein 5 precursor, putative 173 8e-44
At1g18250 putative protein 170 7e-43
At1g20030 thaumatin-like protein, predicted GPI-anchored protein... 169 2e-42
At2g17860 putative thaumatin-like pathogenesis-related protein 167 5e-42
At4g38670 thaumatin-like protein 166 1e-41
At5g02140 thaumatin-like protein 162 1e-40
At5g38280 receptor serine/threonine kinase PR5K 157 6e-39
At4g24180 thaumatin-like protein 152 1e-37
At1g70250 hypothetical protein 147 5e-36
At5g40020 thaumatin-like protein 140 8e-34
>At4g11650 osmotin precursor like protein
Length = 244
Score = 361 bits (927), Expect = e-100
Identities = 162/219 (73%), Positives = 183/219 (82%), Gaps = 7/219 (3%)
Query: 7 SSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMAR 66
S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W L V AGT MAR
Sbjct: 12 SALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWRLDVAAGTKMAR 65
Query: 67 IWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGF 126
IWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N DFYDISLVDGF
Sbjct: 66 IWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGQPPNTLAEYALNQFNNLDFYDISLVDGF 125
Query: 127 NIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPT 186
NIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++CCTNGQG+C T
Sbjct: 126 NIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYCCTNGQGSCSDT 185
Query: 187 NFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+SRFFK RC D+YSYPQDDPTSTFTC +NY+VVFCP
Sbjct: 186 EYSRFFKQRCPDAYSYPQDDPTSTFTC-TNTNYRVVFCP 223
>At1g75050 thaumatin-like protein
Length = 257
Score = 206 bits (524), Expect = 9e-54
Identities = 106/227 (46%), Positives = 127/227 (55%), Gaps = 21/227 (9%)
Query: 18 VTLTRAANFEIVNNCPYTVWAAASPG-------GGRRLDRGQTWNLGVNAGTAMARIWGR 70
+ ++ A F + N+CPYTVW G GG L G + L AG + R W R
Sbjct: 30 IAVSAATVFTLQNSCPYTVWPGILSGNDNTLGDGGFPLTPGASVQLTAPAGWS-GRFWAR 88
Query: 71 TGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPM 130
TGCNFD +G G C TGDC G L C G GVPP TLAEF L G +DFYD+SLVDG+N+ M
Sbjct: 89 TGCNFDASGHGNCGTGDCGGVLKCNGGGVPPVTLAEFTLVGDGGKDFYDVSLVDGYNVEM 148
Query: 131 DFFPL--NGGCHKISCTADINGQCPNELRAP-------GGCNNPCTVYKTNEFCCTNGQG 181
P +G CH C AD+N CPNELR C + C + + EFCCT
Sbjct: 149 GIKPQGGSGDCHYAGCVADVNAVCPNELRLMDPHTGIIAACKSACAAFNSEEFCCTGAHA 208
Query: 182 ---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
TC PT++S FK C +YSY DD TSTFTC GSNY + FCP
Sbjct: 209 TPQTCSPTHYSAMFKSACPGAYSYAYDDATSTFTC-TGSNYLISFCP 254
>At1g75040 thaumatin-like protein
Length = 239
Score = 200 bits (509), Expect = 5e-52
Identities = 104/233 (44%), Positives = 136/233 (57%), Gaps = 22/233 (9%)
Query: 10 LLMLTASLVTLTRAANFEIVNNCPYTVWAAASPG-------GGRRLDRGQTWNLGVNAGT 62
L+ +T+ + + A +F + NNCP TVWA G GG L G + L AG
Sbjct: 12 LVFITSGIAVM--ATDFTLRNNCPTTVWAGTLAGQGPKLGDGGFELTPGASRQLTAPAGW 69
Query: 63 AMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISL 122
+ R W RTGCNFD +G GRC TGDC GGL C G GVPP TLAEF L G +DFYD+SL
Sbjct: 70 S-GRFWARTGCNFDASGNGRCVTGDC-GGLRCNGGGVPPVTLAEFTLVGDGGKDFYDVSL 127
Query: 123 VDGFNIPMDFFPL--NGGCHKISCTADINGQCPNELRAPG-----GCNNPCTVYKTNEFC 175
VDG+N+ + P +G C C +D+N CP+ L+ C + C + T+++C
Sbjct: 128 VDGYNVKLGIRPSGGSGDCKYAGCVSDLNAACPDMLKVMDQNNVVACKSACERFNTDQYC 187
Query: 176 CTNGQG---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
C TC PT++SR FK+ C D+YSY DD TSTFTC G+NY++ FCP
Sbjct: 188 CRGANDKPETCPPTDYSRIFKNACPDAYSYAYDDETSTFTC-TGANYEITFCP 239
>At1g75030 thaumatin-like protein
Length = 246
Score = 186 bits (472), Expect = 1e-47
Identities = 100/226 (44%), Positives = 121/226 (53%), Gaps = 22/226 (9%)
Query: 23 AANFEIVNNCPYTVWAAASPG-------GGRRLDRGQTWNLGVNAGTAMARIWGRTGCNF 75
A F + N+C YTVW G GG L G + L G + R W RTGCNF
Sbjct: 23 ATVFTLQNSCAYTVWPGTLSGNSITLGDGGFPLTPGASVQLTAPTGWS-GRFWARTGCNF 81
Query: 76 DGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFAL-NQYGNQDFYDISLVDGFNIPMDFFP 134
D +G G C TGDC G L C G GVPP TLAEF + + DFYD+SLVDG+N+ M P
Sbjct: 82 DASGHGTCVTGDCGGVLKCTGGGVPPATLAEFTVGSSNAGMDFYDVSLVDGYNVKMGIKP 141
Query: 135 LN--GGCHKISCTADINGQCPNELR-------APGGCNNPCTVYKTNEFCCTNGQG---T 182
G C C +DIN CP+ELR + C + C + + EFCCT T
Sbjct: 142 QGGFGNCKYAGCVSDINEICPSELRIMDPNSGSVAACKSACAAFSSPEFCCTGAHATPQT 201
Query: 183 CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGS 228
C PT +S FK+ C +YSY DD +STFTC GSNY + FCP S
Sbjct: 202 CSPTYYSSMFKNACPSAYSYAYDDASSTFTC-TGSNYLITFCPTQS 246
>At5g24620 thaumatin-like protein
Length = 420
Score = 181 bits (459), Expect = 3e-46
Identities = 98/243 (40%), Positives = 132/243 (53%), Gaps = 29/243 (11%)
Query: 10 LLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGR--------RLDRGQTWNLGVNAG 61
+L+ + L +++ ++ F I NNCP+T+W G G RLD GQ+ + G
Sbjct: 11 VLVYVSLLFSVSYSSTFVITNNCPFTIWPGTLAGSGTQPLPTTGFRLDVGQSVRIPTALG 70
Query: 62 TAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWG-VPPNTLAEFALNQ-YGNQDFYD 119
+ RIW RTGCNFD G G+C TGDC G L C G G PP +L E L ++DFYD
Sbjct: 71 WS-GRIWARTGCNFDANGAGKCMTGDCGGKLECAGNGAAPPTSLFEITLGHGSDDKDFYD 129
Query: 120 ISLVDGFNIPMDFFPLNGG----CHKISCTADINGQCPNELRAPG----------GCNNP 165
+SLVDG+N+P+ P GG C+ C ADIN CP EL+ G C +
Sbjct: 130 VSLVDGYNLPIVALPTGGGLVGACNATGCVADINISCPKELQVMGEEEAERGGVVACKSA 189
Query: 166 CTVYKTNEFCCTN---GQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVV 222
C + +++CC+ TC P+ +S FK C +YSY DD TSTFTC A S Y ++
Sbjct: 190 CEAFGLDQYCCSGQFANPTTCRPSLYSSIFKRACPRAYSYAFDDGTSTFTCKA-SEYAII 248
Query: 223 FCP 225
FCP
Sbjct: 249 FCP 251
>At4g38660 thaumatin-like protein
Length = 345
Score = 177 bits (450), Expect = 3e-45
Identities = 106/239 (44%), Positives = 133/239 (55%), Gaps = 23/239 (9%)
Query: 4 LTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWA-----AASPG---GGRRLDRGQTWN 55
LTLS +L+L + T + + F N C YTVW A SP G L +G + +
Sbjct: 13 LTLSFTVLLLAS---TGSYGSTFTFANRCGYTVWPGILSNAGSPTLSTTGFELPKGTSRS 69
Query: 56 LGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQYG 113
L G + R W RTGC FD +G G C+TGDC + + C G G PP TLAEF L G
Sbjct: 70 LQAPTGWS-GRFWARTGCKFDSSGSGTCKTGDCGSNAVECAGLGAAPPVTLAEFTLGT-G 127
Query: 114 NQDFYDISLVDGFNIPM--DFFPLNGGCHKISCTADINGQCPNELRAPGG--CNNPCTVY 169
DFYD+SLVDG+NIPM + +G C CT D+N QCP ELR G C + C +
Sbjct: 128 GDDFYDVSLVDGYNIPMIVEVAGGSGQCASTGCTTDLNIQCPAELRFGDGDACKSACLAF 187
Query: 170 KTNEFCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
++ E+CC+ T C P+ +S FK C SYSY DD TSTFTC AG +Y V FCP
Sbjct: 188 RSPEYCCSGAYATPSSCRPSVYSEMFKAACPRSYSYAYDDATSTFTC-AGGDYTVTFCP 245
>At1g77700 thaumatin-like protein
Length = 356
Score = 177 bits (450), Expect = 3e-45
Identities = 97/233 (41%), Positives = 120/233 (50%), Gaps = 20/233 (8%)
Query: 26 FEIVNNCPYTVWAAASPG-----GGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDGAGR 80
F I+N+C T+W A +PG GG L GQ+ G + RIWGRTGC FD G
Sbjct: 91 FTIINSCDQTIWPAITPGENFNGGGFELKPGQSIVFHAPVGWS-GRIWGRTGCKFDSTGT 149
Query: 81 GRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLN--GG 138
G C+TG C L C G PP +LAEF L DFYD+SLVDGFN+PM P+N G
Sbjct: 150 GTCETGSCGSTLKCSASGKPPASLAEFTL---AALDFYDVSLVDGFNLPMSVTPMNGKGN 206
Query: 139 CHKISCTADINGQCPNELRAPG-----GCNNPCTVYKTNEFCCTNGQG---TCGPTNFSR 190
C C AD+ CP EL C + C V+ +E+CC G C PT +S+
Sbjct: 207 CSVAGCVADLRPHCPQELAVKSNGKVISCRSACDVFDRDEYCCRGVYGNPVVCQPTYYSK 266
Query: 191 FFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGSPHVTLMMPGNDTRGY 243
FK C +YSY DDPTS TC A S+Y + FC T + +D Y
Sbjct: 267 IFKQACPTAYSYAYDDPTSIMTCTA-SDYVISFCSSRFVLFTFLTTSHDYFNY 318
>At1g75800 thaumatin, putative
Length = 330
Score = 176 bits (446), Expect = 1e-44
Identities = 96/247 (38%), Positives = 127/247 (50%), Gaps = 26/247 (10%)
Query: 4 LTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAAS--------PGGGRRLDRGQTWN 55
L L + L+ + V+ R+ +F +VN C YTVW P G L +G+
Sbjct: 3 LPLPLIFLIFSHLFVSGVRSTSFIMVNKCEYTVWPGLLSNAGVPPLPTTGFVLQKGEERT 62
Query: 56 LGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQYG 113
+ R WGRT C+ D G+ C TGDC +G L C G G PP TLAEF L+
Sbjct: 63 ISAPTSWG-GRFWGRTQCSTDTDGKFTCLTGDCGSGTLECSGSGATPPATLAEFTLDGSN 121
Query: 114 NQDFYDISLVDGFNIPMDFFPLNGG---CHKISCTADINGQCPNELRAPG---------G 161
DFYD+SLVDG+N+PM P G C C D+NG CP+EL+ G
Sbjct: 122 GLDFYDVSLVDGYNVPMLVAPQGGSGLNCSSTGCVVDLNGSCPSELKVTSLDGRGKQSMG 181
Query: 162 CNNPCTVYKTNEFCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSN 218
C + C ++T E+CC+ GT C P+++S FK C +YSY DD +STFTC N
Sbjct: 182 CKSACEAFRTPEYCCSGAHGTPDTCKPSSYSLMFKTACPRAYSYAYDDQSSTFTCAESPN 241
Query: 219 YKVVFCP 225
Y + FCP
Sbjct: 242 YVITFCP 248
>At1g73620 thaumatin like protein
Length = 264
Score = 175 bits (443), Expect = 2e-44
Identities = 97/239 (40%), Positives = 124/239 (51%), Gaps = 23/239 (9%)
Query: 7 SSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAM-- 64
SSL L+ LV+ A+ N C YTVW P G+ L G + L N +
Sbjct: 25 SSLFLLPLLLLVSQASASTVIFYNKCTYTVWPGIQPSSGQSLLAGGGFKLSPNRAYTLQL 84
Query: 65 -----ARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQG-WGVPPNTLAEFALNQYGNQDFY 118
R WGR GC+FD +GRGRC TGDC G C G GVPP TLAE L + DFY
Sbjct: 85 PPLWSGRFWGRHGCSFDRSGRGRCATGDCGGSFLCNGAGGVPPATLAEITLGH--DMDFY 142
Query: 119 DISLVDGFNIPMDFFPL--NGGCHKISCTADINGQCPNELRAPG-------GCNNPCTVY 169
D+SLVDG+N+ M P+ G C C +D+N CP L+ C + C+ +
Sbjct: 143 DVSLVDGYNLAMSIMPVKGTGKCTYAGCVSDLNRMCPVGLQVRSRDGTQVVACKSACSAF 202
Query: 170 KTNEFCCTNGQG---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+ +CCT G +C PT +S+ FK C +YSY DDPTS TC + +NY V FCP
Sbjct: 203 NSPRYCCTGLFGNPQSCKPTAYSKIFKVACPKAYSYAYDDPTSIATC-SKANYVVTFCP 260
>At4g36010 thaumatin-like predicted GPI-anchored protein
Length = 301
Score = 174 bits (440), Expect = 5e-44
Identities = 104/252 (41%), Positives = 133/252 (52%), Gaps = 30/252 (11%)
Query: 8 SLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRR--------LDRGQTWNLGVN 59
SL L+L + + F IVN C YTVW G G L+ +T + +
Sbjct: 7 SLFLLLILVFIYGVSSTTFTIVNQCSYTVWPGLLSGAGTSPLPTTGFSLNPTETRVIPIP 66
Query: 60 AGTAMARIWGRTGCNFDGA-GRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQYGNQD 116
A + RIWGRT C D GR C TGDC + + C G G PP TLAEF LN D
Sbjct: 67 AAWS-GRIWGRTLCTQDATTGRFTCITGDCGSSTVECSGSGAAPPATLAEFTLNGANGLD 125
Query: 117 FYDISLVDGFNIPMDFFPLNGG--------CHKISCTADINGQCPNELR-----APG-GC 162
FYD+SLVDG+NIPM P GG C C A++NG CP +L+ A G C
Sbjct: 126 FYDVSLVDGYNIPMTIVPQGGGDAGGVAGNCTTTGCVAELNGPCPAQLKVATTGAEGVAC 185
Query: 163 NNPCTVYKTNEFCCTNGQG---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNY 219
+ C + T E+CC+ G TC P+ +S+FFK+ C +YSY DD TSTFTC G++Y
Sbjct: 186 KSACEAFGTPEYCCSGAFGTPDTCKPSEYSQFFKNACPRAYSYAYDDGTSTFTC-GGADY 244
Query: 220 KVVFCPLGSPHV 231
+ FCP +P V
Sbjct: 245 VITFCPSPNPSV 256
>At1g19320 pathogenesis-related protein 5 precursor, putative
Length = 247
Score = 173 bits (438), Expect = 8e-44
Identities = 96/243 (39%), Positives = 128/243 (52%), Gaps = 27/243 (11%)
Query: 7 SSLLLMLTASLVTLTRAAN---FEIVNNCPYTVW-------AAASPGGGRRLDRGQTWNL 56
+S LL ++ + T T + + F + N+C T+W A GG L G +
Sbjct: 6 TSHLLFISFIIATCTISVSGTTFTLTNHCGSTIWPGILTANGAQLGDGGFALASGSSVTF 65
Query: 57 GVNAGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQG-WGVPPNTLAEFALNQYGN- 114
V+ G + R W RT CNFD +G G+C TGDC L C G G PP TLAEF + G
Sbjct: 66 TVSPGWS-GRFWARTYCNFDASGSGKCGTGDCGSKLKCAGAGGAPPATLAEFTIGSSGKK 124
Query: 115 ---QDFYDISLVDGFNIPMDFFPL--NGGCHKISCTADINGQCPNELRAPG-----GCNN 164
QDFYD+SLVDG+N+ M P +G C C +D+N CP EL+ G C +
Sbjct: 125 NAVQDFYDVSLVDGYNVQMKITPQGGSGDCKTAGCVSDVNAICPKELQVTGPSGVAACKS 184
Query: 165 PCTVYKTNEFCCTNG---QGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKV 221
C + E+CCT TC PTN+S+ FK C +YSY DD +STFTC +NY++
Sbjct: 185 ACEAFNKPEYCCTGAYSTPATCPPTNYSKIFKQACPSAYSYAYDDASSTFTC-TNANYEI 243
Query: 222 VFC 224
FC
Sbjct: 244 SFC 246
>At1g18250 putative protein
Length = 243
Score = 170 bits (430), Expect = 7e-43
Identities = 95/242 (39%), Positives = 131/242 (53%), Gaps = 25/242 (10%)
Query: 6 LSSLLLMLTASLVTLTRAANFEIV--NNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTA 63
++S+ L L A L+ L+ A+ ++ N C + VW P G+ L G + L N +
Sbjct: 1 MASINLFLFAFLLLLSHASASTVIFYNKCKHPVWPGIQPSAGQNLLAGGGFKLPANKAHS 60
Query: 64 M-------ARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQG-WGVPPNTLAEFALNQYGNQ 115
+ R WGR GC FD +GRG C TGDC G L+C G G PP TLAE L
Sbjct: 61 LQLPPLWSGRFWGRHGCTFDRSGRGHCATGDCGGSLSCNGAGGEPPATLAEITLGP--EL 118
Query: 116 DFYDISLVDGFNIPMDFFPLNGG--CHKISCTADINGQCP--NELRAPGG-----CNNPC 166
DFYD+SLVDG+N+ M P+ G C C +D+N CP ++R+ G C + C
Sbjct: 119 DFYDVSLVDGYNLAMSIMPVKGSGQCSYAGCVSDLNQMCPVGLQVRSRNGKRVVACKSAC 178
Query: 167 TVYKTNEFCCTNGQG---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVF 223
+ + + ++CCT G +C PT +S+ FK C +YSY DDPTS TC + +NY V F
Sbjct: 179 SAFNSPQYCCTGLFGNPQSCKPTAYSKIFKVACPKAYSYAYDDPTSIATC-SKANYIVTF 237
Query: 224 CP 225
CP
Sbjct: 238 CP 239
>At1g20030 thaumatin-like protein, predicted GPI-anchored protein
(by homology)
Length = 299
Score = 169 bits (427), Expect = 2e-42
Identities = 93/234 (39%), Positives = 121/234 (50%), Gaps = 28/234 (11%)
Query: 25 NFEIVNNCPYTVW--------AAASPGGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFD 76
+F N C YTVW + P G L +G+T + + R WGRT C+ D
Sbjct: 4 SFTFSNKCDYTVWPGILSNAGVSPLPTTGFVLLKGETRTINAPSSWG-GRFWGRTLCSTD 62
Query: 77 GAGRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFP 134
G+ C TGDC +G + C G G PP TLAEF L+ G DFYD+SLVDG+N+ M P
Sbjct: 63 SDGKFSCATGDCGSGKIECSGAGAAPPATLAEFTLDGSGGLDFYDVSLVDGYNVQMLVVP 122
Query: 135 LNGG---CHKISCTADINGQCPNELRAPG---------GCNNPCTVYKTNEFCCTNGQG- 181
G C C D+NG CP+ELR C + C ++ E+CC+ G
Sbjct: 123 QGGSGQNCSSTGCVVDLNGSCPSELRVNSVDGKEAVAMACKSACEAFRQPEYCCSGAFGS 182
Query: 182 --TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGSPHVTL 233
TC P+ +SR FK C +YSY DD +STFTC NY + FCP SP+ +L
Sbjct: 183 PDTCKPSTYSRIFKSACPRAYSYAYDDKSSTFTCAKSPNYVITFCP--SPNTSL 234
>At2g17860 putative thaumatin-like pathogenesis-related protein
Length = 253
Score = 167 bits (423), Expect = 5e-42
Identities = 99/230 (43%), Positives = 125/230 (54%), Gaps = 29/230 (12%)
Query: 23 AANFEIVNNCPYTVW--------AAASPGGGRRLDRGQTWNLGVNAGTAMARIWGRTGCN 74
+ F IVN C YTVW A P G L+ ++ + + A + RIWGRT CN
Sbjct: 22 STTFTIVNQCNYTVWPGLLSGAGTAPLPTTGFSLNSFESRLISIPAAWS-GRIWGRTLCN 80
Query: 75 FDG-AGRGRCQTGDC-TGGLNCQGWG-VPPNTLAEFALNQYGNQDFYDISLVDGFNIPMD 131
+ G C TGDC + + C G G +PP TLAEF LN N DFYD+SLVDG+NIPM
Sbjct: 81 QNEITGIFTCVTGDCGSSQIECSGAGAIPPATLAEFTLNGAENLDFYDVSLVDGYNIPMT 140
Query: 132 FFPLNGG-----CHKISCTADINGQCPNELR----APG----GCNNPCTVYKTNEFCCTN 178
P G C+ C AD+NG CP +L+ A G C + C + T EFCC
Sbjct: 141 IVPYGGAIGVGKCNATGCAADLNGLCPEQLKVTVEAAGTGAVACKSACEAFGTPEFCCNG 200
Query: 179 GQG---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
G TC P+ +S FFK C +YSY DD TSTFTC +G++Y + FCP
Sbjct: 201 AFGTPDTCQPSEYSVFFKKTCPTAYSYAYDDGTSTFTC-SGADYVITFCP 249
>At4g38670 thaumatin-like protein
Length = 281
Score = 166 bits (420), Expect = 1e-41
Identities = 96/243 (39%), Positives = 132/243 (53%), Gaps = 27/243 (11%)
Query: 9 LLLMLTASLVTLTRAA--NFEIVNNCPYTVW--------AAASPGGGRRLDRGQTWNLGV 58
LL M+ A + +L+ +F+I N C T+W +A P G RL RG++ + +
Sbjct: 6 LLFMILAFVSSLSEVEPISFKITNRCRNTIWPGLLSGANSAPLPTTGFRLSRGKSKTVAI 65
Query: 59 NAGTAMARIWGRTGCNFD-GAGRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQYGNQ 115
+ R+W RT C+ D +G C TGDC +G + C G G PP TLAEF LN G
Sbjct: 66 PESWS-GRLWARTLCSQDRSSGSFVCLTGDCGSGKVECSGSGAKPPATLAEFTLNGTGGL 124
Query: 116 DFYDISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNEL----RAPGG---CNNP 165
DFYD+SLVDG+N+PM P GGC C D+NG CP +L R G C +
Sbjct: 125 DFYDVSLVDGYNLPMLILPKKIVIGGCGATGCLVDLNGACPRDLKLVTRGNGNGVACRSA 184
Query: 166 CTVYKTNEFCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVV 222
C + +CC++ T C P+ +S FFK C +YSY DD TST+TC G++Y ++
Sbjct: 185 CEAFGDPRYCCSDAYATPDTCQPSVYSLFFKHACPRAYSYAYDDKTSTYTCATGADYFII 244
Query: 223 FCP 225
FCP
Sbjct: 245 FCP 247
>At5g02140 thaumatin-like protein
Length = 294
Score = 162 bits (410), Expect = 1e-40
Identities = 93/242 (38%), Positives = 129/242 (52%), Gaps = 21/242 (8%)
Query: 4 LTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTA 63
+T S+ L +L ++ T A IVNNC ++W GGG+ R +++G T
Sbjct: 1 MTKSATLFLLFLTISAFTDGAQLIIVNNCQESIWPGILGGGGQITPRNGGFHMGSGEETI 60
Query: 64 M-------ARIWGRTGCNFDGAGRGRCQTGDCT-GGLNCQG-WGVPPNTLAEFALNQYGN 114
+ RIWGR GC F+ G+G CQTGDC G +NCQG GVPP T+ E L +
Sbjct: 61 IDVPDKWSGRIWGRQGCTFNQNGKGSCQTGDCNDGSINCQGTGGVPPATVVEMTLGSSSS 120
Query: 115 Q-DFYDISLVDGFNIPMDFFPLNG--GCHKISCTADINGQCPN--ELRAPG---GCNNPC 166
FYD+SLVDGFN+P+ P+ G GC +C ++N CP+ E++ G GC + C
Sbjct: 121 PLHFYDVSLVDGFNLPVSMKPIGGGVGCGVAACEVNLNICCPSALEVKRDGKVVGCKSAC 180
Query: 167 TVYKTNEFCCTN---GQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVF 223
++ ++CCT C PT F+ FK C +YSY DD +S C A S Y + F
Sbjct: 181 LAMQSAKYCCTGEYANPQACKPTLFANLFKAICPKAYSYAFDDSSSLNKCRA-SRYVITF 239
Query: 224 CP 225
CP
Sbjct: 240 CP 241
>At5g38280 receptor serine/threonine kinase PR5K
Length = 665
Score = 157 bits (396), Expect = 6e-39
Identities = 90/263 (34%), Positives = 131/263 (49%), Gaps = 28/263 (10%)
Query: 5 TLSSLLLMLTASLVTLTRAANFEIVNNCPYTVW--------AAASPGGGRRLDRGQTWNL 56
+LS + L+++ V+ + NF I N C YTVW A + P G L +G++ +
Sbjct: 6 SLSLMFLLVSHFFVSGVMSRNFTIENKCDYTVWPGFLTMTTAVSLPTNGFSLKKGESRVI 65
Query: 57 GVNAGTAMARIWGRTGCNFDGAGRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQYGN 114
V + R+WGR+ C+ G C TGDC +G + C G PP TL +F L+
Sbjct: 66 NVPPSWS-GRLWGRSFCSTSSTGNFSCATGDCGSGKIECSDSGARPPTTLIDFTLDATDG 124
Query: 115 QDFYDISLVDGFNIPMDFFPLNGG----CHKISCTADINGQCPNELRAPGG--------C 162
QDFYD+S+VDG+N+P+ P G C + C ++N CP+EL+ G C
Sbjct: 125 QDFYDVSVVDGYNLPLVVVPQRLGSGRTCSNVGCVVNLNKTCPSELKVMGSSNKEHPIAC 184
Query: 163 NNPCTVYKTNEFCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNY 219
N C + EFCC G C PT +S FK+ C +YSY D+ +TF C NY
Sbjct: 185 MNACQKFGLPEFCCYGEYGKPAKCQPTLYSTNFKNECPLAYSYAYDNENNTFRCSNSPNY 244
Query: 220 KVVFCPLGSPHVTLMMPGNDTRG 242
+ FCP + ++ P +T G
Sbjct: 245 VITFCP--NDISSMSQPSKETNG 265
>At4g24180 thaumatin-like protein
Length = 255
Score = 152 bits (385), Expect = 1e-37
Identities = 90/235 (38%), Positives = 115/235 (48%), Gaps = 44/235 (18%)
Query: 24 ANFEIVNNCPYTVWAAAS----------------PGGGRRLDRGQTWNLGVNAGTAMARI 67
A IVN C +TVW PGG R +W+ R
Sbjct: 28 ATITIVNRCSFTVWPGILSNSGSGDIGTTGFELVPGGSRSFQAPASWS---------GRF 78
Query: 68 WGRTGCNFDG-AGRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQ-----YGNQDFYD 119
W RTGCNF+ G+G C TGDC + + C G G PP TLAEF + QDFYD
Sbjct: 79 WARTGCNFNSDTGQGTCLTGDCGSNQVECNGAGAKPPATLAEFTIGSGPADPARKQDFYD 138
Query: 120 ISLVDGFNIPMDFFPLNGG----CHKISCTADINGQCPNELRAPGG--CNNPCTVYKTNE 173
+SLVDG+N+PM +GG C C D+N +CP ELR G C + C + + E
Sbjct: 139 VSLVDGYNVPM-LVEASGGSEGTCLTTGCVTDLNQKCPTELRFGSGSACKSACEAFGSPE 197
Query: 174 FCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+CC+ + C P+ +S FK C SYSY DD TSTFTC ++Y + FCP
Sbjct: 198 YCCSGAYASPTECKPSMYSEIFKSACPRSYSYAFDDATSTFTC-TDADYTITFCP 251
>At1g70250 hypothetical protein
Length = 676
Score = 147 bits (371), Expect = 5e-36
Identities = 87/250 (34%), Positives = 122/250 (48%), Gaps = 29/250 (11%)
Query: 4 LTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRR--------LDRGQTWN 55
+ L SL +L L+ + + +F I N C YT+W A G RR L++G+T
Sbjct: 13 IRLKSLQYILNFFLLYVAMSRSFTIENKCQYTIWPATY--GYRRSLETTGFVLEKGETRT 70
Query: 56 LGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDCTGG-LNCQGWGVPPNTLAEFALNQYGN 114
+ + + R WGRT C+ + G C TGDCT G + C G + P T+ EF L YG
Sbjct: 71 IKAPSSW-IGRFWGRTLCSTNSTGGFSCATGDCTSGKIKCLGIPIDPTTVVEFNLASYG- 128
Query: 115 QDFYDISLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRA---------PGGCNNP 165
D+Y +++ +G+N+P+ P N C I C D+N CP+EL P C
Sbjct: 129 VDYYVVNVFNGYNLPLLVTPENKNCRSIECVIDMNETCPSELMVNSSGLGSHHPIACMTT 188
Query: 166 CTVYKTNEFCCTN-------GQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSN 218
C Y+ E CC G C T +SR F + C +YSY D S+FTCP SN
Sbjct: 189 CQRYQLPELCCIGLSSGVVVPPGICKRTIYSRTFNNVCPSAYSYAYDVDNSSFTCPNFSN 248
Query: 219 YKVVFCPLGS 228
+ + FCP S
Sbjct: 249 FVITFCPSSS 258
>At5g40020 thaumatin-like protein
Length = 256
Score = 140 bits (352), Expect = 8e-34
Identities = 82/263 (31%), Positives = 120/263 (45%), Gaps = 43/263 (16%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIV----NNCPYTVWAAASP-------------- 42
M + + S L L + +S + + + E+ N C + +W A +P
Sbjct: 1 MKFQSTSLLQLFIISSCIVYGKVTSHEVTFYVQNKCSFPIWPAVAPNSGHPVLASGGFYL 60
Query: 43 --GGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGW-GV 99
GG +R+D W+ RIW RTGC+F + C+TGDC G L C G G
Sbjct: 61 PCGGIKRIDAPLGWS---------GRIWARTGCDFTSNSKQACETGDCDGHLECNGLIGK 111
Query: 100 PPNTLAEFALN-QYGNQDFYDISLVDGFNIP--MDFFPLNGGCHKISCTADINGQCPNEL 156
PP TL + A+ +FYD+SLVDG+N+P ++ P++ C + C D+ CP EL
Sbjct: 112 PPATLIQIAVQADKSKPNFYDVSLVDGYNLPVVVNSKPVSSKCTILGCHKDLKTTCPEEL 171
Query: 157 RAPG------GCNNPCTVYKTNEFCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDP 207
+ C + C + + FCC N GT C +S FK+ C + YSY + P
Sbjct: 172 QVLNEEGRVVACKSACLAFDNDRFCCRNAYGTPEKCKRNTYSMLFKEACPNYYSYAYETP 231
Query: 208 TSTFTCPAGSNYKVVFCPLGSPH 230
TC A Y + FCP H
Sbjct: 232 PPLVTCSA-KEYLITFCPSNWGH 253
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,393,883
Number of Sequences: 26719
Number of extensions: 308207
Number of successful extensions: 623
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 487
Number of HSP's gapped (non-prelim): 35
length of query: 243
length of database: 11,318,596
effective HSP length: 97
effective length of query: 146
effective length of database: 8,726,853
effective search space: 1274120538
effective search space used: 1274120538
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0304.6


