

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0302a.1
(299 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
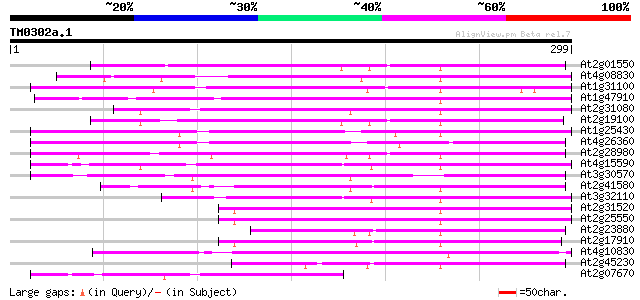
Score E
Sequences producing significant alignments: (bits) Value
At2g01550 putative non-LTR retroelement reverse transcriptase 103 9e-23
At4g08830 putative protein 101 6e-22
At1g31100 hypothetical protein 101 6e-22
At1g47910 reverse transcriptase, putative 99 3e-21
At2g31080 putative non-LTR retroelement reverse transcriptase 98 5e-21
At2g19100 putative non-LTR retroelement reverse transcriptase 97 1e-20
At1g25430 hypothetical protein 96 3e-20
At4g26360 putative protein 95 4e-20
At2g28980 putative non-LTR retroelement reverse transcriptase 95 5e-20
At4g15590 reverse transcriptase like protein 94 1e-19
At3g30570 putative reverse transcriptase 93 2e-19
At2g41580 putative non-LTR retroelement reverse transcriptase 91 8e-19
At3g32110 non-LTR reverse transcriptase, putative 88 5e-18
At2g31520 putative non-LTR retroelement reverse transcriptase 87 1e-17
At2g25550 putative non-LTR retroelement reverse transcriptase 87 1e-17
At2g23880 putative non-LTR retroelement reverse transcriptase 83 2e-16
At2g17910 putative non-LTR retroelement reverse transcriptase 82 3e-16
At4g10830 putative protein 81 6e-16
At2g45230 putative non-LTR retroelement reverse transcriptase 81 6e-16
At2g07670 putative non-LTR retrolelement reverse transcriptase 80 1e-15
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 103 bits (258), Expect = 9e-23
Identities = 75/272 (27%), Positives = 125/272 (45%), Gaps = 21/272 (7%)
Query: 44 YTFKEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFL 103
+ F +KLK LK KL KE +G+L K ++ + + N S + + +
Sbjct: 700 FRFTKKLKALKPKLRGLAKEKMGNLVKRTREAYLSLCQAQ-QSNSQNPSQRAMEIESEAY 758
Query: 104 VEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAV-DGEWVEDP 162
V + ++A + L Q S+ W+K GD N K FH + R N+IR + DG
Sbjct: 759 VRWDRIASIEEKYLKQVSKLHWLKVGDKNNKTFHRAATARAAQNSIREIQKEDGSTATTK 818
Query: 163 KVVKSKMIEFFEA--QFTSNPEVGVFLEGT----PFRSISDEDNLALTQTFNLEEIRTAV 216
+K++ FF+ Q N G+ +E P+ E ++ LT + + +EIR A+
Sbjct: 819 DDIKNETERFFQEFLQLIPNDYEGITVEKLTSLLPYHCSPAEKDM-LTASVSAKEIRGAL 877
Query: 217 WECEGDRSLGPD------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISN 264
+ D+S GPD ++ + + +K F G PKG N++ + LIP
Sbjct: 878 FSMPNDKSPGPDGYTSEFYKRAWDIIGAEFVLAVKSFFEKGFLPKGVNTTILALIPKKLE 937
Query: 265 LMSLNDYISISVIGCMYKIVSKVLTTRLSKVM 296
+ DY IS +YK++SK++ RL V+
Sbjct: 938 AKEMKDYRPISCCNVIYKVISKIIANRLKHVL 969
>At4g08830 putative protein
Length = 947
Score = 101 bits (251), Expect = 6e-22
Identities = 76/301 (25%), Positives = 141/301 (46%), Gaps = 45/301 (14%)
Query: 26 FGECVSTWSSFDIQGWG-AYTFKEK-------LKRLKLKLNQWEKENVGDLDKICKDPVT 77
FGE ++ S D+ G YT+K KRL +L +W ++ GD+ K ++ +
Sbjct: 8 FGEWINRVSLIDMGFSGNKYTWKRGRVENTFVAKRLD-RLKKWNRDVFGDVKKKKEELMR 66
Query: 78 RI----NALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNT 133
I +ALD+ + D L +E + F KSR++W+ GD NT
Sbjct: 67 EIQVVQDALDVHQSDDMLRKEEDLIK-----------------AFDKSREKWIALGDRNT 109
Query: 134 KYFHNSVNWRRRTNAIRGLA-VDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVF--LEGT 190
YFH + RRR N + L +G W+ D K +++ +++++ ++ + + V L
Sbjct: 110 NYFHTTTIIRRRQNRVERLHNEEGTWIFDAKELEALAVQYYQRLYSMDDVLQVVNPLPQD 169
Query: 191 PFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVL 238
++ E+ L+L + F E+ A+ ++ GPD V+ ++R
Sbjct: 170 GLERLTREEVLSLNKPFLATEVEVAIRSMGKYKAPGPDGYQPIFYQSCWEVVGNSVIRFA 229
Query: 239 KDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVMGH 298
DF +G+ P N + + LIP + +N + IS+ ++K+++K++ RL KV+G
Sbjct: 230 LDFFSSGILPPQTNDALVVLIPKVPKPERMNQFRPISLCNAIFKMITKMMVLRLKKVIGK 289
Query: 299 I 299
+
Sbjct: 290 L 290
>At1g31100 hypothetical protein
Length = 1090
Score = 101 bits (251), Expect = 6e-22
Identities = 82/315 (26%), Positives = 146/315 (46%), Gaps = 33/315 (10%)
Query: 12 KPFRNRNCWLQNHAFGECVST-WSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PFR N LQN F V W S ++ G + +KLK LK + + EN +L+K
Sbjct: 158 RPFRFYNFLLQNPDFISLVGELWYSINVVGSSMFKMSKKLKALKNPIRTFSMENFSNLEK 217
Query: 71 ICKDP----VTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWV 126
K+ + R N N + + + QR ++ + K +S Q+SR W+
Sbjct: 218 RVKEAHNLVLYRQNKTLSDPTIPNAALEMEAQR-----KWLILVKAEESFFCQRSRVTWM 272
Query: 127 KDGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIEFFEAQFTSNPEVGV 185
+GDSNT YFH + R+ N I + D G ++ +K IE+F +
Sbjct: 273 GEGDSNTSYFHRMADSRKAVNTIHIIIDDNGVKIDTQLGIKEHCIEYFSNLLGGEVGPPM 332
Query: 186 FLEG-----TPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------ 228
++ PFR D+ L +F+ ++I++A + +++ GPD
Sbjct: 333 LIQEDFDLLLPFRCSHDQKK-ELAMSFSRQDIKSAFFSFPSNKTSGPDGFPVEFFKETWS 391
Query: 229 VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDY--ISISVIG--CMYKIV 284
V+ ++ + +F + V K N++ + LIP I+N +ND+ IS + G +YK++
Sbjct: 392 VIGTEVTDAVSEFFTSSVLLKQWNATTLVLIPKITNASKMNDFRPISCNDFGPITLYKVI 451
Query: 285 SKVLTTRLSKVMGHI 299
+++LT RL ++ +
Sbjct: 452 ARLLTNRLQCLLSQV 466
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 99.0 bits (245), Expect = 3e-21
Identities = 82/301 (27%), Positives = 141/301 (46%), Gaps = 23/301 (7%)
Query: 14 FRNRNCWLQNHAFGECVSTWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKICK 73
FR W+ E +S + D G+ F EKL + +++W K L +
Sbjct: 24 FRFDKRWIGKDGLLEAISQGWNLD-SGFREGQFVEKLTNCRRAISKWRKS----LIPFGR 78
Query: 74 DPVTRINA-LDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSN 132
+ + A LD+ + D + S +E + L E ++ + +QKSR W+K GD+N
Sbjct: 79 QTIEDLKAELDVAQRDDDRSREEITELTLRLKEAYRD---EEQYWYQKSRSLWMKLGDNN 135
Query: 133 TKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIEFFEAQFTS-NPEVGVFLEGT 190
+K+FH RR N I GL + G W + +++ + +F+ FT+ NP+V G
Sbjct: 136 SKFFHALTKQRRARNRITGLHDENGIWSIEDDDIQNIAVSYFQNLFTTANPQVFDEALGE 195
Query: 191 PFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVL 238
I+D N LT E+R A++ +++ GPD ++K D+L ++
Sbjct: 196 VQVLITDRINDLLTADATECEVRAALFMIHPEKAPGPDGMTALFFQKSWAIIKSDLLSLV 255
Query: 239 KDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVMGH 298
F + GV+ K N++ I LIP + + IS+ YK++SK+L RL V+ +
Sbjct: 256 NSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLCNVGYKVISKILCQRLKTVLPN 315
Query: 299 I 299
+
Sbjct: 316 L 316
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 98.2 bits (243), Expect = 5e-21
Identities = 71/263 (26%), Positives = 125/263 (46%), Gaps = 24/263 (9%)
Query: 56 KLNQWEKENVGDL----DKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAK 111
KL +W +E GD+ +K+ D + L + D L+ +E L E V +
Sbjct: 139 KLRKWNREVFGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEV-----LLKEMDLVLE 193
Query: 112 FNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDGE-WVEDPKVVKSKMI 170
++L FQKSR+++++ GD NT +FH S RRR N I L D + WV D +++ +
Sbjct: 194 QEETLWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMAL 253
Query: 171 EFFEAQFTSN--PEVGVFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD 228
+++ ++ EV L F SIS+ + AL Q F E+ +AV ++ GPD
Sbjct: 254 TYYKRLYSLEDVSEVRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPD 313
Query: 229 ------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISV 276
+ + R + +F GV P N + + LI ++ + + +S+
Sbjct: 314 GYQPVFYQQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQFRPVSL 373
Query: 277 IGCMYKIVSKVLTTRLSKVMGHI 299
++KI++K++ TRL V+ +
Sbjct: 374 CNVLFKIITKMMVTRLKNVISKL 396
>At2g19100 putative non-LTR retroelement reverse transcriptase
Length = 1447
Score = 96.7 bits (239), Expect = 1e-20
Identities = 78/279 (27%), Positives = 118/279 (41%), Gaps = 37/279 (13%)
Query: 44 YTFKEKLKRLKLKLNQWEKENVGDL--------DKICKDPVTRINALDIKEGDGNLSSQE 95
+ F +KLK LK L KE +G+L D +CK + +N N +
Sbjct: 693 FRFSKKLKSLKPLLRNLAKERLGNLVKKTREAYDTLCKKQESTLN---------NPTPNA 743
Query: 96 QKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAV- 154
K+ + + VA + L +KS+ W+ GD N K FH +V R N+I +
Sbjct: 744 MKEEVEAHDRWEHVAGLEEKFLKKKSKLHWLDGGDKNNKAFHRAVVTREAQNSISEIQCQ 803
Query: 155 DGEWVEDPKVVKSKMIEFFEA--QFTSNPEVGVFLEGT----PFRSISDEDNLALTQTFN 208
DG +K+ FF Q N GV + PFR E L LT+
Sbjct: 804 DGSVTAKGDEIKAYAERFFREFLQLIPNEYEGVTMADLQDLLPFRCSETEHEL-LTRVVT 862
Query: 209 LEEIRTAVWECEGDRSLGPD------------VLKVDILRVLKDFHRNGVWPKGGNSSFI 256
EEI+ ++ D+S GPD +L + + ++ F G PKG N++ +
Sbjct: 863 AEEIKKVLFSMPNDKSPGPDGFTSEFFKATWEILGNEFILAIQSFFAKGFLPKGINTTIL 922
Query: 257 TLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKV 295
LIP + DY IS +YK++SK++ RL V
Sbjct: 923 ALIPKKKEAKEMKDYRPISCCNVIYKVISKIIANRLKLV 961
>At1g25430 hypothetical protein
Length = 1213
Score = 95.5 bits (236), Expect = 3e-20
Identities = 81/319 (25%), Positives = 143/319 (44%), Gaps = 45/319 (14%)
Query: 12 KPFRNRNCWLQNHAFGECV-STWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PF+ N L+N F V W + ++ G + +KLK LK + + + N +L+K
Sbjct: 247 RPFKFFNYLLKNLDFLNLVRDNWFTLNVVGSSMFRVSKKLKALKKPIKDFSRLNYSELEK 306
Query: 71 ICKDPVT-RINALDIKEGDG---NLSSQEQKQRDDFLVEYWKVAKFNDSLLF-QKSRDRW 125
K+ I D D N S + + +R W + + F QKSR W
Sbjct: 307 RTKEAHDFLIGCQDRTLADPTPINASFELEAERK------WHILTAAEESFFRQKSRISW 360
Query: 126 VKDGDSNTKYFHNSVNWRRRTNAIRGLAV-DGEWVEDPKVVKSKMIEFFEAQFTSNPEVG 184
+GD NTKYFH + R +N+I L +G+ V+ + + +F +
Sbjct: 361 FAEGDGNTKYFHRMADARNSSNSISALYDGNGKLVDSQEGILDLCASYFGSLLGD----- 415
Query: 185 VFLEGTPFRSISDEDNLALT------------QTFNLEEIRTAVWECEGDRSLGPD---- 228
E P+ ++ NL L+ TF+ E+IR A++ ++S GPD
Sbjct: 416 ---EVDPYLMEQNDMNLLLSYRCSPAQVCELESTFSNEDIRAALFSLPRNKSCGPDGFTA 472
Query: 229 --------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCM 280
++ ++ +K+F +G K N++ I LIP I N +D+ IS + +
Sbjct: 473 EFFIDSWSIVGAEVTDAIKEFFSSGCLLKQWNATTIVLIPKIVNPTCTSDFRPISCLNTL 532
Query: 281 YKIVSKVLTTRLSKVMGHI 299
YK+++++LT RL +++ +
Sbjct: 533 YKVIARLLTDRLQRLLSGV 551
>At4g26360 putative protein
Length = 1141
Score = 95.1 bits (235), Expect = 4e-20
Identities = 77/304 (25%), Positives = 138/304 (45%), Gaps = 35/304 (11%)
Query: 12 KPFRNRNCWLQNHAFGECV-STWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PF+ N L+N F V W S ++ G + +KLK LK + + + N +L+K
Sbjct: 239 RPFKFFNFLLKNPEFLNLVWDVWYSTNVVGSSMFRVSKKLKALKKPIKDFSRLNYSNLEK 298
Query: 71 ICKDPVTRINALDIKEGDG----NLSSQEQKQRDDFLVEYWKV-AKFNDSLLFQKSRDRW 125
++ + + D N + + + QR W++ A +S Q+SR W
Sbjct: 299 RTEEAHETLLSFQNLTLDNPSLENAAHELEAQRK------WQILATAEESFFRQRSRVTW 352
Query: 126 VKDGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIEFFEAQFTSNPEVG 184
+GD NT+YFH + R+ N I L D G ++ + + +FE + + +
Sbjct: 353 FAEGDGNTRYFHRMADSRKSVNTITTLVDDSGTQIDSQQGIADHCALYFENLLSDDND-- 410
Query: 185 VFLEGTPFRSISDEDNLALTQT------------FNLEEIRTAVWECEGDRSLGPDVLKV 232
P+ D+ NL LT F+ E+I+ A + +++ GPD V
Sbjct: 411 ------PYSLEQDDMNLLLTYRCPYSQVADLEAMFSDEDIKAAFFGLPSNKACGPDGFPV 464
Query: 233 DILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRL 292
+++F +G K N++ I LIP N +D+ IS + +YK+++++LT RL
Sbjct: 465 TA--AVREFFISGNLLKQWNATTIVLIPKFPNASCTSDFRPISCMNTLYKVIARLLTDRL 522
Query: 293 SKVM 296
K++
Sbjct: 523 QKLL 526
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 94.7 bits (234), Expect = 5e-20
Identities = 83/311 (26%), Positives = 141/311 (44%), Gaps = 31/311 (9%)
Query: 12 KPFRNRNCWLQNHAFGECV-STWSS---FDIQGWGAYTFKEKLKRLKLKLNQWEKENVGD 67
KPF+ N + F V S W+S + Y F +KLK LK L + KE +GD
Sbjct: 546 KPFKFVNVLTKLPQFLPVVESHWASSAPLYVSTSALYRFSKKLKTLKPHLRELGKEKLGD 605
Query: 68 LDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEY--WK-VAKFNDSLLFQKSRDR 124
L K ++ + L ++ L++ Q+ + L Y W +++ + L QKS+
Sbjct: 606 LPKRTREA----HILLCEKQATTLANPSQETIAEELKAYTDWTHLSELEEGFLKQKSKLH 661
Query: 125 WVKDGDSNTKYFHNSVNWRRRTNAIRGL-AVDGEWVEDPKVVKSKMIEFFEAQFT--SNP 181
W+ GD N YFH + R+ N+IR + + E ++ + +K + FF S
Sbjct: 662 WMNVGDGNNSYFHKAAQVRKMRNSIREIRGPNAETLQTSEEIKGEAERFFNEFLNRQSGD 721
Query: 182 EVGVFLEGT----PFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD--------- 228
G+ +E +R + N+ LT+ EEI+ ++ ++S GPD
Sbjct: 722 FHGISVEDLRNLMSYRCSVTDQNI-LTREVTGEEIQKVLFAMPNNKSPGPDGYTSEFFKA 780
Query: 229 ---VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVS 285
+ D + ++ F G PKG N++ + LIP + + DY IS +YK++S
Sbjct: 781 TWSLTGPDFIAAIQSFFVKGFLPKGLNATILALIPKKDEAIEMKDYRPISCCNVLYKVIS 840
Query: 286 KVLTTRLSKVM 296
K+L RL ++
Sbjct: 841 KILANRLKLLL 851
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 93.6 bits (231), Expect = 1e-19
Identities = 74/308 (24%), Positives = 142/308 (46%), Gaps = 32/308 (10%)
Query: 12 KPFRNRNCWLQNHAFGECVSTWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDL--- 68
+PFR WL + F E ++ +S+D + L RL+ +L +W KE G++
Sbjct: 53 RPFRFEAAWLSHEGFKELLT--ASWDT----GLSTPVALNRLRWQLKKWNKEVFGNIHVR 106
Query: 69 -DKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVK 127
+K+ D + L++ + D L + D L E+ + ++L FQKSR++ +
Sbjct: 107 KEKVVSDLKAVQDLLEVVQTDDLLMKE-----DTLLKEFDVLLHQEETLWFQKSREKLLA 161
Query: 128 DGDSNTKYFHNSVNWRRRTNAIRGLA-VDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVF 186
GD NT +FH S RRR N I L + WV + + ++ ++++ + S +V V
Sbjct: 162 LGDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLY-SLEDVSVV 220
Query: 187 LEGTP---FRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLK 231
P F ++ E+ L + F +E+ AV ++ GPD +
Sbjct: 221 RGTLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVG 280
Query: 232 VDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTR 291
+ + + +F +GV PK N + L+ ++ + + +S+ ++KI++K++ R
Sbjct: 281 ESVSKFVMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIR 340
Query: 292 LSKVMGHI 299
L V+ +
Sbjct: 341 LKNVISKL 348
>At3g30570 putative reverse transcriptase
Length = 1099
Score = 93.2 bits (230), Expect = 2e-19
Identities = 69/292 (23%), Positives = 131/292 (44%), Gaps = 34/292 (11%)
Query: 12 KPFRNRNCWLQNHAFGECV-STWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PFR WL + F E + ++W G E LK ++ L +W KE GD+ +
Sbjct: 291 RPFRFEAAWLSHPGFKELLLASWK-------GNIPTPEALKGIQTTLRKWNKEVFGDIQE 343
Query: 71 ICKDPVTRINALDIKEGDGNLSSQEQ---KQRDDFLVEYWKVAKFNDSLLFQKSRDRWVK 127
+ + I + D S+Q ++ + ++E+ V + ++L KSR++W
Sbjct: 344 KKEKLMVEIKTVQ----DSLYSNQTDDLLRRESELIIEFDVVLEQEETLWLHKSREKWFV 399
Query: 128 DGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIEFFEAQFTSN--PEVG 184
GD NT +FH S RRR N I L + G W+ + ++ I++++ ++ + EV
Sbjct: 400 QGDRNTSFFHTSTIIRRRHNRIEMLKNEKGGWISNTNELEKLAIDYYKRLYSLDDVDEVV 459
Query: 185 VFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPDVLKVDILRVLKDFHRN 244
L F ++ E+ + L + F+ E+ +A + I R + DF +
Sbjct: 460 ERLPAGRFLGLNQEEQIRLNKPFSAMEVESA----------------LKITRFVLDFFLS 503
Query: 245 GVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVM 296
G P+ N + + LI + + + IS+ + +I++ + RL V+
Sbjct: 504 GNLPQETNDAMVVLIAKVGKPEKITQFRPISLCNVLLEIITNTMVERLKSVI 555
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 90.9 bits (224), Expect = 8e-19
Identities = 75/275 (27%), Positives = 130/275 (47%), Gaps = 46/275 (16%)
Query: 49 KLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQ------------ 96
+L + ++QW++EN D K +T++ KE S Q
Sbjct: 3 RLVECRKAISQWKREN----DFNAKSRITKLRRELDKEKSATFPSWTQISLLQDVLGDAY 58
Query: 97 KQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDG 156
++ +DF W++ KSRD+W+ GD N+K+F +V R +N++R L +
Sbjct: 59 REEEDF----WRL----------KSRDKWMVGGDKNSKFFQATVKANRVSNSLRFLVDEN 104
Query: 157 EWVEDPKVVKSKM-IEFFEAQFTSN--PEVGVFLEGTPFRSISDEDNLALTQTFNLEEIR 213
+ K K+ + FFE F+S+ + LEG R ++++ N LT+ N +EI
Sbjct: 105 GNEQTVNREKGKIAVTFFEDLFSSSYPSSMDSVLEGFNKR-VTEDMNQDLTKKVNEQEIY 163
Query: 214 TAVWECEGDRSLGPD------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPN 261
AV+ + + GPD ++K I+ ++ F + G+ P+ N + + LIP
Sbjct: 164 KAVFSINAESAPGPDGFTALFFQRQWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPK 223
Query: 262 ISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVM 296
I+ + D IS+ MYKI+SK+L+ RL K +
Sbjct: 224 ITKPARMADIRPISLCSVMYKIISKILSARLKKYL 258
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 88.2 bits (217), Expect = 5e-18
Identities = 62/235 (26%), Positives = 117/235 (49%), Gaps = 24/235 (10%)
Query: 82 LDIKEGDGNLSSQEQKQRD-DFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSV 140
LD+ + D L +E+ +D D ++E +V + QKSR++W GD NTK+FH S
Sbjct: 845 LDLHQSDDLLKKEEELLKDFDVVLEQEEV------VWMQKSREKWFVHGDRNTKFFHTST 898
Query: 141 NWRRRTNAIRGLA-VDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVFLEGTP---FRSIS 196
RRR N I L DG W+ + + +++ I++++ + S ++ +E P F ++S
Sbjct: 899 IIRRRRNQIEMLQDNDGRWLSNAQELETHAIDYYKRLY-SLDDLDAVVEQLPQEGFTALS 957
Query: 197 DEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVLKDFHRN 244
+ D +LT+ F+ E+ A+ ++ GPD V+ + + + DF +
Sbjct: 958 EADFSSLTKPFSPLEVEGAIRSMGKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFFSS 1017
Query: 245 GVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVMGHI 299
G +P+ N + LI + + + IS+ ++K ++KV+ RL V+ +
Sbjct: 1018 GSFPQETNDVLVVLIAKVLKPEKITQFRPISLCNVLFKTITKVMVGRLKGVINKL 1072
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 87.0 bits (214), Expect = 1e-17
Identities = 59/204 (28%), Positives = 101/204 (48%), Gaps = 16/204 (7%)
Query: 112 FNDSLLF--QKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSK 168
F+D ++ QKSR++W+K+GD NT YFH R N + + D G K + +
Sbjct: 485 FSDEEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNH 544
Query: 169 MIEFFEAQFTSNPEVGVFLEGTPFRS-ISDEDNLALTQTFNLEEIRTAVWECEGDRSLGP 227
+FF F++N ++ F+S +++ NL LT+ F+ EI A+ + D++ GP
Sbjct: 545 AQDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGP 604
Query: 228 D------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISIS 275
D ++ D++ +K F N + I +IP I+N +L+DY I+
Sbjct: 605 DGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIA 664
Query: 276 VIGCMYKIVSKVLTTRLSKVMGHI 299
+ +YK++SK L RL + I
Sbjct: 665 LCNVLYKVISKCLVNRLKSHLNSI 688
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 87.0 bits (214), Expect = 1e-17
Identities = 59/204 (28%), Positives = 101/204 (48%), Gaps = 16/204 (7%)
Query: 112 FNDSLLF--QKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSK 168
F+D ++ QKSR++W+K+GD NT YFH R N + + D G K + +
Sbjct: 711 FSDEEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNH 770
Query: 169 MIEFFEAQFTSNPEVGVFLEGTPFRS-ISDEDNLALTQTFNLEEIRTAVWECEGDRSLGP 227
+FF F++N ++ F+S +++ NL LT+ F+ EI A+ + D++ GP
Sbjct: 771 AQDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGP 830
Query: 228 D------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISIS 275
D ++ D++ +K F N + I +IP I+N +L+DY I+
Sbjct: 831 DGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIA 890
Query: 276 VIGCMYKIVSKVLTTRLSKVMGHI 299
+ +YK++SK L RL + I
Sbjct: 891 LCNVLYKVISKCLVNRLKSHLNSI 914
>At2g23880 putative non-LTR retroelement reverse transcriptase
Length = 1216
Score = 82.8 bits (203), Expect = 2e-16
Identities = 53/187 (28%), Positives = 92/187 (48%), Gaps = 20/187 (10%)
Query: 129 GDSNTKYFHNSVNWRRRTNAIRGLAV-DGEWVEDPKVVKSKMIEFFEAQFTSNPE--VGV 185
GD N K FH ++ R N+IR + DG V + ++++ + +F+ + P G+
Sbjct: 90 GDRNNKTFHRAITTREAVNSIREIVTRDGLVVTSQQDIQTEAVNYFQDFLQTIPADYEGM 149
Query: 186 FLEGT----PFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------V 229
+E PFR S++D+ LT+ EEI+ ++ D+S GPD +
Sbjct: 150 CVEELENLLPFRC-SEDDHRLLTRVVTGEEIKKVIFSMPKDKSPGPDGYTSEFYKASWEI 208
Query: 230 LKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLT 289
+ +++ ++ F G PKG NS+ + LIP + DY IS +YK +SK+L
Sbjct: 209 IGDEVIIAIQSFFAKGFLPKGVNSTILALIPKKKEAREIKDYRPISCCNVLYKAISKILA 268
Query: 290 TRLSKVM 296
RL +++
Sbjct: 269 NRLKRIL 275
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 82.4 bits (202), Expect = 3e-16
Identities = 65/200 (32%), Positives = 96/200 (47%), Gaps = 18/200 (9%)
Query: 112 FNDSLLF--QKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDGEWVEDPKVVKSKM 169
+ D LF QKSR +W+ GD NT +FH +V+ R N + L + + K K+
Sbjct: 307 YGDEELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKI 366
Query: 170 IE-FFEAQFTSNPEV--GVFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLG 226
FFE FTS + LEG + ++ E N L Q E+ AV+ + + G
Sbjct: 367 ASSFFENLFTSTYILTHNNHLEGLQAK-VTSEMNHNLIQEVTELEVYNAVFSINKESAPG 425
Query: 227 PD------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISI 274
PD ++K IL + F GV P+ N + I LIP I++ ++D I
Sbjct: 426 PDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPI 485
Query: 275 SVIGCMYKIVSKVLTTRLSK 294
S+ +YKI+SK+LT RL K
Sbjct: 486 SLCSVLYKIISKILTQRLKK 505
>At4g10830 putative protein
Length = 1294
Score = 81.3 bits (199), Expect = 6e-16
Identities = 70/269 (26%), Positives = 117/269 (43%), Gaps = 29/269 (10%)
Query: 45 TFKEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLV 104
T ++ + +LK K N + + L ++ +N + + RD+
Sbjct: 638 TCRQAMAKLKHKSNLNSRIRINQLQAALDKAMSSVNRTERRTISHIQRELTVAYRDE--E 695
Query: 105 EYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLA-VDGEWVEDPK 163
YW+ QKSR++W+K+GD NT++FH R N + + +G K
Sbjct: 696 RYWQ----------QKSRNQWMKEGDRNTEFFHACTKTRFSVNRLVTIKDEEGMIYRGDK 745
Query: 164 VVKSKMIEFFEAQFTSNPEVGVFLEGTPFRSISDED-NLALTQTFNLEEIRTAVWECEGD 222
+ EFF + SN ++ F+ I E N LT+ + EI A+ D
Sbjct: 746 EIGVHAQEFFTKVYESNGRPVSIIDFAGFKPIVTEQINDDLTKDLSDLEIYNAICHIGDD 805
Query: 223 RSLGPDVLKV------------DILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLND 270
++ GPD L D+++ +K F R + N + I +IP I+N +L+D
Sbjct: 806 KAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSD 865
Query: 271 YISISVIGCMYKIVSKVLTTRLSKVMGHI 299
Y I++ +YKI+SK L RL GH+
Sbjct: 866 YRPIALCNVLYKIISKCLVERLK---GHL 891
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 81.3 bits (199), Expect = 6e-16
Identities = 56/196 (28%), Positives = 95/196 (47%), Gaps = 22/196 (11%)
Query: 119 QKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDG--EWVEDPKVVKSKMIEFFEAQ 176
+KSR W+++GD NTKYFH + RR N I+ L + EW D + ++ E + +
Sbjct: 338 EKSRIMWMRNGDRNTKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDL--GRVAEAYFKK 395
Query: 177 FTSNPEVGVFLEG----TPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD---- 228
++ +VG +E TP +SD+ N L EE++ A + + GPD
Sbjct: 396 LFASEDVGYTVEELENLTPL--VSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNG 453
Query: 229 --------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCM 280
+ I +++ F R+G +G N + I LIP I + D+ IS+ +
Sbjct: 454 FLYQQFWETMGDQITEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVI 513
Query: 281 YKIVSKVLTTRLSKVM 296
YK++ K++ RL K++
Sbjct: 514 YKVIGKLMANRLKKIL 529
>At2g07670 putative non-LTR retrolelement reverse transcriptase
Length = 913
Score = 80.1 bits (196), Expect = 1e-15
Identities = 53/172 (30%), Positives = 88/172 (50%), Gaps = 16/172 (9%)
Query: 12 KPFRNRNCWLQNHAFGECVSTWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKI 71
+PFR WL + F + + +S++ +G L LK KL +W +E GD+++
Sbjct: 713 RPFRFEAAWLTHSGFKDLLQ--ASWNTEGETPVA----LAALKSKLKKWNREVFGDVNRR 766
Query: 72 CKDPVTRINA----LDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVK 127
+ + I L+I + D LS +E+ + E+ V + + L FQKSR++WV+
Sbjct: 767 KESLMNEIKVVQELLEINQTDNLLSKEEE-----LIKEFDVVLEQEEVLWFQKSREKWVE 821
Query: 128 DGDSNTKYFHNSVNWRRRTNAIRGL-AVDGEWVEDPKVVKSKMIEFFEAQFT 178
GD NTKYFH RRR N I L A DG WV + ++ ++++ ++
Sbjct: 822 LGDRNTKYFHTMTVVRRRRNRIEMLKADDGSWVSQQQELEKMAVDYYSRLYS 873
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,037,754
Number of Sequences: 26719
Number of extensions: 306240
Number of successful extensions: 1198
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1068
Number of HSP's gapped (non-prelim): 68
length of query: 299
length of database: 11,318,596
effective HSP length: 99
effective length of query: 200
effective length of database: 8,673,415
effective search space: 1734683000
effective search space used: 1734683000
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0302a.1


