

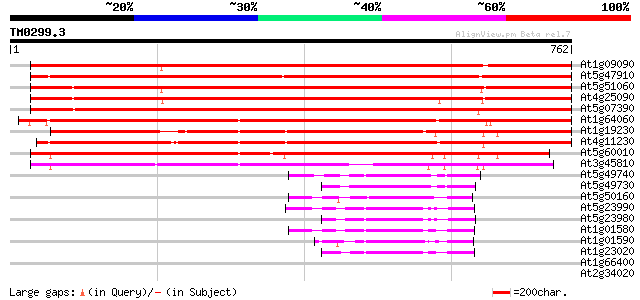
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0299.3
(762 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09090 respiratory burst oxidase protein B like 996 0.0
At5g47910 respiratory burst oxidase protein 926 0.0
At5g51060 respiratory burst oxidase protein 911 0.0
At4g25090 respiratory burst oxidase - like protein 857 0.0
At5g07390 respiratory burst oxidase protein A 842 0.0
At1g64060 cytochrome b245 beta chain homolog RbohAp108 825 0.0
At1g19230 hypothetical protein 725 0.0
At4g11230 respiratory burst oxidase homolog F - like protein 699 0.0
At5g60010 respiratory burst oxidase protein - like 664 0.0
At3g45810 respiratory burst oxidase - like protein 600 e-172
At5g49740 FRO1-like protein; NADPH oxidase-like 96 7e-20
At5g49730 FRO2-like protein; NADPH oxidase-like 92 1e-18
At5g50160 FRO1 and FRO2-like protein 90 4e-18
At5g23990 FRO2 homolog 84 2e-16
At5g23980 FRO2 homolog 83 7e-16
At1g01580 hypothetical protein 81 2e-15
At1g01590 hypothetical protein 76 6e-14
At1g23020 putative superoxide-generating NADPH oxidase flavocyto... 75 1e-13
At1g66400 calmodulin-related protein 38 0.024
At2g34020 unknown protein 37 0.041
>At1g09090 respiratory burst oxidase protein B like
Length = 843
Score = 996 bits (2575), Expect = 0.0
Identities = 479/742 (64%), Positives = 585/742 (78%), Gaps = 13/742 (1%)
Query: 29 KVDRYKS-GATHALQCFRVMTKN-VVFEGWSEVEKRFDGLAIEGKLLKTRFNECIGMSEL 86
++DR KS GA AL+ R + KN V GW EV RFD LA+EGKL K++F CIGM E
Sbjct: 74 RLDRSKSFGAMFALRGLRFIAKNDAVGRGWDEVAMRFDKLAVEGKLPKSKFGHCIGMVES 133
Query: 87 KDFACKLFDALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRINE 146
+F +LF+AL R RG TS+SITK EL E WEQIT SFD R+Q FFD+VDKN DGRI
Sbjct: 134 SEFVNELFEALVRRRGTTSSSITKTELFEFWEQITGNSFDDRLQIFFDMVDKNLDGRITG 193
Query: 147 EQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQS-- 204
++VKEII+LSAS+NKLSKI+E EYA+LIMEELD DNLGYIEL++LE LLLQ +QS
Sbjct: 194 DEVKEIIALSASANKLSKIKENVDEYAALIMEELDRDNLGYIELHNLETLLLQVPSQSNN 253
Query: 205 ---TTDSKVVSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFT 261
+ + + +++MLSQK +PTK+RNP+KR ++YF +NWKRIWV+ LW+SIC+ LFT
Sbjct: 254 SPSSANKRALNKMLSQKLIPTKDRNPVKRFAMNISYFFLENWKRIWVLTLWISICITLFT 313
Query: 262 WKFFQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKL-GMVVP 320
WKF QYK + VF+VMGYCV +AKG AETLKFNMALIL P+CRNTITWLR+K+KL G VVP
Sbjct: 314 WKFLQYKRKTVFEVMGYCVTVAKGSAETLKFNMALILLPVCRNTITWLRTKSKLIGSVVP 373
Query: 321 FDDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWF 380
FDDN+NFHKV+AFGIA+GIGLHA++HL CDFPRLLH E+EP+K FFG +P+NY WF
Sbjct: 374 FDDNINFHKVVAFGIAVGIGLHAISHLACDFPRLLHAKNVEFEPMKKFFGDERPENYGWF 433
Query: 381 LKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVL 440
+KGT+GWTGV MVVLM + ++LAQ W RRNR LPK LK+LTGFNAFWYSHHL +IVYVL
Sbjct: 434 MKGTDGWTGVTMVVLMLVAYVLAQSWFRRNRANLPKSLKRLTGFNAFWYSHHLFVIVYVL 493
Query: 441 LVIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFA 500
L++HG+F+YLSK+WY TTWMYLAVP++LY ERLIRAFR K V++ V+ Y GNV +
Sbjct: 494 LIVHGYFVYLSKEWYHKTTWMYLAVPVLLYAFERLIRAFRPGAKAVKVLKVAVYPGNVLS 553
Query: 501 LKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAI 560
L MSKP+GFKY SGQYIY+NCSD+SP +WHPFSITSA DDYLSVHIRTLGDWTSQL+++
Sbjct: 554 LYMSKPKGFKYTSGQYIYINCSDVSPLQWHPFSITSASGDDYLSVHIRTLGDWTSQLKSL 613
Query: 561 LEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGA 620
K CQ S Q + A+ + R PRL IDGPYG+PAQDY+NY+VLLLVGLGIGA
Sbjct: 614 YSKVCQLPSTSQSGLFIADIGQANNITRFPRLLIDGPYGAPAQDYRNYDVLLLVGLGIGA 673
Query: 621 TPLISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFKGL 680
TPLISI++ +LNNIK Q +E G + K + ATK+AYFYWVTREQGS EWF +
Sbjct: 674 TPLISIIRDVLNNIKNQNSIERG-----TNQHIKNYVATKRAYFYWVTREQGSLEWFSEV 728
Query: 681 MNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKTHFGR 740
MNE+AE D EG+IELHNYCT VY+EGD SALITMLQSL+ +K G+D VS TRV+THF R
Sbjct: 729 MNEVAEYDSEGMIELHNYCTSVYEEGDARSALITMLQSLHHAKSGIDIVSGTRVRTHFAR 788
Query: 741 PNWRNVYEHVSLKHPDKRVGKY 762
PNWR+V++HV++ H ++RVG +
Sbjct: 789 PNWRSVFKHVAVNHVNQRVGVF 810
>At5g47910 respiratory burst oxidase protein
Length = 921
Score = 926 bits (2392), Expect = 0.0
Identities = 450/739 (60%), Positives = 573/739 (76%), Gaps = 12/739 (1%)
Query: 29 KVDRYKSGATHALQCFRVMTKNVVFEGWSEVEKRFDGLAIE--GKLLKTRFNECIGMS-E 85
+ DR S A HAL+ + + W V++RFD L+ + G LL +F EC+GM+ E
Sbjct: 157 RFDRTSSAAIHALKGLKFIATKTA--AWPAVDQRFDKLSADSNGLLLSAKFWECLGMNKE 214
Query: 86 LKDFACKLFDALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRIN 145
KDFA +LF ALAR ++ +ITK++LR WEQI+++SFD+++Q FFD+VDK+ DGR+
Sbjct: 215 SKDFADQLFRALARRNNVSGDAITKEQLRIFWEQISDESFDAKLQVFFDMVDKDEDGRVT 274
Query: 146 EEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQST 205
EE+V EIISLSAS+NKLS IQ+++ EYA+LIMEELDPDN G+I + +LEMLLLQA QS
Sbjct: 275 EEEVAEIISLSASANKLSNIQKQAKEYAALIMEELDPDNAGFIMIENLEMLLLQAPNQSV 334
Query: 206 T--DSKVVSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFTWK 263
DS+++SQMLSQK P KE NP+ R + + YFI DNW+R+W+M LWL IC LFT+K
Sbjct: 335 RMGDSRILSQMLSQKLRPAKESNPLVRWSEKIKYFILDNWQRLWIMMLWLGICGGLFTYK 394
Query: 264 FFQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFDD 323
F QYKN+A + VMGYCV +AKGGAETLKFNMALIL P+CRNTITWLR+KTKLG VVPFDD
Sbjct: 395 FIQYKNKAAYGVMGYCVCVAKGGAETLKFNMALILLPVCRNTITWLRNKTKLGTVVPFDD 454
Query: 324 NVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKG 383
++NFHKVIA GI +G+ LHA AHLTCDFPRL+ YEP++ +FG ++P +YWWF+KG
Sbjct: 455 SLNFHKVIASGIVVGVLLHAGAHLTCDFPRLIAADEDTYEPMEKYFG-DQPTSYWWFVKG 513
Query: 384 TEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVI 443
EGWTG++MVVLMAI F LA PW RRN+L LP LKKLTGFNAFWY+HHL IIVY LL++
Sbjct: 514 VEGWTGIVMVVLMAIAFTLATPWFRRNKLNLPNFLKKLTGFNAFWYTHHLFIIVYALLIV 573
Query: 444 HGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKM 503
HG LYL+K WY+ TTWMYLAVP++LY SERL+RAFRS K V++ V+ Y GNV +L M
Sbjct: 574 HGIKLYLTKIWYQKTTWMYLAVPILLYASERLLRAFRSSIKPVKMIKVAVYPGNVLSLHM 633
Query: 504 SKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEK 563
+KPQGFKY SGQ++ VNC +SPFEWHPFSITSAP DDYLSVHIRTLGDWT +L+ + +
Sbjct: 634 TKPQGFKYKSGQFMLVNCRAVSPFEWHPFSITSAPGDDYLSVHIRTLGDWTRKLRTVFSE 693
Query: 564 ACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPL 623
C+P + + +L+A+ D + P P++ IDGPYG+PAQDYK Y+V+LLVGLGIGATP+
Sbjct: 694 VCKPPTAGKSGLLRADG-GDGNLP-FPKVLIDGPYGAPAQDYKKYDVVLLVGLGIGATPM 751
Query: 624 ISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFKGLMNE 683
ISILK ++NN+K + +E+ N K F T++AYFYWVTREQGSFEWFKG+M+E
Sbjct: 752 ISILKDIINNMKGPD--RDSDIENNNSNNNSKGFKTRKAYFYWVTREQGSFEWFKGIMDE 809
Query: 684 IAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKTHFGRPNW 743
I+E D+EG+IELHNYCT VY+EGD ALI MLQSL +K G+D VS TRVK+HF +PNW
Sbjct: 810 ISELDEEGIIELHNYCTSVYEEGDARVALIAMLQSLQHAKNGVDVVSGTRVKSHFAKPNW 869
Query: 744 RNVYEHVSLKHPDKRVGKY 762
R VY+ ++++HP KR+G +
Sbjct: 870 RQVYKKIAVQHPGKRIGVF 888
>At5g51060 respiratory burst oxidase protein
Length = 905
Score = 911 bits (2354), Expect = 0.0
Identities = 442/746 (59%), Positives = 563/746 (75%), Gaps = 15/746 (2%)
Query: 29 KVDRYKSGATHALQCFRVMTKNVVFEGWSEVEKRFDGLAIE--GKLLKTRFNECIGMSEL 86
K+DR KS A+ AL+ + ++K GWS VEKRF+ + G LL+T+F ECIGM+
Sbjct: 130 KLDRTKSAASQALKGLKFISKTDGGAGWSAVEKRFNQITATTGGLLLRTKFGECIGMTS- 188
Query: 87 KDFACKLFDALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRINE 146
KDFA +LFDALAR R IT I D+L+E WEQI +QSFDSR++TFFD+VDK+ADGR+ E
Sbjct: 189 KDFALELFDALARRRNITGEVIDGDQLKEFWEQINDQSFDSRLKTFFDMVDKDADGRLTE 248
Query: 147 EQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQS-- 204
++V+EIISLSAS+N LS IQ+R+ EYA+LIMEELDPDN+GYI L SLE LLLQA+ QS
Sbjct: 249 DEVREIISLSASANNLSTIQKRADEYAALIMEELDPDNIGYIMLESLETLLLQAATQSVI 308
Query: 205 --TTDSKVVSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFTW 262
T + K +S M+SQ+ PT RNP+KR + L +F+ DNW+R WV+ LW + LFT+
Sbjct: 309 TSTGERKNLSHMMSQRLKPTFNRNPLKRWYRGLRFFLLDNWQRCWVIVLWFIVMAILFTY 368
Query: 263 KFFQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFD 322
K+ QY+ V+ VMG CV +AKG AET+K NMALIL P+CRNTITWLR+KT+LG VVPFD
Sbjct: 369 KYIQYRRSPVYPVMGDCVCMAKGAAETVKLNMALILLPVCRNTITWLRNKTRLGRVVPFD 428
Query: 323 DNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLK 382
DN+NFHKVIA GI +G+ +HA AHL CDFPRLLH T Y PL+ FFG +P +YW F+
Sbjct: 429 DNLNFHKVIAVGIIVGVTMHAGAHLACDFPRLLHATPEAYRPLRQFFGDEQPKSYWHFVN 488
Query: 383 GTEGWTGVLMVVLMAIPFILAQPWARRNRLK-LPKPLKKLTGFNAFWYSHHLLIIVYVLL 441
EG TG++MV+LMAI F LA PW RR +L LP PLKKL FNAFWY+HHL +IVY+LL
Sbjct: 489 SVEGITGLVMVLLMAIAFTLATPWFRRGKLNYLPGPLKKLASFNAFWYTHHLFVIVYILL 548
Query: 442 VIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFAL 501
V HG++LYL++ W+ TTWMYL VP++LY ERLIRAFRS K V I+ V+ Y GNV A+
Sbjct: 549 VAHGYYLYLTRDWHNKTTWMYLVVPVVLYACERLIRAFRSSIKAVTIRKVAVYPGNVLAI 608
Query: 502 KMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAIL 561
+S+PQ FKY SGQY++VNC+ +SPFEWHPFSITSAP+DDYLSVHIR LGDWT L+ +
Sbjct: 609 HLSRPQNFKYKSGQYMFVNCAAVSPFEWHPFSITSAPQDDYLSVHIRVLGDWTRALKGVF 668
Query: 562 EKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGAT 621
+ C+P +L+A+ L + P P++ IDGPYG+PAQDYK YEV+LLVGLGIGAT
Sbjct: 669 SEVCKPPPAGVSGLLRADMLHGANNPDFPKVLIDGPYGAPAQDYKKYEVVLLVGLGIGAT 728
Query: 622 PLISILKGMLNNIKQQQD-----LENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEW 676
P+ISI+K ++NNIK ++ +ENG S + +KK+ F T++AYFYWVTREQGSF+W
Sbjct: 729 PMISIVKDIVNNIKAKEQAQLNRMENGT--SEPQRSKKESFRTRRAYFYWVTREQGSFDW 786
Query: 677 FKGLMNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKT 736
FK +MNE+AE D VIE+HNYCT VY+EGD SALI MLQSL +K G+D VS TRV +
Sbjct: 787 FKNIMNEVAERDANRVIEMHNYCTSVYEEGDARSALIHMLQSLNHAKNGVDIVSGTRVMS 846
Query: 737 HFGRPNWRNVYEHVSLKHPDKRVGKY 762
HF +PNWRNVY+ +++ HP+ +VG +
Sbjct: 847 HFAKPNWRNVYKRIAMDHPNTKVGVF 872
>At4g25090 respiratory burst oxidase - like protein
Length = 863
Score = 857 bits (2214), Expect = 0.0
Identities = 416/754 (55%), Positives = 557/754 (73%), Gaps = 23/754 (3%)
Query: 29 KVDRYKSGATHALQCFRVMTKNVVFEGWSEVEKRFDGLAI--EGKLLKTRFNECIGMSEL 86
++DR KS A AL+ ++++K W+ VEKR+ + +G LL+++F ECIGM+
Sbjct: 80 RLDRSKSTAGQALKGLKIISKTDGNAAWTVVEKRYLKITANTDGLLLRSKFGECIGMNS- 138
Query: 87 KDFACKLFDALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRINE 146
K+FA +LFDALAR + IT+ EL++ WEQI ++SFDSR+ TFFDL+DK++DGR+ E
Sbjct: 139 KEFALELFDALARKSHLKGDVITETELKKFWEQINDKSFDSRLITFFDLMDKDSDGRLTE 198
Query: 147 EQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQSTT 206
++V+EII LS+S+N LS IQ ++ EYA++IMEELDPD++GYI + SL+ LLLQA +S +
Sbjct: 199 DEVREIIKLSSSANHLSCIQNKADEYAAMIMEELDPDHMGYIMMESLKKLLLQAETKSVS 258
Query: 207 ------DSKVVSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALF 260
+ K +S ML++ PT++ N ++R L +F+ D+W+R+WV+ALWL+I LF
Sbjct: 259 TDINSEERKELSDMLTESLKPTRDPNHLRRWYCQLRFFVLDSWQRVWVIALWLTIMAILF 318
Query: 261 TWKFFQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVP 320
+K+ QYKNRAV++V+G CV +AKG AETLK NMALIL P+CRNTITWLR+KT+LG+ VP
Sbjct: 319 AYKYIQYKNRAVYEVLGPCVCLAKGAAETLKLNMALILLPVCRNTITWLRNKTRLGVFVP 378
Query: 321 FDDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWF 380
FDDN+NFHKVIA GIAIG+ +H+V+HL CDFP L+ T AEY PL FFG+ +P Y F
Sbjct: 379 FDDNLNFHKVIAVGIAIGVAIHSVSHLACDFPLLIAATPAEYMPLGKFFGEEQPKRYLHF 438
Query: 381 LKGTEGWTGVLMVVLMAIPFILAQPWARRNRL--KLPKPLKKLTGFNAFWYSHHLLIIVY 438
+K TEG TG++MV LM I F LA PW RR +L KLP PLKKL FNAFWY+HHL +IVY
Sbjct: 439 VKSTEGITGLVMVFLMVIAFTLAMPWFRRGKLEKKLPGPLKKLASFNAFWYTHHLFVIVY 498
Query: 439 VLLVIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNV 498
+LLV+HG+++YL+K+WYK TTWMYLAVP+ LY ERLIRAFRS + V++ ++ Y G V
Sbjct: 499 ILLVLHGYYIYLNKEWYKKTTWMYLAVPVALYAYERLIRAFRSSIRTVKVLKMAAYPGKV 558
Query: 499 FALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQ 558
L+MSKP FKYMSGQY++VNC +SPFEWHPFSITS P+DDYLSVHI+ LGDWT +Q
Sbjct: 559 LTLQMSKPTNFKYMSGQYMFVNCPAVSPFEWHPFSITSTPQDDYLSVHIKALGDWTEAIQ 618
Query: 559 AILEKACQPSSDDQKCILQANTLP-------DTSKPRIPRLWIDGPYGSPAQDYKNYEVL 611
+ ++ + + I Q + P + PR P++ IDGPYG+PAQDYK YEV+
Sbjct: 619 GVFSESREKIVEKHHVIFQVSKPPPVGDMLNGANSPRFPKIMIDGPYGAPAQDYKKYEVV 678
Query: 612 LLVGLGIGATPLISILKGMLNNIKQQQDL---ENGVLESRVKTNKKKHFATKQAYFYWVT 668
LL+GLGIGATP+ISI+K ++NN + ++ L E G + + K+ F T++AYFYWVT
Sbjct: 679 LLIGLGIGATPMISIIKDIINNTETKEQLSQMEKG--SPQEQQGNKETFKTRRAYFYWVT 736
Query: 669 REQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDK 728
+EQG+F+WFK +MNEIAE DK VIELHN+CT VY+EGD SALI MLQSL +K GLD
Sbjct: 737 KEQGTFDWFKNIMNEIAERDKSKVIELHNHCTSVYEEGDVRSALIRMLQSLNYAKNGLDI 796
Query: 729 VSETRVKTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
V+ TRV +HF RPNW+NVY+ +++ HP VG +
Sbjct: 797 VAGTRVMSHFARPNWKNVYKQIAMDHPGANVGVF 830
>At5g07390 respiratory burst oxidase protein A
Length = 902
Score = 842 bits (2176), Expect = 0.0
Identities = 414/747 (55%), Positives = 543/747 (72%), Gaps = 15/747 (2%)
Query: 29 KVDRYKSGATHALQCFRVMTKNVVFEGWSEVEKRFDGLAI--EGKLLKTRFNECIGMSEL 86
K+ R KS A AL+ + +TK GW EVEKRF + + G L ++RF ECIGM
Sbjct: 125 KLRRSKSRAELALKGLKFITKTDGVTGWPEVEKRFYVMTMTTNGLLHRSRFGECIGMKST 184
Query: 87 KDFACKLFDALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRINE 146
+FA LFDALAR ++ SI +EL+E W+QIT+Q FDSR++TFF +VDK++DGR+NE
Sbjct: 185 -EFALALFDALARRENVSGDSININELKEFWKQITDQDFDSRLRTFFAMVDKDSDGRLNE 243
Query: 147 EQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQSTT 206
+V+EII+LSAS+N+L I+ ++ EYA+LIMEELDP + GYI + +LE+LLLQA Q
Sbjct: 244 AEVREIITLSASANELDNIRRQADEYAALIMEELDPYHYGYIMIENLEILLLQAPMQDVR 303
Query: 207 DS--KVVSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFTWKF 264
D K +S+MLSQ + + RN R + + YF+ DNWKR+WVMALW+ LFTWKF
Sbjct: 304 DGEGKKLSKMLSQNLMVPQSRNLGARFCRGMKYFLFDNWKRVWVMALWIGAMAGLFTWKF 363
Query: 265 FQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFDDN 324
+Y+ R+ ++VMG CV IAKG AETLK NMA+IL P+CRNTITWLR+KTKL +VPFDD+
Sbjct: 364 MEYRKRSAYEVMGVCVCIAKGAAETLKLNMAMILLPVCRNTITWLRTKTKLSAIVPFDDS 423
Query: 325 VNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKGT 384
+NFHKVIA GI++G+G+HA +HL CDFPRL+ +YEP++ +FG + Y F++
Sbjct: 424 LNFHKVIAIGISVGVGIHATSHLACDFPRLIAADEDQYEPMEKYFGP-QTKRYLDFVQSV 482
Query: 385 EGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIH 444
EG TG+ MVVLM I F LA W RRN+L LP PLKK+TGFNAFWYSHHL +IVY LLV+H
Sbjct: 483 EGVTGIGMVVLMTIAFTLATTWFRRNKLNLPGPLKKITGFNAFWYSHHLFVIVYSLLVVH 542
Query: 445 GHFLYLS-KKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKM 503
G ++YL + WYK TTWMYL VP++LY ERLIRAFRS + V + V+ GNV +L +
Sbjct: 543 GFYVYLIIEPWYKKTTWMYLMVPVVLYLCERLIRAFRSSVEAVSVLKVAVLPGNVLSLHL 602
Query: 504 SKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEK 563
S+P F+Y SGQY+Y+NCS +S EWHPFSITSAP DDYLSVHIR LGDWT QL+++ +
Sbjct: 603 SRPSNFRYKSGQYMYLNCSAVSTLEWHPFSITSAPGDDYLSVHIRVLGDWTKQLRSLFSE 662
Query: 564 ACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPL 623
C+P D+ + +A++ P PR+ IDGPYG+PAQDYK +EV+LLVGLGIGATP+
Sbjct: 663 VCKPRPPDEHRLNRADSKHWDYIPDFPRILIDGPYGAPAQDYKKFEVVLLVGLGIGATPM 722
Query: 624 ISILKGMLNNIK--------QQQDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFE 675
ISI+ ++NN+K +Q + N V + K + F TK+AYFYWVTREQGSF+
Sbjct: 723 ISIVSDIINNLKGVEEGSNRRQSPIHNMVTPPVSPSRKSETFRTKRAYFYWVTREQGSFD 782
Query: 676 WFKGLMNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVK 735
WFK +M+E+ E D++ VIELHNYCT VY+EGD SALITMLQSL +K+G+D VS TRV
Sbjct: 783 WFKNVMDEVTETDRKNVIELHNYCTSVYEEGDARSALITMLQSLNHAKHGVDVVSGTRVM 842
Query: 736 THFGRPNWRNVYEHVSLKHPDKRVGKY 762
+HF RPNWR+V++ +++ HP RVG +
Sbjct: 843 SHFARPNWRSVFKRIAVNHPKTRVGVF 869
>At1g64060 cytochrome b245 beta chain homolog RbohAp108
Length = 944
Score = 825 bits (2130), Expect = 0.0
Identities = 415/783 (53%), Positives = 539/783 (68%), Gaps = 38/783 (4%)
Query: 12 LDTRFTCSNSNGGC----------------DKLKVDRYKSGATHALQCFRVMT---KNVV 52
L T T +N +GG + ++DR +S A AL+ R ++ KNV
Sbjct: 135 LTTTSTAANQSGGAGGGLVNSALEARALRKQRAQLDRTRSSAQRALRGLRFISNKQKNV- 193
Query: 53 FEGWSEVEKRFDGLAIEGKLLKTRFNECIGMSELKDFACKLFDALARCRGITSTSITKDE 112
+GW++V+ F+ G + ++ F +CIGM + K+FA +LFDAL+R R + I DE
Sbjct: 194 -DGWNDVQSNFEKFEKNGYIYRSDFAQCIGMKDSKEFALELFDALSRRRRLKVEKINHDE 252
Query: 113 LREIWEQITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEY 172
L E W QI ++SFDSR+Q FFD+VDKN DGRI EE+VKEII LSAS+NKLS+++E++ EY
Sbjct: 253 LYEYWSQINDESFDSRLQIFFDIVDKNEDGRITEEEVKEIIMLSASANKLSRLKEQAEEY 312
Query: 173 ASLIMEELDPDNLGYIELYSLEMLLLQASAQSTTDSKV--VSQMLSQKFVPTKERNPIKR 230
A+LIMEELDP+ LGYIEL+ LE LLLQ + SQ LSQ + ++ I R
Sbjct: 313 AALIMEELDPERLGYIELWQLETLLLQKDTYLNYSQALSYTSQALSQNLQGLRGKSRIHR 372
Query: 231 GLQALAYFIQDNWKRIWVMALWLSICVALFTWKFFQYKNRAVFDVMGYCVVIAKGGAETL 290
Y +Q+NWKRIWV++LW+ I + LF WKFFQYK + F VMGYC++ AKG AETL
Sbjct: 373 MSSDFVYIMQENWKRIWVLSLWIMIMIGLFLWKFFQYKQKDAFHVMGYCLLTAKGAAETL 432
Query: 291 KFNMALILFPICRNTITWLRSKTKLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLTCD 350
KFNMALILFP+CRNTITWLRS T+L VPFDDN+NFHK IA I + + LH HL CD
Sbjct: 433 KFNMALILFPVCRNTITWLRS-TRLSYFVPFDDNINFHKTIAGAIVVAVILHIGDHLACD 491
Query: 351 FPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKGTEGWTGVLMVVLMAIPFILAQPWARRN 410
FPR++ T +Y + + K Y+ +KG EG TG+LMV+LM I F LA W RRN
Sbjct: 492 FPRIVRATEYDYNRYLFHYFQTKQPTYFDLVKGPEGITGILMVILMIISFTLATRWFRRN 551
Query: 411 RLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMILY 470
+KLPKP +LTGFNAFWYSHHL +IVY+LL++HG FLY +K WY TTWMYLAVP++LY
Sbjct: 552 LVKLPKPFDRLTGFNAFWYSHHLFVIVYILLILHGIFLYFAKPWYVRTTWMYLAVPVLLY 611
Query: 471 ESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWH 530
ER +R FRS VR+ V+ Y GNV L+MSKP F+Y SGQY++V C +SPFEWH
Sbjct: 612 GGERTLRYFRSGSYSVRLLKVAIYPGNVLTLQMSKPTQFRYKSGQYMFVQCPAVSPFEWH 671
Query: 531 PFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIP 590
PFSITSAPEDDY+S+HIR LGDWT +L+ + + C+P + +L+A+ +T+K +P
Sbjct: 672 PFSITSAPEDDYISIHIRQLGDWTQELKRVFSEVCEPPVGGKSGLLRAD---ETTKKSLP 728
Query: 591 RLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQDLENGVLE---- 646
+L IDGPYG+PAQDY+ Y+VLLLVGLGIGATP ISILK +LNNI + ++ + + +
Sbjct: 729 KLLIDGPYGAPAQDYRKYDVLLLVGLGIGATPFISILKDLLNNIVKMEEHADSISDFSRS 788
Query: 647 SRVKT-------NKKKHFATKQAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYC 699
S T +K+ T AYFYWVTREQGSF+WFKG+MNE+AE D+ GVIE+HNY
Sbjct: 789 SEYSTGSNGDTPRRKRILKTTNAYFYWVTREQGSFDWFKGVMNEVAELDQRGVIEMHNYL 848
Query: 700 TGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDKRV 759
T VY+EGD SALITM+Q+L +K G+D VS TRV+THF RPNW+ V +S KH + R+
Sbjct: 849 TSVYEEGDARSALITMVQALNHAKNGVDIVSGTRVRTHFARPNWKKVLTKLSSKHCNARI 908
Query: 760 GKY 762
G +
Sbjct: 909 GVF 911
>At1g19230 hypothetical protein
Length = 926
Score = 725 bits (1871), Expect = 0.0
Identities = 368/723 (50%), Positives = 485/723 (66%), Gaps = 45/723 (6%)
Query: 56 WSEVEKRFDGLAIEGKLLKTRFNECIGMSELKDFACKLFDALARCRGITSTSITKDELRE 115
W +VEKRF+ L+ G L + F EC+GM + KDFA +FDALAR R ITKDEL +
Sbjct: 200 WKKVEKRFESLSKNGLLARDDFGECVGMVDSKDFAVSVFDALARRRRQKLEKITKDELHD 259
Query: 116 IWEQITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASL 175
W QI++QSFD+R+Q FFD+ D N DG+I E++KE++ LSAS+NKL+K++E++ EYASL
Sbjct: 260 FWLQISDQSFDARLQIFFDMADSNEDGKITREEIKELLMLSASANKLAKLKEQAEEYASL 319
Query: 176 IMEELDPDNLGYIELYSLEMLLLQASAQSTTDSKVVSQMLSQKFVPTKERNPIKRGLQAL 235
IMEELDP+N GYIEL+ LE LLLQ A R L
Sbjct: 320 IMEELDPENFGYIELWQLETLLLQRDAYMN----------------------YSRPLSTT 357
Query: 236 AYFIQDNWKRIWVMALWLSICVALFTWKFFQYKNRAVFDVMGYCVVIAKGGAETLKFNMA 295
+ + +NW+R WV+ +W+ + LF WKF +Y+ +A F VMGYC+ AKG AETLK NMA
Sbjct: 358 SGGV-NNWQRSWVLLVWVMLMAILFVWKFLEYREKAAFKVMGYCLTTAKGAAETLKLNMA 416
Query: 296 LILFPICRNTITWLRSKTKLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLL 355
L+L P+CRNT+TWLRS T+ VPFDDN+NFHK+IA IAIGI +HA HL CDFPR++
Sbjct: 417 LVLLPVCRNTLTWLRS-TRARACVPFDDNINFHKIIACAIAIGILVHAGTHLACDFPRII 475
Query: 356 HTTAAEYEPLKPFFGKNKPDNYWWFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLP 415
+++ ++ + F KP + + G EG TG+ MV+L I F LA RRNR++LP
Sbjct: 476 NSSPEQFVLIASAFNGTKP-TFKDLMTGAEGITGISMVILTTIAFTLASTHFRRNRVRLP 534
Query: 416 KPLKKLTGFNAFWYSHHLLIIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMILYESERL 475
PL +LTGFNAFWY+HHLL++VY++L++HG FL+ + KWY+ TTWMY++VP++LY +ER
Sbjct: 535 APLDRLTGFNAFWYTHHLLVVVYIMLIVHGTFLFFADKWYQKTTWMYISVPLVLYVAERS 594
Query: 476 IRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSIT 535
+RA RS V+I VS G V +L MSKP GFKY SGQYI++ C IS FEWHPFSIT
Sbjct: 595 LRACRSKHYSVKILKVSMLPGEVLSLIMSKPPGFKYKSGQYIFLQCPTISRFEWHPFSIT 654
Query: 536 SAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCIL---QANTLPDTSKPRIPRL 592
SAP DD LSVHIRTLGDWT +L+ +L D C++ + + + P+L
Sbjct: 655 SAPGDDQLSVHIRTLGDWTEELRRVL----TVGKDLSTCVIGRSKFSAYCNIDMINRPKL 710
Query: 593 WIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQ-DLE--------NG 643
+DGPYG+PAQDY++Y+VLLL+GLGIGATP ISILK +LNN + +Q D E N
Sbjct: 711 LVDGPYGAPAQDYRSYDVLLLIGLGIGATPFISILKDLLNNSRDEQTDNEFSRSDFSWNS 770
Query: 644 VLES-RVKTNKKKHFATK---QAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYC 699
S T H K +A+FYWVTRE GS EWF+G+M EI++ D G IELHNY
Sbjct: 771 CTSSYTTATPTSTHGGKKKAVKAHFYWVTREPGSVEWFRGVMEEISDMDCRGQIELHNYL 830
Query: 700 TGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDKRV 759
T VY EGD S LI M+Q+L +K+G+D +S TRV+THF RPNW+ V+ ++ KHP+ V
Sbjct: 831 TSVYDEGDARSTLIKMVQALNHAKHGVDILSGTRVRTHFARPNWKEVFSSIARKHPNSTV 890
Query: 760 GKY 762
G +
Sbjct: 891 GVF 893
>At4g11230 respiratory burst oxidase homolog F - like protein
Length = 942
Score = 699 bits (1803), Expect = 0.0
Identities = 378/754 (50%), Positives = 483/754 (63%), Gaps = 40/754 (5%)
Query: 37 ATHALQCFRVMTKNVVFEGWSEVEKRFDGLAIEGKLLKTRFNECIGMSE--LKDFACKLF 94
A H L+ +V W++V+ F L+ +G L K+ F CIG+ K+FA +LF
Sbjct: 168 AIHGLKFISSKENGIV--DWNDVQNNFAHLSKDGYLFKSDFAHCIGLENENSKEFADELF 225
Query: 95 DALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIIS 154
DAL R R I I EL E W QIT++SFDSR+Q FF++ + + + II
Sbjct: 226 DALCRRRRIMVDKINLQELYEFWYQITDESFDSRLQIFFNMYCYQLSSNLVKHIDQHIII 285
Query: 155 LSASSNKLSKIQERSGEYASLIMEELDPDNL--GYIELYSLEMLLLQA--SAQSTTDSKV 210
LSAS+N LS+++ER+ EYA+LIMEEL PD L YIEL LE+LLL+ S +
Sbjct: 286 LSASANNLSRLRERAEEYAALIMEELAPDGLYSQYIELKDLEILLLEKDISHSYSLPFSQ 345
Query: 211 VSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFTWKFFQYKNR 270
S+ LSQ K+R R + L Y +QDNWKRIWV+ LW I LF WK +QYK++
Sbjct: 346 TSRALSQNL---KDRR--WRMSRNLLYSLQDNWKRIWVLTLWFVIMAWLFMWKCYQYKHK 400
Query: 271 AVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFDDNVNFHKV 330
F VMGYC+V+AKG AETLKFNMALIL P+CRNTIT+LRS T L VPFDD +NFHK
Sbjct: 401 DAFHVMGYCLVMAKGAAETLKFNMALILLPVCRNTITYLRS-TALSHSVPFDDCINFHKT 459
Query: 331 IAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEP-LKPFFGKNKPDNYWWFLKGTEGWTG 389
I+ I + LHA +HL CDFPR+L +T +Y+ L +FG +P Y+ + G TG
Sbjct: 460 ISVAIISAMLLHATSHLACDFPRILASTDTDYKRYLVKYFGVTRP-TYFGLVNTPVGITG 518
Query: 390 VLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHGHFLY 449
++MV M I F LA RRN KLPKP KLTG+NAFWYSHHLL+ VYVLLVIHG LY
Sbjct: 519 IIMVAFMLIAFTLASRRCRRNLTKLPKPFDKLTGYNAFWYSHHLLLTVYVLLVIHGVSLY 578
Query: 450 LSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGF 509
L KWY+ T WMYLAVP++LY ER+ R FRS V I V Y GNV L+MSKP F
Sbjct: 579 LEHKWYRKTVWMYLAVPVLLYVGERIFRFFRSRLYTVEICKVVIYPGNVVVLRMSKPTSF 638
Query: 510 KYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSS 569
Y SGQY++V C +S FEWHPFSITS+P DDYLS+HIR GDWT ++ C
Sbjct: 639 DYKSGQYVFVQCPSVSKFEWHPFSITSSPGDDYLSIHIRQRGDWTEGIKKAFSVVCHAPE 698
Query: 570 DDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKG 629
+ +L+A+ ++ P L IDGPYG+PAQD+ Y+V+LLVGLGIGATP +SIL+
Sbjct: 699 AGKSGLLRADV---PNQRSFPELLIDGPYGAPAQDHWKYDVVLLVGLGIGATPFVSILRD 755
Query: 630 MLNN-IKQQQDLE--------------------NGVLESRVKTNKKKHFATKQAYFYWVT 668
+LNN IKQQ+ E N SR+ ++K TK AYFYWVT
Sbjct: 756 LLNNIIKQQEQAECISGSCSNSNISSDHSFSCLNSEAASRIPQTQRKTLNTKNAYFYWVT 815
Query: 669 REQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDK 728
REQGSF+WFK +MNEIA++D++GVIE+HNY T VY+EGD S L+TM+Q+L +K G+D
Sbjct: 816 REQGSFDWFKEIMNEIADSDRKGVIEMHNYLTSVYEEGDTRSNLLTMIQTLNHAKNGVDI 875
Query: 729 VSETRVKTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
S T+V+THFGRP W+ V +S KH + R+G +
Sbjct: 876 FSGTKVRTHFGRPKWKKVLSKISTKHRNARIGVF 909
>At5g60010 respiratory burst oxidase protein - like
Length = 839
Score = 664 bits (1713), Expect = 0.0
Identities = 342/733 (46%), Positives = 471/733 (63%), Gaps = 32/733 (4%)
Query: 29 KVDRYKSGATHALQCFRVMTKNVVF---EGWSEVEKRFDGLAIEGKLLKTRFNECIGMSE 85
+V+R S A LQ R + + V + W +E RF+ +++GKL K +F CIGM +
Sbjct: 96 RVERTTSSAARGLQSLRFLDRTVTGRERDAWRSIENRFNQFSVDGKLPKEKFGVCIGMGD 155
Query: 86 LKDFACKLFDALARCRGI-TSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRI 144
+FA ++++AL R R I T I K++L+ WE + + D R+Q FFD+ DKN DG++
Sbjct: 156 TMEFAAEVYEALGRRRQIETENGIDKEQLKLFWEDMIKKDLDCRLQIFFDMCDKNGDGKL 215
Query: 145 NEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQS 204
EE+VKE+I LSAS+N+L +++ + YASLIMEELDPD+ GYIE++ LE+LL +
Sbjct: 216 TEEEVKEVIVLSASANRLGNLKKNAAAYASLIMEELDPDHKGYIEMWQLEILLTGMVTNA 275
Query: 205 TTDSKVVSQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFTWKF 264
T+ SQ L++ +P + R P+ + + A + +NWK++WV+ALW I V LF WK+
Sbjct: 276 DTEKMKKSQTLTRAMIPERYRTPMSKYVSVTAELMHENWKKLWVLALWAIINVYLFMWKY 335
Query: 265 FQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPFDDN 324
++ ++++ G CV AKG AETLK NMALIL P+CR T+T LRS T L VVPFDDN
Sbjct: 336 EEFMRNPLYNITGRCVCAAKGAAETLKLNMALILVPVCRKTLTILRS-TFLNRVVPFDDN 394
Query: 325 VNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGK---NKPDNYWWFL 381
+NFHKVIA+ IA LH H+ C++PRL ++ Y+ + G N +Y +
Sbjct: 395 INFHKVIAYMIAFQALLHTALHIFCNYPRL---SSCSYDVFLTYAGAALGNTQPSYLGLM 451
Query: 382 KGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLL 441
+ TGVLM+ M F LA + RRN +KLPKP L GFNAFWY+HHLL++ Y+LL
Sbjct: 452 LTSVSITGVLMIFFMGFSFTLAMHYFRRNIVKLPKPFNVLAGFNAFWYAHHLLVLAYILL 511
Query: 442 VIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLI-RAFRSCFKYVRIQSVSWYTGNVFA 500
+IHG++L + K WY+ TTWMYLAVPM+ Y SERL R + V + Y+GNV A
Sbjct: 512 IIHGYYLIIEKPWYQKTTWMYLAVPMLFYASERLFSRLLQEHSHRVNVIKAIVYSGNVLA 571
Query: 501 LKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAI 560
L ++KP GFKY SG Y++V C D+S FEWHPFSITSAP DDYLSVHIR LGDWT++L++
Sbjct: 572 LYVTKPPGFKYKSGMYMFVKCPDLSKFEWHPFSITSAPGDDYLSVHIRALGDWTTELRSR 631
Query: 561 LEKACQPSSDDQK----CILQANTLPDTSKPRI-------PRLWIDGPYGSPAQDYKNYE 609
K C+P+ K +++ T P I P+++I GPYG+PAQ+Y+ ++
Sbjct: 632 FAKTCEPTQAAAKPKPNSLMRMETRAAGVNPHIEESQVLFPKIFIKGPYGAPAQNYQKFD 691
Query: 610 VLLLVGLGIGATPLISILKGMLNNIK-----QQQDLENGVLESRVKTNKKKHFATK---- 660
+LLLVGLGIGATP ISILK MLN++K Q E V + + K
Sbjct: 692 ILLLVGLGIGATPFISILKDMLNHLKPGIPRSGQKYEGSVGGESIGGDSVSGGGGKKFPQ 751
Query: 661 QAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLY 720
+AYF+WVTREQ SF+WFKG+M++IAE DK VIE+HNY T +Y+ GD SALI M+Q L
Sbjct: 752 RAYFFWVTREQASFDWFKGVMDDIAEYDKTHVIEMHNYLTSMYEAGDARSALIAMVQKLQ 811
Query: 721 QSKYGLDKVSETR 733
+K G+D VSE+R
Sbjct: 812 HAKNGVDIVSESR 824
>At3g45810 respiratory burst oxidase - like protein
Length = 835
Score = 600 bits (1547), Expect = e-172
Identities = 326/753 (43%), Positives = 448/753 (59%), Gaps = 78/753 (10%)
Query: 29 KVDRYKSGATHALQCFRVMTKNVVF---EGWSEVEKRFDGLAIEGKLLKTRFNECIGMSE 85
+V+R S A LQ R + + V + W +E RF+ A++G+L K +F CIGM +
Sbjct: 106 RVERTTSSAARGLQSLRFLDRTVTGRERDSWRSIENRFNQFAVDGRLPKDKFGVCIGMGD 165
Query: 86 LKDFACKLFDALARCRGI-TSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRI 144
+FA K+++AL R R I T I K++L+ WE + + D R+Q FFD+ DK+ DG++
Sbjct: 166 TLEFAAKVYEALGRRRQIKTENGIDKEQLKLFWEDMIKKDLDCRLQIFFDMCDKDGDGKL 225
Query: 145 NEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQASAQS 204
EE+VKE+I LSAS+N+L +++ + YASLIMEELDP+ GYIE++ LE+LL S
Sbjct: 226 TEEEVKEVIVLSASANRLVNLKKNAASYASLIMEELDPNEQGYIEMWQLEVLL--TGIVS 283
Query: 205 TTDS-KVV--SQMLSQKFVPTKERNPIKRGLQALAYFIQDNWKRIWVMALWLSICVALFT 261
DS KVV SQ L++ +P + R P + + A + ++WK+IWV+ LWL++ V LF
Sbjct: 284 NADSHKVVRKSQQLTRAMIPKRYRTPTSKYVVVTAELMYEHWKKIWVVTLWLAVNVVLFM 343
Query: 262 WKFFQYKNRAVFDVMGYCVVIAKGGAETLKFNMALILFPICRNTITWLRSKTKLGMVVPF 321
WK+ ++ ++++ G C+ AKG AE LK NMALIL P+ R T+T+LRS T L ++PF
Sbjct: 344 WKYEEFTTSPLYNITGRCLCAAKGTAEILKLNMALILVPVLRRTLTFLRS-TFLNHLIPF 402
Query: 322 DDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFL 381
DDN+NFHK+IA IA+ LH H+ C++PRL Y K Y +
Sbjct: 403 DDNINFHKLIAVAIAVISLLHTALHMLCNYPRLSSCPYNFYSDYAGNLLGAKQPTYLGLM 462
Query: 382 KGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLL 441
TGVLM++ M I F LA + RRN +KLP P +L GFN+FWY+HHLL+I Y LL
Sbjct: 463 LTPVSVTGVLMIIFMGISFTLAMHYFRRNIVKLPIPFNRLAGFNSFWYAHHLLVIAYALL 522
Query: 442 VIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFAL 501
+IHG+ L + K WY+ T + Y+GNV AL
Sbjct: 523 IIHGYILIIEKPWYQKTAIV--------------------------------YSGNVLAL 550
Query: 502 KMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAIL 561
M+KPQGFKY SG Y++V C DIS FEWHPFSITSAP D+YLSVHIR LGDWTS+L+
Sbjct: 551 YMTKPQGFKYKSGMYMFVKCPDISKFEWHPFSITSAPGDEYLSVHIRALGDWTSELRNRF 610
Query: 562 EKACQP---SSDDQKCILQANTLPDTSKPRI-------PRLWIDGPYGSPAQDYKNYEVL 611
+ C+P S +++ T + P + PR++I GPYG+PAQ Y+ +++L
Sbjct: 611 AETCEPHQKSKPSPNDLIRMETRARGANPHVEESQALFPRIFIKGPYGAPAQSYQKFDIL 670
Query: 612 LLVGLGIGATPLISILKGMLNNI-----KQQQDLE---------------------NGVL 645
LL+GLGIGATP ISILK MLNN+ K Q E G +
Sbjct: 671 LLIGLGIGATPFISILKDMLNNLKPGIPKTGQKYEGSVGGESLGGSSVYGGSSVNGGGSV 730
Query: 646 ESRVKTNKKKHFATKQAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKE 705
+ ++AYFYWVTREQ SFEWFKG+M++IA DK VIE+HNY T +Y+
Sbjct: 731 NGGGSVSGGGRKFPQRAYFYWVTREQASFEWFKGVMDDIAVYDKTNVIEMHNYLTSMYEA 790
Query: 706 GDDGSALITMLQSLYQSKYGLDKVSETRVKTHF 738
GD SALI M+Q L +K G+D VSE+RV + F
Sbjct: 791 GDARSALIAMVQKLQHAKNGVDIVSESRVCSFF 823
>At5g49740 FRO1-like protein; NADPH oxidase-like
Length = 739
Score = 95.9 bits (237), Expect = 7e-20
Identities = 71/264 (26%), Positives = 127/264 (47%), Gaps = 35/264 (13%)
Query: 379 WFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVY 438
W G GV+ +V + ++ + R+N +L F+Y+H L I+
Sbjct: 250 WKATGIAVLPGVISLVAGLLMWVTSLHTVRKNYFEL------------FFYTHQLYIVFV 297
Query: 439 VLLVIH-GHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGN 497
V L +H G +L+ +A + L+ +R +R ++S + V + S
Sbjct: 298 VFLALHVGDYLF-----------SIVAGGIFLFILDRFLRFYQSR-RTVDVISAKSLPCG 345
Query: 498 VFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPED--DYLSVHIRTLGDWTS 555
L +SKP +Y + +I++ ++S +WHPFS++S+P D +++V I+ LG WT+
Sbjct: 346 TLELVLSKPPNMRYNALSFIFLQVKELSWLQWHPFSVSSSPLDGNHHVAVLIKVLGGWTA 405
Query: 556 QLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVG 615
+L+ L + + DQ + S P+I ++GPYG + + YE L+LV
Sbjct: 406 KLRDQLSTLYEAENQDQ-------LISPESYPKITTC-VEGPYGHESPYHLAYENLVLVA 457
Query: 616 LGIGATPLISILKGMLNNIKQQQD 639
GIG TP +IL +L+ + +D
Sbjct: 458 GGIGITPFFAILSDILHRKRDGKD 481
>At5g49730 FRO2-like protein; NADPH oxidase-like
Length = 738
Score = 91.7 bits (226), Expect = 1e-18
Identities = 62/212 (29%), Positives = 107/212 (50%), Gaps = 23/212 (10%)
Query: 424 FNAFWYSHHLLIIVYVLLVIH-GHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSC 482
F F+Y+H L I+ V L +H G +++ +A + L+ +R +R F
Sbjct: 282 FELFFYTHQLYIVFIVFLALHVGDYMF-----------SIVAGGIFLFILDRFLR-FCQS 329
Query: 483 FKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPED-- 540
+ V + S L +SKP +Y + +I++ ++S +WHPFS++S+P D
Sbjct: 330 RRTVDVISAKSLPCGTLELVLSKPPNMRYNALSFIFLQVRELSWLQWHPFSVSSSPLDGN 389
Query: 541 DYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGS 600
+++V I+ LG WT++L+ L + + DQ + S P+I ++GPYG
Sbjct: 390 HHVAVLIKVLGGWTAKLRDQLSNLYEAENQDQ-------LISPQSYPKITTC-VEGPYGH 441
Query: 601 PAQDYKNYEVLLLVGLGIGATPLISILKGMLN 632
+ + YE L+LV GIG TP +IL +L+
Sbjct: 442 ESPYHLAYENLVLVAGGIGITPFFAILSDILH 473
>At5g50160 FRO1 and FRO2-like protein
Length = 728
Score = 90.1 bits (222), Expect = 4e-18
Identities = 69/256 (26%), Positives = 113/256 (43%), Gaps = 49/256 (19%)
Query: 379 WFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVY 438
W G G++ +V + +I + P RR F F+Y+HHL I+
Sbjct: 230 WQRTGRVYVAGLISLVTGLLMWITSLPQIRRKN------------FEVFYYTHHLYIVFL 277
Query: 439 VLLVIHG---HFLYLSKKWYKNTTWMYLAVP-MILYESERLIRAFRSCFKYVRIQSVSWY 494
V + H HF Y +P M L+ ++++R +S + I S + +
Sbjct: 278 VAFLFHAGDRHF--------------YWVLPGMFLFGLDKILRIVQSRSESC-ILSANLF 322
Query: 495 TGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDY--LSVHIRTLGD 552
+ L + K Y +I++N +S F+WHPFSI S+ D LS+ ++ GD
Sbjct: 323 SCKAIELVLPKDPMLNYAPSSFIFLNIPLVSRFQWHPFSIISSSSVDKHSLSIMMKCEGD 382
Query: 553 WTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLL 612
WT+ + +E+A + I++ ++GPYG + D+ Y+ L
Sbjct: 383 WTNSVYNKIEEAANCENKINNIIVR----------------VEGPYGPASVDFLRYDNLF 426
Query: 613 LVGLGIGATPLISILK 628
LV GIG TP +SILK
Sbjct: 427 LVAGGIGITPFLSILK 442
>At5g23990 FRO2 homolog
Length = 707
Score = 84.3 bits (207), Expect = 2e-16
Identities = 70/260 (26%), Positives = 112/260 (42%), Gaps = 49/260 (18%)
Query: 375 DNYWWFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLL 434
+ + W G + +V+ ++ + P RR + F F+Y+HHL
Sbjct: 230 ETFAWNATYVPNLAGTIAMVIGIAIWVTSLPSFRRKK------------FEIFFYTHHLY 277
Query: 435 IIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMI-LYESERLIRAFRSCFKYVRIQSVSW 493
+ V IH +W + +P I L+ +R +R +S K R+ S
Sbjct: 278 GLYIVFYAIH-----------VGDSWFCMILPNIFLFFIDRYLRFLQST-KRSRLVSAKI 325
Query: 494 YTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAP--EDDYLSVHIRTLG 551
+ L +K G Y ++++ IS +WHPF+ITS+ E D LSV IR G
Sbjct: 326 LPSDNLELTFAKTSGLHYTPTSILFLHVPSISKLQWHPFTITSSSNLEKDTLSVVIRKQG 385
Query: 552 DWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVL 611
WT +L L SS D L+ +T +GPYG + D ++ L
Sbjct: 386 SWTQKLYTHLS-----SSIDS---LEVST--------------EGPYGPNSFDVSRHDSL 423
Query: 612 LLVGLGIGATPLISILKGML 631
+LVG G G TP IS+++ ++
Sbjct: 424 ILVGGGSGVTPFISVIRELI 443
>At5g23980 FRO2 homolog
Length = 699
Score = 82.8 bits (203), Expect = 7e-16
Identities = 65/212 (30%), Positives = 97/212 (45%), Gaps = 37/212 (17%)
Query: 424 FNAFWYSHHLLIIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMI-LYESERLIRAFRSC 482
F F+Y+HHL + V VIH +W + +P I L+ +R +R +S
Sbjct: 259 FEIFFYTHHLYGLYIVFYVIH-----------VGDSWFCMILPNIFLFFIDRYLRFLQST 307
Query: 483 FKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAP--ED 540
K R+ S + L SK G Y ++++ IS +WHPF+ITS+ E
Sbjct: 308 -KRSRLVSARILPSDNLELTFSKTPGLHYTPTSILFLHVPSISKIQWHPFTITSSSNLEK 366
Query: 541 DYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGS 600
D LSV IR G WT +L L SS D L+ +T +GPYG
Sbjct: 367 DTLSVVIRRQGSWTQKLYTHLS-----SSIDS---LEVST--------------EGPYGP 404
Query: 601 PAQDYKNYEVLLLVGLGIGATPLISILKGMLN 632
+ D + L+LV G G TP IS+++ +++
Sbjct: 405 NSFDVSRHNSLILVSGGSGITPFISVIRELIS 436
>At1g01580 hypothetical protein
Length = 725
Score = 81.3 bits (199), Expect = 2e-15
Identities = 64/256 (25%), Positives = 112/256 (43%), Gaps = 47/256 (18%)
Query: 379 WFLKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVY 438
W KG G + + + + P RR F F+Y+H+L I+
Sbjct: 253 WDTKGVSNLAGEIALAAGLVMWATTYPKIRRRF------------FEVFFYTHYLYIVFM 300
Query: 439 VLLVIHGHFLYLSKKWYKNTTWMYLAVP-MILYESERLIRAFRSCFKYVRIQSVSWYTGN 497
+ V+H ++ ++A+P ++ +R +R +S + VR+ + +
Sbjct: 301 LFFVLH-----------VGISFSFIALPGFYIFLVDRFLRFLQSR-ENVRLLAARILPSD 348
Query: 498 VFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAP--EDDYLSVHIRTLGDWTS 555
L SK Y ++VN IS +WHPF+ITS+ E + LS+ I+ G W++
Sbjct: 349 TMELTFSKNSKLVYSPTSIMFVNIPSISKLQWHPFTITSSSKLEPEKLSIVIKKEGKWST 408
Query: 556 QLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVG 615
+L L SS DQ L + ++GPYG + D+ +E L++V
Sbjct: 409 KLHQRL------SSSDQIDRLAVS--------------VEGPYGPASADFLRHEALVMVC 448
Query: 616 LGIGATPLISILKGML 631
G G TP IS+++ ++
Sbjct: 449 GGSGITPFISVIRDLI 464
>At1g01590 hypothetical protein
Length = 704
Score = 76.3 bits (186), Expect = 6e-14
Identities = 64/221 (28%), Positives = 103/221 (45%), Gaps = 39/221 (17%)
Query: 415 PKPLKKLTGFNAFWYSHHLLIIVYVLLVIH---GHFLYLSKKWYKNTTWMYLAVPMILYE 471
PK ++L F F+YSH+L I+ + V H H L +Y ++
Sbjct: 270 PKIRRRL--FEVFFYSHYLYIVFMLFFVFHVGISHALIPLPGFY-------------IFL 314
Query: 472 SERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHP 531
+R +R +S V++ S + L SK Y ++VN IS +WHP
Sbjct: 315 VDRFLRFLQSR-NNVKLVSARVLPCDTVELNFSKNPMLMYSPTSTMFVNIPSISKLQWHP 373
Query: 532 FSITSAP--EDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRI 589
F+I S+ E + LSV I++ G W+++L +L + SSD Q N L +
Sbjct: 374 FTIISSSKLEPETLSVMIKSQGKWSTKLYDMLSSS---SSD------QINRLAVS----- 419
Query: 590 PRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGM 630
++GPYG + D+ +E L++V G G TP ISI++ +
Sbjct: 420 ----VEGPYGPSSTDFLRHESLVMVSGGSGITPFISIVRDL 456
>At1g23020 putative superoxide-generating NADPH oxidase
flavocytochrome
Length = 693
Score = 75.5 bits (184), Expect = 1e-13
Identities = 58/211 (27%), Positives = 96/211 (45%), Gaps = 35/211 (16%)
Query: 424 FNAFWYSHHLLIIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVP-MILYESERLIRAFRSC 482
F F+Y+H+L ++ + V H Y ++ P ++ +R +R +S
Sbjct: 276 FEVFFYTHYLYMVFMLFFVFHVGISYA-----------LISFPGFYIFMVDRFLRFLQSR 324
Query: 483 FKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAP--ED 540
V++ S L SK Y ++VN IS +WHPF+ITS+ E
Sbjct: 325 -NNVKLVSARVLPCETVELNFSKNPMLMYSPTSILFVNIPSISKLQWHPFTITSSSKLEP 383
Query: 541 DYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGS 600
LSV I++ G W+S+L +L +S +Q L + ++GPYG
Sbjct: 384 KKLSVMIKSQGKWSSKLHHML------ASSNQIDHLAVS--------------VEGPYGP 423
Query: 601 PAQDYKNYEVLLLVGLGIGATPLISILKGML 631
+ DY ++ L++V G G TP ISI++ +L
Sbjct: 424 ASTDYLRHDSLVMVSGGSGITPFISIIRDLL 454
>At1g66400 calmodulin-related protein
Length = 157
Score = 37.7 bits (86), Expect = 0.024
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 9/82 (10%)
Query: 129 VQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYI 188
++ F DKN DG+I+ +++K++I + + S E +M+E D D G+I
Sbjct: 16 IKKVFQRFDKNNDGKISIDELKDVIGALSPN--------ASQEETKAMMKEFDLDGNGFI 67
Query: 189 ELYSLEMLLLQASAQSTTDSKV 210
+L + L Q S QS+ +S +
Sbjct: 68 DLDEF-VALFQISDQSSNNSAI 88
Score = 29.3 bits (64), Expect = 8.6
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 19/96 (19%)
Query: 113 LREIWEQITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEY 172
L +I +Q +N S ++ FDL D + +GRI S+N+L + + GE
Sbjct: 75 LFQISDQSSNNSAIRDLKEAFDLYDLDRNGRI-------------SANELHSVMKNLGEK 121
Query: 173 ASL-----IMEELDPDNLGYIELYSL-EMLLLQASA 202
S+ ++ ++D D G ++ +M+++ SA
Sbjct: 122 CSIQDCQRMINKVDSDGDGCVDFEEFKKMMMINGSA 157
>At2g34020 unknown protein
Length = 610
Score = 37.0 bits (84), Expect = 0.041
Identities = 18/61 (29%), Positives = 34/61 (55%), Gaps = 3/61 (4%)
Query: 129 VQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYI 188
+++ FD +D+N DG+I Q+ E+ L+ K++ E+AS ++ E D D G +
Sbjct: 329 LKSLFDKIDRNKDGKI---QISELKDLTVEFGVFGKMKCDINEFASTLLAEFDKDKNGEL 385
Query: 189 E 189
+
Sbjct: 386 D 386
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,554,435
Number of Sequences: 26719
Number of extensions: 759810
Number of successful extensions: 2040
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1929
Number of HSP's gapped (non-prelim): 58
length of query: 762
length of database: 11,318,596
effective HSP length: 107
effective length of query: 655
effective length of database: 8,459,663
effective search space: 5541079265
effective search space used: 5541079265
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0299.3


