

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0303.2
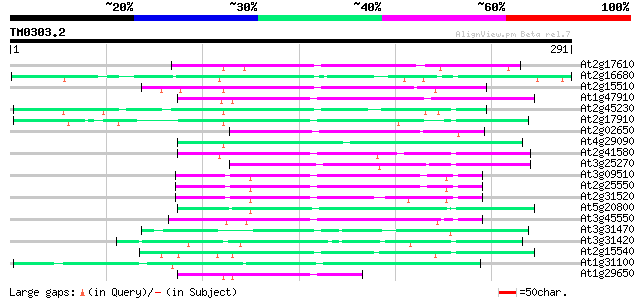
(291 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g17610 putative non-LTR retroelement reverse transcriptase 77 8e-15
At2g16680 putative non-LTR retroelement reverse transcriptase 75 3e-14
At2g15510 putative non-LTR retroelement reverse transcriptase 69 4e-12
At1g47910 reverse transcriptase, putative 64 1e-10
At2g45230 putative non-LTR retroelement reverse transcriptase 64 1e-10
At2g17910 putative non-LTR retroelement reverse transcriptase 61 6e-10
At2g02650 putative reverse transcriptase 61 6e-10
At4g29090 putative protein 60 1e-09
At2g41580 putative non-LTR retroelement reverse transcriptase 60 1e-09
At3g25270 hypothetical protein 60 2e-09
At3g09510 putative non-LTR reverse transcriptase 59 4e-09
At2g25550 putative non-LTR retroelement reverse transcriptase 59 4e-09
At2g31520 putative non-LTR retroelement reverse transcriptase 57 9e-09
At5g20800 putative protein 57 2e-08
At3g45550 putative protein 55 3e-08
At3g31470 hypothetical protein 52 3e-07
At3g31420 hypothetical protein 52 3e-07
At2g15540 putative non-LTR retroelement reverse transcriptase 52 4e-07
At1g31100 hypothetical protein 51 6e-07
At1g29650 reverse transcriptase, putative 50 1e-06
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 77.4 bits (189), Expect = 8e-15
Identities = 57/197 (28%), Positives = 90/197 (44%), Gaps = 23/197 (11%)
Query: 85 GVPDAWVWMPSNVGIYSVNSAYHFL------QEPTLPDTDEA-----LVSIWHSFAPSNV 133
G DA W+ ++ G YSV S YH L Q +LP +E +IW AP +
Sbjct: 451 GANDAITWIYTHDGNYSVKSGYHLLRKLSQQQHASLPSPNEVSAQTVFTNIWKQNAPPKI 510
Query: 134 KGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVW-QGC 192
K F WR + + NL +R++IT D C C + E HLLF C S +W Q
Sbjct: 511 KHFWWRSAHNALPTAGNLKRRRLIT---DDTCQRCGEASEDVNHLLFQCRVSKEIWEQAH 567
Query: 193 LHWLGCSFIQPSSARDHLSHFRFGINKLQQ--RLALSIWLAVIWSVWLARNEVVFRGGAA 250
+ + +S +L I KL Q R +S++ + W +W RN+++F
Sbjct: 568 IKLCPGDSLMSNSFNQNLE----SIQKLNQSARKDVSLFPFIGWRIWKMRNDLIFNNKRW 623
Query: 251 NIYSVLE--LVKRRSWQ 265
+I ++ L+ ++ W+
Sbjct: 624 SIPDSIQKALIDQQQWK 640
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 75.5 bits (184), Expect = 3e-14
Identities = 80/319 (25%), Positives = 125/319 (39%), Gaps = 55/319 (17%)
Query: 2 KILKSGDVTDFWHDDWLGTGCLRGAF--YRLYNLSRQKWNCVNTLGSWEHGRWHWKFLWR 59
+++ +G+ T+ W D WL G R + L N+ + + ++ L W+ K L
Sbjct: 876 RLIGNGEQTNVWIDKWLFDGHSRRPMNLHSLMNIHMKVSHLIDPLTR----NWNLKKL-- 929
Query: 60 RPLVGRELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYH----------FL 109
EL EK + + L D++ W +N G+Y+V S Y F
Sbjct: 930 -----TELFHEKDVQLIMHQRPLISS-EDSYCWAGTNNGLYTVKSGYERSSRETFKNLFK 983
Query: 110 QEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCN 169
+ P + +W +K F W+ L A + L R + T AD C C
Sbjct: 984 EADVYPSVNPLFDKVWSLETVPKIKVFMWKALKGALAVEDRLRSRGIRT--ADG-CLFCK 1040
Query: 170 DYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQP------SSARDHLSHF-----RFGIN 218
+ +ET HLLF C F+ VW S IQ +S +++H FGI
Sbjct: 1041 EEIETINHLLFQCPFARQVW-------ALSLIQAPATGFGTSIFSNINHVIQNSQNFGIP 1093
Query: 219 KLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMK---GFS 275
+ + +S WL +W +W RN+ +F+G ++ W A K G S
Sbjct: 1094 RHMR--TVSPWL--LWEIWKNRNKTLFQGTGLTSSEIVAKAYEECNLWINAQEKSSGGVS 1149
Query: 276 YSSSDWFSNP---LLCILG 291
S W P L C +G
Sbjct: 1150 PSEHKWNPPPAGELKCNIG 1168
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 68.6 bits (166), Expect = 4e-12
Identities = 54/200 (27%), Positives = 86/200 (43%), Gaps = 25/200 (12%)
Query: 69 WEKGLLDAL----GTVQLTKGVP-----DAWVWMPSNVGIYSVNSAYHFL---------- 109
W++ LL+ L +TK P D WVW+ + G YSV S Y
Sbjct: 748 WKRDLLEELFYPTDVELITKRNPVVNMDDFWVWLHAKTGEYSVKSGYWLAFQSNKLDLIQ 807
Query: 110 QEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCN 169
Q LP T+ +W + + +K F WR+L + +++R + D +C +C
Sbjct: 808 QANLLPSTNGLKEQVWSTKSSPKIKMFLWRILSAALPVADQIIRRGMSI---DPRCQICG 864
Query: 170 DYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINK--LQQRLALS 227
D E+ H+LF+C+ + VW Q +S ++ F F + K L L
Sbjct: 865 DEGESTNHVLFTCSMARQVWALSGVPTPEFGFQNASIFANI-QFLFELKKMILVPDLVKR 923
Query: 228 IWLAVIWSVWLARNEVVFRG 247
W V+W +W RN++ F G
Sbjct: 924 SWPWVLWRLWKNRNKLFFDG 943
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 63.9 bits (154), Expect = 1e-10
Identities = 50/192 (26%), Positives = 77/192 (40%), Gaps = 13/192 (6%)
Query: 88 DAWVWMPSNVGIYSVNSAYHF----LQEPTL---PDTDEALVSIWHSFAPSNVKGFAWRV 140
D W + G Y+V S YH L E T PD IW P ++ F W++
Sbjct: 785 DTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWKVQCPPKLRHFLWQI 844
Query: 141 LLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSF 200
L ENL KR + L D C C E+ H LF C + +W
Sbjct: 845 LSGCVPVSENLRKRGI---LCDKGCVSCGASEESINHTLFQCHPARQIWALSQIPTAPGI 901
Query: 201 IQPSSARDHLSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVK 260
+S +L H + ++ + + + +IW +W ARNE VF + +L L
Sbjct: 902 FPSNSIFTNLDHLFW---RIPSGVDSAPYPWIIWYIWKARNEKVFENVDKDPMEILLLAV 958
Query: 261 RRSWQWNKANMK 272
+ + W +A ++
Sbjct: 959 KEAQSWQEAQVE 970
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 63.5 bits (153), Expect = 1e-10
Identities = 68/269 (25%), Positives = 103/269 (38%), Gaps = 42/269 (15%)
Query: 3 ILKSGDVTDFWHDDWLGTGCLRGA--FYRLYNLSRQKWNCVNTLGSW--EHGR-WHWKFL 57
++ +G+ + W D W+G + A R + +S+ N ++ + GR W+W +
Sbjct: 925 VIGNGETINVWTDPWIGAKPAKAAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLV 984
Query: 58 WRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFL-------- 109
L+ + E L G K D + W S G YSV S Y +
Sbjct: 985 ---SLLFPDNTQENILALRPGG----KETRDRFTWEYSRSGHYSVKSGYWVMTEIINQRN 1037
Query: 110 --QEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCAL 167
QE P D IW P + F WR + + + NL R + + C
Sbjct: 1038 NPQEVLQPSLDPIFQQIWKLDVPPKIHHFLWRCVNNCLSVASNLAYRHLAR---EKSCVR 1094
Query: 168 CNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFR-----FGINKLQ- 221
C + ET HLLF C F+ L W P S FR ++K Q
Sbjct: 1095 CPSHGETVNHLLFKCPFAR------LTWAISPLPAPPGGEWAESLFRNMHHVLSVHKSQP 1148
Query: 222 ---QRLALSIWLAVIWSVWLARNEVVFRG 247
AL W ++W +W RN++VF+G
Sbjct: 1149 EESDHHALIPW--ILWRLWKNRNDLVFKG 1175
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 61.2 bits (147), Expect = 6e-10
Identities = 68/305 (22%), Positives = 110/305 (35%), Gaps = 79/305 (25%)
Query: 3 ILKSGDVTDFWHDDWLGTGCLRGAFYR------------LYNLSRQKWNCVNTLGSWEHG 50
++ +G T W D WL G R R L + + + WN +N L
Sbjct: 903 VIGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSRNWN-LNML----RD 957
Query: 51 RWHWK----FLWRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAY 106
+ WK L +RPL +E D++ W+ S+ G+YSV + Y
Sbjct: 958 LFPWKDVEIILKQRPLFFKE---------------------DSFCWLHSHNGLYSVKTGY 996
Query: 107 HFL----------QEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQV 156
FL + P + IW+ ++ F W+ L + L R +
Sbjct: 997 EFLSKQVHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALHGAIPVEDRLRTRGI 1056
Query: 157 ITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQ-GCLHWLGCSF-----------IQPS 204
+D C +C+ ET H+LF C + VW L G F I +
Sbjct: 1057 ---RSDDGCLMCDTENETINHILFECPLARQVWAITHLSSAGSEFSNSVYTNMSRLIDLT 1113
Query: 205 SARDHLSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSW 264
D H RF +S W ++W +W RN ++F G + ++++
Sbjct: 1114 QQNDLPHHLRF----------VSPW--ILWFLWKNRNALLFEGKGSITTTLVDKAYEAYH 1161
Query: 265 QWNKA 269
+W A
Sbjct: 1162 EWFSA 1166
>At2g02650 putative reverse transcriptase
Length = 365
Score = 61.2 bits (147), Expect = 6e-10
Identities = 38/134 (28%), Positives = 64/134 (47%), Gaps = 6/134 (4%)
Query: 115 PDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVET 174
P + E +IW +K F WR + A+ L R + AD C C ET
Sbjct: 28 PGSTEVKQAIWKLHVAPKIKHFLWRCVTGALATNTRLRSRNID---ADPICQRCCIEEET 84
Query: 175 GFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSIWLA--V 232
H++F+C ++ SVW+ +G + PSS D+L+ ++K Q +L +L +
Sbjct: 85 IHHIMFNCPYTQSVWRSANIIIGNQWGPPSSFEDNLNRL-IQLSKTQTTNSLDRFLPFWI 143
Query: 233 IWSVWLARNEVVFR 246
+W +W +RN +F+
Sbjct: 144 MWRLWKSRNVFLFQ 157
>At4g29090 putative protein
Length = 575
Score = 60.5 bits (145), Expect = 1e-09
Identities = 50/190 (26%), Positives = 74/190 (38%), Gaps = 14/190 (7%)
Query: 88 DAWVWMPSNVGIYSVNSAYHFL----------QEPTLPDTDEALVSIWHSFAPSNVKGFA 137
D++ W ++ G Y+V S Y L QE + P + IW S ++ F
Sbjct: 211 DSYTWDYTSSGDYTVKSGYWVLTQIINKRSSPQEVSEPSLNPIYQKIWKSQTSPKIQHFL 270
Query: 138 WRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLG 197
W+ L + L R + A +C C + V HLLF CTF+ W +
Sbjct: 271 WKCLSNSLPVAGALAYRHLSKESACIRCPSCKETVN---HLLFKCTFARLTWAISSIPIP 327
Query: 198 CSFIQPSSARDHLSH-FRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVL 256
S +L F G Q A + ++W +W RNE+VFRG N VL
Sbjct: 328 LGGEWADSIYVNLYWVFNLGNGNPQWEKASQLVPWLLWRLWKNRNELVFRGREFNAQEVL 387
Query: 257 ELVKRRSWQW 266
+ +W
Sbjct: 388 RRAEDDLEEW 397
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 60.5 bits (145), Expect = 1e-09
Identities = 50/198 (25%), Positives = 81/198 (40%), Gaps = 23/198 (11%)
Query: 88 DAWVWMPSNVGIYSVNSAYH----------FLQEPTLPDTDEALVSIWHSFAPSNVKGFA 137
D++ W +N G+Y+V S Y F + P + IW+ + +K F
Sbjct: 729 DSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKIWNLNSAPKIKVFL 788
Query: 138 WRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVW-----QGC 192
W+VL A + L R V L + C++C + ET H+LF C + VW Q
Sbjct: 789 WKVLKGAVAVEDRLRTRGV---LIEDGCSMCPEKNETLNHILFQCPLARQVWALTPMQSP 845
Query: 193 LHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANI 252
H G S + +H+ +S W +IW +W RN+ +F G +
Sbjct: 846 NHGFGDSIF---TNVNHVIGNCHNTELSPHLRYVSPW--IIWILWKNRNKRLFEGIGSVS 900
Query: 253 YSVLELVKRRSWQWNKAN 270
S++ +W KA+
Sbjct: 901 LSIVGKALEDCKEWLKAH 918
>At3g25270 hypothetical protein
Length = 343
Score = 59.7 bits (143), Expect = 2e-09
Identities = 42/159 (26%), Positives = 70/159 (43%), Gaps = 9/159 (5%)
Query: 115 PDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVET 174
P E IW +K F W++L A+ +NL +R + QC C ET
Sbjct: 9 PGKAEIKAKIWKLKTAPKIKHFLWKLLSGALATGDNLKRRHIRNH---PQCHRCCQEDET 65
Query: 175 GFHLLFSCTFSLSVWQ--GCLHW-LGCSFIQPSSARDHLSHFRFGINKLQQRLALSIWLA 231
HL F C ++ VW+ G H L + I + + L N+ Q L+IW
Sbjct: 66 SQHLFFDCFYAQQVWRASGIPHQELRTTGITMETKMELLLSSCLA-NRQPQLFNLAIW-- 122
Query: 232 VIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKAN 270
++W +W +RN++VF+ + + + L+ + +W N
Sbjct: 123 ILWRLWKSRNQLVFQQKSISWQNTLQRARNDVQEWEDTN 161
>At3g09510 putative non-LTR reverse transcriptase
Length = 484
Score = 58.5 bits (140), Expect = 4e-09
Identities = 48/176 (27%), Positives = 74/176 (41%), Gaps = 27/176 (15%)
Query: 87 PDAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALVS-----------IWHSFAPSNVKG 135
PD +W + G Y+V S Y L P T+ ++ IW+ +K
Sbjct: 116 PDKIIWNYNTTGEYTVRSGYWLLTHD--PSTNIPAINPPHGSIDLKTRIWNLPIMPKLKH 173
Query: 136 FAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHW 195
F WR L A+ E L R + D C C+ E+ H LF+C F+ W+
Sbjct: 174 FLWRALSQALATTERLTTRGM---RIDPSCPRCHRENESINHALFTCPFATMAWRLSDSS 230
Query: 196 LGCSFIQPSSARDHLSHFRFGINKLQQRLA------LSIWLAVIWSVWLARNEVVF 245
L + + + +++S+ +N +Q L +WL IW +W ARN VVF
Sbjct: 231 LIRNQLMSNDFEENISNI---LNFVQDTTMSDFHKLLPVWL--IWRIWKARNNVVF 281
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 58.5 bits (140), Expect = 4e-09
Identities = 48/176 (27%), Positives = 74/176 (41%), Gaps = 27/176 (15%)
Query: 87 PDAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALVS-----------IWHSFAPSNVKG 135
PD +W + G Y+V S Y L P T+ ++ IW+ +K
Sbjct: 1382 PDKIIWNYNTTGEYTVRSGYWLLTHD--PSTNIPAINPPHGSIDLKTRIWNLPIMPKLKH 1439
Query: 136 FAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHW 195
F WR L A+ E L R + D C C+ E+ H LF+C F+ W+
Sbjct: 1440 FLWRALSQALATTERLTTRGM---RIDPSCPRCHRENESINHALFTCPFATMAWRLSDSS 1496
Query: 196 LGCSFIQPSSARDHLSHFRFGINKLQQRLA------LSIWLAVIWSVWLARNEVVF 245
L + + + +++S+ +N +Q L +WL IW +W ARN VVF
Sbjct: 1497 LIRNQLMSNDFEENISNI---LNFVQDTTMSDFHKLLPVWL--IWRIWKARNNVVF 1547
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 57.4 bits (137), Expect = 9e-09
Identities = 50/178 (28%), Positives = 72/178 (40%), Gaps = 31/178 (17%)
Query: 87 PDAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALVS-----------IWHSFAPSNVKG 135
PD +W + G Y+V S Y L P T+ ++ IW+ +K
Sbjct: 1156 PDKIIWNYNTTGEYTVRSGYWLLTHD--PSTNIPAINPPHGSIDLKTRIWNLPIMPKLKH 1213
Query: 136 FAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHW 195
F WR L A+ E L R + D C C+ E+ H LF+C F+ W W
Sbjct: 1214 FLWRALSQALATTERLTTRGM---RIDPICPRCHRENESINHALFTCPFATMAW-----W 1265
Query: 196 LGCSFIQPSS--ARDHLSHFRFGINKLQQRLA------LSIWLAVIWSVWLARNEVVF 245
L S + + + D + +N +Q L +WL IW +W ARN VVF
Sbjct: 1266 LSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHKLLPVWL--IWRIWKARNNVVF 1321
>At5g20800 putative protein
Length = 359
Score = 56.6 bits (135), Expect = 2e-08
Identities = 45/194 (23%), Positives = 75/194 (38%), Gaps = 16/194 (8%)
Query: 88 DAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALVS---------IWHSFAPSNVKGFAW 138
D W + G Y+ S YH + + DT+ + + W P ++ F W
Sbjct: 83 DTLGWHFTKSGKYTFKSGYHTSRIEYI-DTNLSFIGPEITLLKAYSWKVQCPPKLRHFLW 141
Query: 139 RVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGC 198
++L RENL KR + D CA C +T H LF C + +W
Sbjct: 142 QILASCVPVRENLRKRGIN---CDIGCARCGATEKTINHTLFQCHPARQIWALSQIPTVI 198
Query: 199 SFIQPSSARDHLSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLEL 258
S ++L H + + + W +IW +W ARN +F + +L L
Sbjct: 199 GIFPSDSIYENLDHLFWRVQSAVDSFSYP-W--IIWYIWKARNAKLFDNVDKDPLDILSL 255
Query: 259 VKRRSWQWNKANMK 272
++ + W A ++
Sbjct: 256 AEKEAQSWKSAQVE 269
>At3g45550 putative protein
Length = 851
Score = 55.5 bits (132), Expect = 3e-08
Identities = 46/176 (26%), Positives = 74/176 (41%), Gaps = 19/176 (10%)
Query: 83 TKGVPDAWVWMPSNVGIYSVNSAYHFLQE------PTLPDTDEAL---VSIWHSFAPSNV 133
T+ D +W ++ G Y+V S Y PT+ ++ IW+ +
Sbjct: 617 TRSKTDRLIWSYNSTGDYTVRSGYWLSTHDPSNTIPTMAKPHGSVDLKTKIWNLPIMPKL 676
Query: 134 KGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCL 193
K F WR+L + + L R + D C C E+ H LF+C F+ W+
Sbjct: 677 KHFLWRILSKALPTTDRLTTRGM---RIDPGCPRCRRENESINHALFTCPFATMAWRLSD 733
Query: 194 HWLGCSFIQPSSARDHLSHFRFGINKL----QQRLALSIWLAVIWSVWLARNEVVF 245
L S I ++ D++S+ + Q+L + WL +W +W ARN VVF
Sbjct: 734 TPLYRSSILSNNIEDNISNILLLLQNTTITDSQKL-IPFWL--LWRIWKARNNVVF 786
>At3g31470 hypothetical protein
Length = 410
Score = 52.4 bits (124), Expect = 3e-07
Identities = 47/210 (22%), Positives = 84/210 (39%), Gaps = 42/210 (20%)
Query: 69 WEKGLLDALGTVQLTK-GVPDAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALVSIWHS 127
+E+ +LD ++L+K G D + W+PS G+YS Y+ +
Sbjct: 119 YEEAILD----IRLSKCGASDGFAWLPSKTGLYSAKLGYYA------------------T 156
Query: 128 FAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLS 187
AP + G + ENL R + + DA C C + ET HL C F+
Sbjct: 157 IAPDHPGGQTTEAMRGALPVGENLRARGINS---DASCPFCGER-ETTLHLFAKCDFASW 212
Query: 188 VWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALS--------IWLAVIWSVWLA 239
+W+ Q + + + R I + + + L ++ +IW++W+
Sbjct: 213 IWRNAP-------FQRNLDPNAIPSLRALIEQAKILICLPPCGIGFGPLFPWIIWAIWIN 265
Query: 240 RNEVVFRGGAANIYSVLELVKRRSWQWNKA 269
RN+ +F ++ L V + +WN A
Sbjct: 266 RNQRIFNEKSSTPAETLTQVLALAQEWNLA 295
>At3g31420 hypothetical protein
Length = 1491
Score = 52.4 bits (124), Expect = 3e-07
Identities = 58/253 (22%), Positives = 92/253 (35%), Gaps = 52/253 (20%)
Query: 56 FLWRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWV------------------------ 91
F+W+ L GR L +KGL +G L D W+
Sbjct: 1064 FVWKSILHGRNL-LKKGLRCCVGDGSLINAWLDPWLPLHSPRAPYKQEDAPEQLLVCSTA 1122
Query: 92 ------WMPSNVGIYSVNSAYHFLQEPTLPDTD---------EALVSIWHSFAPSNVKGF 136
W + G+Y+V SAY LP T + +W + +K F
Sbjct: 1123 RDDLIGWHYTKDGMYTVKSAYWLATH--LPGTTGTHPPPGDIKLKQLLWKTKTAPKIKHF 1180
Query: 137 AWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWL 196
W++L A+ E L R + +C C D ET HL F C ++ + W+G L
Sbjct: 1181 CWKILSGAIATGEMLRYRHINKQSICKRC--CRDE-ETSQHLFFECDYAKATWRGA--GL 1235
Query: 197 GCSFIQPSSA---RDHLSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIY 253
Q S + F F + L W ++W +W +RN + F+
Sbjct: 1236 PNLIFQDSIVTLEEKFRAMFTFNPSTTNYWRQLPFW--ILWRLWRSRNILTFQQKHIPWE 1293
Query: 254 SVLELVKRRSWQW 266
++L K+ + +W
Sbjct: 1294 VTVQLAKQDALEW 1306
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 52.0 bits (123), Expect = 4e-07
Identities = 53/229 (23%), Positives = 87/229 (37%), Gaps = 31/229 (13%)
Query: 68 EWEKGLLDAL---GTVQLTKGV-------PDAWVWMPSNVGIYSVNSAY---HFLQEPTL 114
+W+ G L L + L G+ D + W + G Y+V S Y L PT
Sbjct: 851 QWKLGRLQELVDPSDIPLILGIRPSRTYKSDDFSWSFTKSGNYTVKSGYWAARDLSRPTC 910
Query: 115 ------PDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALC 168
P +W K F W+ L A+ + L R + T + C C
Sbjct: 911 DLPFQGPSVSALQAQVWKIKTTRKFKHFEWQCLSGCLATNQRLFSRHIGT---EKVCPRC 967
Query: 169 NDYVETGFHLLFSCTFSLSVWQGCLHWLGCS-FIQPSSARDHLSHFRFGINK----LQQR 223
E+ HLLF C S +W L + S +I P ++ + F K +
Sbjct: 968 GAEEESINHLLFLCPPSRQIW--ALSPIPSSEYIFPRNSLFYNFDFLLSRGKEFDIAEDI 1025
Query: 224 LALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMK 272
+ + W ++W +W +RN +F + +L+ + + W +AN K
Sbjct: 1026 MEIFPW--ILWYIWKSRNRFIFENVIESPQVILDFAIQEANVWKQANSK 1072
>At1g31100 hypothetical protein
Length = 1090
Score = 51.2 bits (121), Expect = 6e-07
Identities = 60/245 (24%), Positives = 94/245 (37%), Gaps = 15/245 (6%)
Query: 3 ILKSGDVTDFWHDDWLGTGCLRGAFYRLYNLSRQKWNCVNTLGSWEHGRWHWKFLWRRPL 62
+L G FW D+W G L ++L+ S + C+ +L P
Sbjct: 817 VLGDGKKISFWFDNWSPLGPL----FKLFGSSGPRALCIPIQAKVADACSDVGWLISPPR 872
Query: 63 VGRELEWEKGLLDALGTVQLT--KGVPDAWVWMPSNVGIYSVNSAYHF-LQEPTLPDTDE 119
+ L LL L T+ L PD +VW+ + + ++A + P P D
Sbjct: 873 TDQAL----ALLIHLTTIALPCFDSSPDTFVWIVDDFTCHGFSAARTWEAMRPKKPVKDW 928
Query: 120 ALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLL 179
S+W + W L+R +R+ L V T+ C LC+ E+ HLL
Sbjct: 929 TK-SVWFKGSVPKHAFNMWVSHLNRLPTRQRLAAWGVTTT---TDCCLCSSRPESRDHLL 984
Query: 180 FSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSIWLAVIWSVWLA 239
C FS +W+ L S +S + LS R +K L A ++ +W
Sbjct: 985 LYCVFSAVIWKLVFFRLTPSQAIFNSWAELLSWTRINSSKAPSLLRKIAAQASVFHLWKQ 1044
Query: 240 RNEVV 244
RN V+
Sbjct: 1045 RNNVL 1049
>At1g29650 reverse transcriptase, putative
Length = 1557
Score = 50.1 bits (118), Expect = 1e-06
Identities = 32/106 (30%), Positives = 48/106 (45%), Gaps = 13/106 (12%)
Query: 88 DAWVWMPSNVGIYSVNSAYHFL---QEPTL-------PDTDEALVSIWHSFAPSNVKGFA 137
D WVW+ S G YSV S Y + +P L P T+ IW + +K F
Sbjct: 1274 DFWVWLHSKSGEYSVKSGYWLVFQSNKPELLFKACRQPSTNGLKEKIWSTSTFPKIKLFL 1333
Query: 138 WRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCT 183
WR+L + +L+R + D C +C E+ H+LF+C+
Sbjct: 1334 WRILSAALPVADQILRRGM---NVDPCCQICGQEGESINHVLFTCS 1376
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.138 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,491,418
Number of Sequences: 26719
Number of extensions: 318686
Number of successful extensions: 1004
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 840
Number of HSP's gapped (non-prelim): 118
length of query: 291
length of database: 11,318,596
effective HSP length: 98
effective length of query: 193
effective length of database: 8,700,134
effective search space: 1679125862
effective search space used: 1679125862
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0303.2


