

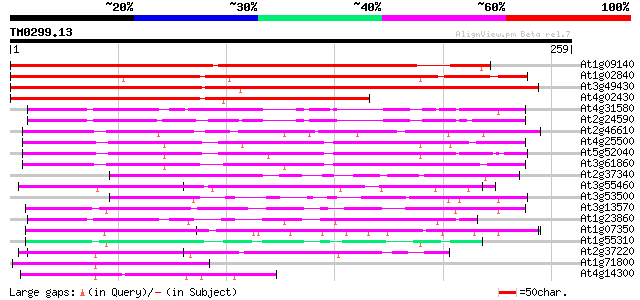
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0299.13
(259 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09140 SF2/ASF splicing modulator, Srp30 like protein 324 3e-89
At1g02840 ribonucleoprotein SF-2 like protein 322 2e-88
At3g49430 PRE-MRNA SPLICING FACTOR SF2-like protein 298 1e-81
At4g02430 unknown protein 259 8e-70
At4g31580 splicing factor 9G8-like SR protein / SRZ-22 73 1e-13
At2g24590 RSZp22 splicing factor like protein 73 2e-13
At2g46610 arginine/serine-rich splicing factor like protein 72 2e-13
At4g25500 splicing factor At-SRp40 72 3e-13
At5g52040 arginine/serine-rich splicing factor RSP41 homolog 70 1e-12
At3g61860 ARGININE/SERINE-RICH SPLICING FACTOR RSP31 69 3e-12
At2g37340 unknown protein 66 2e-11
At3g55460 RNA binding protein like 66 2e-11
At3g53500 splicing factor - like protein 61 5e-10
At3g13570 serine/arginine-rich protein 60 9e-10
At1g23860 9G8-like splicing factor / SRZ-21 60 1e-09
At1g07350 transformer-SR ribonucleoprotein like protein 57 1e-08
At1g55310 Serine/arginine-rich protein 55 5e-08
At2g37220 putative RNA-binding protein 53 2e-07
At1g71800 cleavage stimulation factor like protein 53 2e-07
At4g14300 ribonucleoprotein like protein 50 1e-06
>At1g09140 SF2/ASF splicing modulator, Srp30 like protein
Length = 253
Score = 324 bits (831), Expect = 3e-89
Identities = 169/223 (75%), Positives = 186/223 (82%), Gaps = 18/223 (8%)
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAED 60
MSSR +RTIYVGNLPGDIR EVED+FYK+GPIVDIDLKIPPRPPGYAFV+FED RDA+D
Sbjct: 1 MSSRWNRTIYVGNLPGDIRKCEVEDLFYKYGPIVDIDLKIPPRPPGYAFVEFEDPRDADD 60
Query: 61 AIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPP 120
AIY RDGYDFDG RLRVE+AHGGR S SVDRYS S S+SR SRRSDYRVLVTGLPP
Sbjct: 61 AIYGRDGYDFDGCRLRVEIAHGGRRFSPSVDRYS--SSYSASRAPSRRSDYRVLVTGLPP 118
Query: 121 SASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRA 180
SASWQDLKDHMRKAGDVCFS+VF +R GM+G+VDY+NYDDMKYA+RKLD +EFRNAFS A
Sbjct: 119 SASWQDLKDHMRKAGDVCFSEVFPDRKGMSGVVDYSNYDDMKYAIRKLDATEFRNAFSSA 178
Query: 181 YIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRS-RSRSRS 222
YIRVREY+ SRS+SRSP S+S RSRSRS
Sbjct: 179 YIRVREYE---------------SRSVSRSPDDSKSYRSRSRS 206
>At1g02840 ribonucleoprotein SF-2 like protein
Length = 283
Score = 322 bits (824), Expect = 2e-88
Identities = 179/267 (67%), Positives = 199/267 (74%), Gaps = 35/267 (13%)
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQ--------- 51
MSSRSSRT+YVGNLPGDIR REVED+F K+GP+V IDLK+PPRPPGYAFV+
Sbjct: 1 MSSRSSRTVYVGNLPGDIREREVEDLFSKYGPVVQIDLKVPPRPPGYAFVEVSLPPLILA 60
Query: 52 --------FEDARDAEDAIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGS--- 100
F+DARDAEDAI+ RDGYDFDG RLRVELAHGGR SS D S++GG
Sbjct: 61 VTCVMILDFDDARDAEDAIHGRDGYDFDGHRLRVELAHGGRRSSD--DTRGSFNGGGRGG 118
Query: 101 -------SSRGASRRSDYRVLVTGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIV 153
SRG SRRS++RVLVTGLP SASWQDLKDHMRK GDVCFSQV+R+ G TG+V
Sbjct: 119 GRGRGDGGSRGPSRRSEFRVLVTGLPSSASWQDLKDHMRKGGDVCFSQVYRDARGTTGVV 178
Query: 154 DYTNYDDMKYALRKLDDSEFRNAFSRAYIRVREYD-RDYSRSPSRDSRRSYSRSISRSPY 212
DYT Y+DMKYAL+KLDD+EFRNAFS Y+RVREYD R SRSPSR RSYS+S SRS
Sbjct: 179 DYTCYEDMKYALKKLDDTEFRNAFSNGYVRVREYDSRKDSRSPSRG--RSYSKSRSRSRG 236
Query: 213 LSRSRSRSRSYSGRSISLSPTAKRSRR 239
S SRSRSRS RS S SP AK SRR
Sbjct: 237 RSVSRSRSRS---RSRSRSPKAKSSRR 260
>At3g49430 PRE-MRNA SPLICING FACTOR SF2-like protein
Length = 300
Score = 298 bits (764), Expect = 1e-81
Identities = 159/256 (62%), Positives = 183/256 (71%), Gaps = 13/256 (5%)
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAED 60
MS R SR+IYVGNLPGDIR E+EDIFYK+G IVDI+LK+PPRPP Y FV+FE +RDAED
Sbjct: 1 MSGRFSRSIYVGNLPGDIREHEIEDIFYKYGRIVDIELKVPPRPPCYCFVEFEHSRDAED 60
Query: 61 AIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGA------------SRR 108
AI RDGY+ DG RLRVELAHGGRG SSS DR Y GG S G SR
Sbjct: 61 AIKGRDGYNLDGCRLRVELAHGGRGQSSS-DRRGGYGGGGSGYGGGGGGGGSARFGVSRH 119
Query: 109 SDYRVLVTGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKL 168
S++RV+V GLP SASWQDLKDHMRKAGDVCF++V R+ G G+VDYTNYDDMKYA+RKL
Sbjct: 120 SEFRVIVRGLPSSASWQDLKDHMRKAGDVCFAEVTRDSDGTYGVVDYTNYDDMKYAIRKL 179
Query: 169 DDSEFRNAFSRAYIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSI 228
DD+EFRN ++R +IRV++Y+ SRS S RS SRS SRS RS SRSRS S
Sbjct: 180 DDTEFRNPWARGFIRVKKYESSRSRSRSPSRSRSRSRSRSRSRGRGRSHSRSRSLSRSKS 239
Query: 229 SLSPTAKRSRRLLSLS 244
+K RR LS S
Sbjct: 240 PRKDLSKSPRRSLSRS 255
Score = 30.0 bits (66), Expect = 1.3
Identities = 20/51 (39%), Positives = 26/51 (50%)
Query: 174 RNAFSRAYIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYS 224
R + SR+ + R D +SP R RS SRS SRS S+S + R S
Sbjct: 249 RRSLSRSISKSRSPSPDKKKSPPRAMSRSKSRSRSRSRSPSKSPPKVREGS 299
>At4g02430 unknown protein
Length = 294
Score = 259 bits (663), Expect = 8e-70
Identities = 131/177 (74%), Positives = 144/177 (81%), Gaps = 13/177 (7%)
Query: 1 MSSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAED 60
MSSRSSRTIYVGNLPGDIR REVED+F K+GP+V IDLKIPPRPPGYAFV+FEDARDA+D
Sbjct: 1 MSSRSSRTIYVGNLPGDIREREVEDLFSKYGPVVQIDLKIPPRPPGYAFVEFEDARDADD 60
Query: 61 AIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYS-----------GGSSSRGASRRS 109
AIY RDGYDFDG LRVELAHGGR SS D SYS GG RG SRRS
Sbjct: 61 AIYGRDGYDFDGHHLRVELAHGGRRSSH--DARGSYSGRGRGGRGGGDGGGRERGPSRRS 118
Query: 110 DYRVLVTGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALR 166
+YRV+V+GLP SASWQDLKDHMRK G+VCFSQVFR+ G TGIVDYT+Y+DMKYA+R
Sbjct: 119 EYRVVVSGLPSSASWQDLKDHMRKGGEVCFSQVFRDGRGTTGIVDYTSYEDMKYAVR 175
>At4g31580 splicing factor 9G8-like SR protein / SRZ-22
Length = 200
Score = 73.2 bits (178), Expect = 1e-13
Identities = 81/231 (35%), Positives = 99/231 (42%), Gaps = 54/231 (23%)
Query: 9 IYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRDGY 68
+YVGNL + RE+ED F FG + + + RPPGYAF+ FED RDA DAI DG
Sbjct: 4 VYVGNLDPRVTERELEDEFRAFGVVRSV--WVARRPPGYAFLDFEDPRDARDAIRALDGK 61
Query: 69 DFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASWQDLK 128
+ RVE +H RG R GG RG SD + G
Sbjct: 62 N----GWRVEQSH-NRGERGGGGRGGDRGGGGGGRGGRGGSDLKCYECG----------- 105
Query: 129 DHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIRVREYD 188
+ G F++ R RGG TG R + SR+ R
Sbjct: 106 ----ETGH--FARECRNRGG-TG---------------------RRRSKSRSRTPPR--- 134
Query: 189 RDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYS-GRSISLSPTAKRSR 238
Y RSPS RRSYS +RSP R RS S + GRS S SP R+R
Sbjct: 135 --YRRSPSY-GRRSYSPR-ARSPPPPRRRSPSPPPARGRSYSRSPPPYRAR 181
>At2g24590 RSZp22 splicing factor like protein
Length = 196
Score = 72.8 bits (177), Expect = 2e-13
Identities = 77/230 (33%), Positives = 99/230 (42%), Gaps = 56/230 (24%)
Query: 9 IYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRDGY 68
+YVGNL + RE+ED F FG I + + RPPGYAF+ FED+RDA DAI DG
Sbjct: 4 VYVGNLDPRVTERELEDEFRSFGVIRSV--WVARRPPGYAFLDFEDSRDARDAIREVDGK 61
Query: 69 DFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASWQDLK 128
+G+R+ GG G R GG RG SD + G
Sbjct: 62 --NGWRVEQSHNRGGGGG-----RGGGRGGGDGGRGRG-GSDLKCYECG----------- 102
Query: 129 DHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIRVREYD 188
++G F++ R RGG G R + SR+ R
Sbjct: 103 ----ESGH--FARECRSRGGSGG---------------------RRRSRSRSRSPPR--- 132
Query: 189 RDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSISLSPTAKRSR 238
Y +SP+ RRSYS +RSP R RS S GR+ S SP R+R
Sbjct: 133 --YRKSPTYGGRRSYSPR-ARSP--PPPRRRSPSPRGRNYSRSPPPYRAR 177
>At2g46610 arginine/serine-rich splicing factor like protein
Length = 250
Score = 72.4 bits (176), Expect = 2e-13
Identities = 83/265 (31%), Positives = 115/265 (43%), Gaps = 43/265 (16%)
Query: 7 RTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRD 66
R +YVGN D R ++E +F KFG + +D+K GYAFV FED RDAEDAI D
Sbjct: 2 RHVYVGNFDYDTRHSDLERLFSKFGRVKRVDMK-----SGYAFVYFEDERDAEDAIRRTD 56
Query: 67 G--YDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASW 124
+ + +L VE A +G +R G + S ++R + V P +
Sbjct: 57 NTTFGYGRRKLSVEWAKDFQG-----ERGKPRDGKAVS---NQRPTKTLFVINFDPIRTR 108
Query: 125 Q-DLKDHMRKAGDV---------CFSQVFRERGGMTGIVDYTNYD-------DMKYALRK 167
+ D++ H G V F Q F + T +D T+ ++YALR+
Sbjct: 109 ERDMERHFEPYGKVLNVRMRRNFAFVQ-FATQEDATKALDSTHNSKLLDKVVSVEYALRE 167
Query: 168 LDDSEFRNAFSRAYIRVREYDRDYSRSPSRDSRR----SYSRSISRSPYLSRSR---SRS 220
+ E R A SR R R Y R PS D R Y R +PY R R
Sbjct: 168 AGEREDRYAGSR---RRRSPSPVYRRRPSPDYTRRRSPEYDRYKGPAPYERRKSPDYGRR 224
Query: 221 RSYSGRSISLSPTAKRSRRLLSLSR 245
S GR+ + SP RSR + +R
Sbjct: 225 SSDYGRARARSPGYDRSRSPIQRAR 249
>At4g25500 splicing factor At-SRp40
Length = 350
Score = 72.0 bits (175), Expect = 3e-13
Identities = 70/238 (29%), Positives = 102/238 (42%), Gaps = 25/238 (10%)
Query: 7 RTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRD 66
+ ++ GN D R ++E +F K+G + +D+K G+AFV ED RDAEDAI D
Sbjct: 2 KPVFCGNFEYDAREGDLERLFRKYGKVERVDMKA-----GFAFVYMEDERDAEDAIRALD 56
Query: 67 GYDF--DGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGAS--RRSDYRVLVTGLPPSA 122
++F G RLRVE RG SGG S R +S R S ++ +
Sbjct: 57 RFEFGRKGRRLRVEWTKSERGGDKR-------SGGGSRRSSSSMRPSKTLFVINFDADNT 109
Query: 123 SWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYI 182
+DL+ H G + ++ R + Y +D AL ++S+ +
Sbjct: 110 RTRDLEKHFEPYGKIVNVRIRRN----FAFIQYEAQEDATRALDASNNSKLMDKVISVEY 165
Query: 183 RVREYD-RDYSRSPSRDSRRS-YSRSISRSPYLSRSRSRSRSYSGRSISLSPTAKRSR 238
V++ D R SP R RS R S SPY R R GR S ++ R
Sbjct: 166 AVKDDDARGNGHSPERRRDRSPERRRRSPSPY---KRERGSPDYGRGASPVAAYRKER 220
>At5g52040 arginine/serine-rich splicing factor RSP41 homolog
Length = 356
Score = 69.7 bits (169), Expect = 1e-12
Identities = 69/237 (29%), Positives = 100/237 (42%), Gaps = 24/237 (10%)
Query: 7 RTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRD 66
+ ++ GN D R ++E +F K+G + +D+K G+AFV ED RDAEDAI D
Sbjct: 2 KPVFCGNFEYDARESDLERLFRKYGKVERVDMKA-----GFAFVYMEDERDAEDAIRALD 56
Query: 67 GYDF--DGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGL-PPSAS 123
+++ G RLRVE RG + SGGS + R + V +
Sbjct: 57 RFEYGRTGRRLRVEWTKNDRGGAGR-------SGGSRRSSSGLRPSKTLFVINFDAQNTR 109
Query: 124 WQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIR 183
+DL+ H G + ++ R + Y +D AL + S+ +
Sbjct: 110 TRDLERHFEPYGKIVNVRIRRN----FAFIQYEAQEDATRALDATNSSKLMDKVISVEYA 165
Query: 184 VREYD-RDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSISLSPTAKRSRR 239
V++ D R SP R RS R RSP R S Y GR SP A + R
Sbjct: 166 VKDDDSRGNGYSPERRRDRSPDRR-RRSPSPYRRERGSPDY-GR--GASPVAHKRER 218
>At3g61860 ARGININE/SERINE-RICH SPLICING FACTOR RSP31
Length = 264
Score = 68.6 bits (166), Expect = 3e-12
Identities = 65/235 (27%), Positives = 98/235 (41%), Gaps = 25/235 (10%)
Query: 7 RTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRD 66
R ++VGN + R ++E +F K+G + +D+K GYAFV FED RDAEDAI D
Sbjct: 2 RPVFVGNFEYETRQSDLERLFDKYGRVDRVDMK-----SGYAFVYFEDERDAEDAIRKLD 56
Query: 67 GYDF--DGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASW 124
+ F + RL VE A G RG G + ++ + + V P +
Sbjct: 57 NFPFGYEKRRLSVEWAKGERGRPR----------GDAKAPSNLKPTKTLFVINFDPIRTK 106
Query: 125 Q-DLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAYIR 183
+ D++ H G V ++ R V + +D AL S+ +
Sbjct: 107 EHDIEKHFEPYGKVTNVRIRRN----FSFVQFETQEDATKALEATQRSKILDRVVSVEYA 162
Query: 184 VREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSISLSPTAKRSR 238
+++ D R+ R RRS S R P R R G+ SP R+R
Sbjct: 163 LKDDDERDDRNGGRSPRRSLSPVYRRRPSPDYGR---RPSPGQGRRPSPDYGRAR 214
>At2g37340 unknown protein
Length = 249
Score = 66.2 bits (160), Expect = 2e-11
Identities = 66/192 (34%), Positives = 83/192 (42%), Gaps = 32/192 (16%)
Query: 47 YAFVQFEDARDAEDAIYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGAS 106
YAFV+F D RDA+DA +Y DG DFDG R+ VE + G S D G
Sbjct: 5 YAFVEFGDPRDADDARHYLDGRDFDGSRITVEFSRGAPRGSRDFDSRGPPPGAGRCFNCG 64
Query: 107 RRSDYRVLVTGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALR 166
+ T D K+ + G ERG + + K + +
Sbjct: 65 VDGHWARDCTA-------GDWKNKCYRCG---------ERGHIE--------RNCKNSPK 100
Query: 167 KLDDSEFRNAFSRAYIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRS---RSRSY 223
KL S ++SR+ +R R R RSPSR RS S S SRSP R RS RSRS
Sbjct: 101 KLRRS---GSYSRSPVRSRSPRR--RRSPSRSLSRSRSYSRSRSPVRRRERSVEERSRSP 155
Query: 224 SGRSISLSPTAK 235
SLSP A+
Sbjct: 156 KRMDDSLSPRAR 167
>At3g55460 RNA binding protein like
Length = 262
Score = 65.9 bits (159), Expect = 2e-11
Identities = 62/219 (28%), Positives = 94/219 (42%), Gaps = 12/219 (5%)
Query: 5 SSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDLK---IPPRPPGYAFVQFEDARDAEDA 61
S ++ V N+P D R E+ + F +FGP+ D+ + +P G+AFV+F DA DA +A
Sbjct: 45 SHGSLLVRNIPLDCRPEELREPFERFGPVRDVYIPRDYYSGQPRGFAFVEFVDAYDAGEA 104
Query: 62 IYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPS 121
+ F G + V +A R + R + + G+ RS R + S
Sbjct: 105 QRSMNRRSFAGREITVVVASESRKRPEEM-RVKTRTRSREPSGSRDRSHGRSRSRSISRS 163
Query: 122 ASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAY 181
S + D + +S R RGG + NY Y+ + A + A
Sbjct: 164 RSPRRPSDSRSRYRSRSYSPAPRRRGGPPRGEEDENYSRRSYS------PGYEGAAAAAP 217
Query: 182 IRVREYDRDYSRSP--SRDSRRSYSRSISRSPYLSRSRS 218
R R D + P + RR R++SRSP SRSRS
Sbjct: 218 DRDRNGDNEIREKPGYEAEDRRRGGRAVSRSPSGSRSRS 256
Score = 47.0 bits (110), Expect = 1e-05
Identities = 53/172 (30%), Positives = 72/172 (41%), Gaps = 33/172 (19%)
Query: 81 HGGRGSSSSVDR----YSSYSGGSSSRGASRRSDYRVLVTGLPPSASWQDLKDHMRKAGD 136
+GGRG S Y GG RG+S S +LV +P ++L++ + G
Sbjct: 16 YGGRGRSPPPPPPRRGYGGGGGGGGRRGSSHGS---LLVRNIPLDCRPEELREPFERFGP 72
Query: 137 VCFSQVFRERGGMTG------IVDYTNYDDMKYALRKLDD-------------SEFRNAF 177
V V+ R +G V++ + D A R ++ SE R
Sbjct: 73 V--RDVYIPRDYYSGQPRGFAFVEFVDAYDAGEAQRSMNRRSFAGREITVVVASESRKRP 130
Query: 178 SRAYIRVREYDRDYSRSPSRDSRRSYSRSISRS-----PYLSRSRSRSRSYS 224
++ R R+ S S R RS SRSISRS P SRSR RSRSYS
Sbjct: 131 EEMRVKTRTRSREPSGSRDRSHGRSRSRSISRSRSPRRPSDSRSRYRSRSYS 182
>At3g53500 splicing factor - like protein
Length = 243
Score = 61.2 bits (147), Expect = 5e-10
Identities = 69/200 (34%), Positives = 85/200 (42%), Gaps = 62/200 (31%)
Query: 47 YAFVQFEDARDAEDAIYYRDGYDFDGFRLRVELAHGG-RGSSSSVDRYSSYSGGSSSRGA 105
YAFV+F D RDA+DA YY DG DFDG R+ VE + G RGS + SRG
Sbjct: 5 YAFVEFSDPRDADDARYYLDGRDFDGSRITVEASRGAPRGSRD-----------NGSRG- 52
Query: 106 SRRSDYRVLVTGLPPSASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYAL 165
PP S + CF+ G+ G +
Sbjct: 53 -------------PPPGSGR------------CFN------CGVDG-----------HWA 70
Query: 166 RKLDDSEFRNAFSRAYIRVREYDRDYSRSPSRDSRR---SYSRS--ISRSPYLSRSRSRS 220
R +++N R R +R+ SPS R SYSRS SRSP RS SRS
Sbjct: 71 RDCTAGDWKNKCYRCGER-GHIERNCKNSPSPKKARQGGSYSRSPVKSRSPRRRRSPSRS 129
Query: 221 RSYS-GRSISLSPTAKRSRR 239
RSYS GRS S S + R +
Sbjct: 130 RSYSRGRSYSRSRSPVRREK 149
>At3g13570 serine/arginine-rich protein
Length = 262
Score = 60.5 bits (145), Expect = 9e-10
Identities = 71/244 (29%), Positives = 101/244 (41%), Gaps = 50/244 (20%)
Query: 8 TIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPR------PPGYAFVQFEDARDAEDA 61
++ V NL D R ++ F +FGP+ DI L PR P G+ F+QF D DA +A
Sbjct: 38 SLLVRNLRHDCRQEDLRRPFEQFGPVKDIYL---PRDYYTGDPRGFGFIQFMDPADAAEA 94
Query: 62 IYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPS 121
+ DGY G L V A R + + GG S+R RR PP
Sbjct: 95 KHQMDGYLLLGRELTVVFAEENR--KKPTEMRTRDRGGRSNRFQDRRRS--------PPR 144
Query: 122 ASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAY 181
S R++ R RG Y + ++ R + +
Sbjct: 145 YSRSPPPRRGRRSRS-------RSRG-------YNSPPAKRHQSRSVSPQD--------- 181
Query: 182 IRVREYDRDYSRSPSRDSRRSYS------RSISRSPYLSRSRSRSRSYS-GRSISLSPTA 234
R E +R YSRSP + R S +S SRSP S S ++RSY+ ++ S SP
Sbjct: 182 -RRYEKERSYSRSPPHNGSRVRSGSPGRVKSHSRSPRRSVSPRKNRSYTPEQARSQSPVP 240
Query: 235 KRSR 238
++SR
Sbjct: 241 RQSR 244
Score = 27.3 bits (59), Expect = 8.4
Identities = 26/70 (37%), Positives = 32/70 (45%), Gaps = 12/70 (17%)
Query: 189 RDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSG------RSISLSPTAKRSRRLLS 242
+D RSP R SR R RSRSRSR Y+ +S S+SP +R + S
Sbjct: 136 QDRRRSPPRYSRSPPPRR------GRRSRSRSRGYNSPPAKRHQSRSVSPQDRRYEKERS 189
Query: 243 LSRCGLLRGS 252
SR GS
Sbjct: 190 YSRSPPHNGS 199
>At1g23860 9G8-like splicing factor / SRZ-21
Length = 187
Score = 60.1 bits (144), Expect = 1e-09
Identities = 70/217 (32%), Positives = 92/217 (42%), Gaps = 48/217 (22%)
Query: 9 IYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYRDGY 68
+YVGNL + RE+ED F FG + ++ + RPPGYAF++F+D RDA DAI D
Sbjct: 4 VYVGNLDPRVTERELEDEFKAFGVLRNV--WVARRPPGYAFLEFDDERDALDAISALDRK 61
Query: 69 DFDGFRLRVELAH---GGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPSASWQ 125
+ RVEL+H GGRG GG RG G+ S ++
Sbjct: 62 N----GWRVELSHKDKGGRG------------GGGGRRG------------GIEDSKCYE 93
Query: 126 --DLKDHMRKAGDVCFSQVFRERGGM---TGIVDYTNYDDMKYALRKLDDSEFRNAFSRA 180
+L R+ R RG + + D YA R + R+ R
Sbjct: 94 CGELGHFARECR--------RGRGSVRRRSPSPRRRRSPDYGYARRSISPRGRRSPPRRR 145
Query: 181 YIRVREYDRDYSRSPS-RDSRRSYSRSISRSPYLSRS 216
+ R YSRSP R SRR R SPY RS
Sbjct: 146 SVTPPRRGRSYSRSPPYRGSRRDSPRR-RDSPYGRRS 181
Score = 28.5 bits (62), Expect = 3.7
Identities = 26/65 (40%), Positives = 30/65 (46%), Gaps = 8/65 (12%)
Query: 196 SRDSRRSYSRSISRSPYLSRSRSRSRSYSGRSIS----LSPTAKRS----RRLLSLSRCG 247
+R+ RR RSP R RS Y+ RSIS SP +RS RR S SR
Sbjct: 101 ARECRRGRGSVRRRSPSPRRRRSPDYGYARRSISPRGRRSPPRRRSVTPPRRGRSYSRSP 160
Query: 248 LLRGS 252
RGS
Sbjct: 161 PYRGS 165
Score = 28.1 bits (61), Expect = 4.9
Identities = 24/54 (44%), Positives = 27/54 (49%), Gaps = 7/54 (12%)
Query: 193 RSPSRDSRRSYSRSISR---SPYLSRSRSRSRSYS----GRSISLSPTAKRSRR 239
RSPS RRS +R SP RS R RS + GRS S SP + SRR
Sbjct: 114 RSPSPRRRRSPDYGYARRSISPRGRRSPPRRRSVTPPRRGRSYSRSPPYRGSRR 167
>At1g07350 transformer-SR ribonucleoprotein like protein
Length = 382
Score = 56.6 bits (135), Expect = 1e-08
Identities = 73/259 (28%), Positives = 106/259 (40%), Gaps = 26/259 (10%)
Query: 8 TIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPP---RPPGYAFVQFEDARDAEDAIYY 64
++YV L + R++ED F K G + D+ L + P G+ F+ + DA I
Sbjct: 76 SLYVTGLSHRVTERDLEDHFAKEGKVTDVHLVLDPWTRESRGFGFISMKSVGDANRCIRS 135
Query: 65 RDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDY---RVLVTGLPPS 121
D G + VE A RG + + + Y G ++RG + Y R + S
Sbjct: 136 LDHSVLQGRVITVEKARRRRGRTPTPGK---YLGLRTARGRHKSPSYSPRRSVSCSRSRS 192
Query: 122 ASWQDLK------DHMRKAGDVCFSQVFRERGGMTGIVD------YTNYDDMKYA--LRK 167
S+ + + R+ +S +R R + Y D Y+ R+
Sbjct: 193 RSYSSDRGRSYSPSYGRRGRSSSYSPFYRRRRFYSPSRSPSPDDRYNRRRDRSYSPYYRR 252
Query: 168 LDDSEFRNAFSRAYIRVREYDRDYSRSPSRD-SRRSYSRSISRSPYL-SRSRSRSRSYSG 225
D S + RA R Y R Y RS SR S R +R S SPY R RS S Y G
Sbjct: 253 RDRSRSYSRNCRARDRSPYYMRRY-RSRSRSYSPRYRARDRSCSPYYRGRDRSYSPHYQG 311
Query: 226 RSISLSPTAKRSRRLLSLS 244
R S SP ++ RR S+S
Sbjct: 312 RDRSYSPESRYYRRHRSVS 330
Score = 48.9 bits (115), Expect = 3e-06
Identities = 63/194 (32%), Positives = 75/194 (38%), Gaps = 38/194 (19%)
Query: 87 SSSVDRYSSYSGGSSSRGASRRSDYRV-----------LVTGLPPSASWQDLKDHMRKAG 135
S S+ Y G S SR SR V VTGL + +DL+DH K G
Sbjct: 40 SPSLSPYDKRRGRSVSRSLSRSPTRSVSSDAENPGNSLYVTGLSHRVTERDLEDHFAKEG 99
Query: 136 DVCFSQVF-----RERGGMTGIVDYTNYDDMKYALRKLDDSEF---------------RN 175
V + RE G G + + D +R LD S R
Sbjct: 100 KVTDVHLVLDPWTRESRGF-GFISMKSVGDANRCIRSLDHSVLQGRVITVEKARRRRGRT 158
Query: 176 AFSRAYIRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYS----GRSISLS 231
Y+ +R R +SPS RRS S S SRS S R RS S S GRS S S
Sbjct: 159 PTPGKYLGLRTA-RGRHKSPSYSPRRSVSCSRSRSRSYSSDRGRSYSPSYGRRGRSSSYS 217
Query: 232 PTAKRSRRLLSLSR 245
P +R RR S SR
Sbjct: 218 PFYRR-RRFYSPSR 230
Score = 29.3 bits (64), Expect = 2.2
Identities = 29/53 (54%), Positives = 32/53 (59%), Gaps = 8/53 (15%)
Query: 183 RVREYD-RDYSRSPSRDSRRS-YSRSISRSPYLSRSRSRSRSYSGRSISLSPT 233
RV EY R +RSP SRRS YS S+S PY R R RS S RS+S SPT
Sbjct: 17 RVLEYPLRLENRSPMSYSRRSRYSPSLS--PYDKR---RGRSVS-RSLSRSPT 63
>At1g55310 Serine/arginine-rich protein
Length = 220
Score = 54.7 bits (130), Expect = 5e-08
Identities = 65/219 (29%), Positives = 86/219 (38%), Gaps = 51/219 (23%)
Query: 8 TIYVGNLPGDIRLREVEDIFYKFGPIVDIDLKIPPR------PPGYAFVQFEDARDAEDA 61
++ V NL D R ++ F +FGP+ DI L PR P G+ FVQF D DA DA
Sbjct: 37 SLLVRNLRHDCRQEDLRKSFEQFGPVKDIYL---PRDYYTGDPRGFGFVQFMDPADAADA 93
Query: 62 IYYRDGYDFDGFRLRVELAHGGRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTGLPPS 121
++ DGY G L V A R + + + RG R D R PP
Sbjct: 94 KHHMDGYLLLGRELTVVFAEENRKKPTEM--------RARERGGGRFRDRR----RTPPR 141
Query: 122 ASWQDLKDHMRKAGDVCFSQVFRERGGMTGIVDYTNYDDMKYALRKLDDSEFRNAFSRAY 181
+ R+ R R G DY + ++ R + E R
Sbjct: 142 YYSRSRSPPPRRGRS-------RSRSG-----DYYSPPPRRHHPRSISPREER------- 182
Query: 182 IRVREYD--RDYSRSPSRDSRRSYSRSISRSPYLSRSRS 218
YD R YSRSP+ D SR S +P +SRS
Sbjct: 183 -----YDGRRSYSRSPASDG----SRGRSLTPVRGKSRS 212
Score = 30.4 bits (67), Expect = 0.99
Identities = 28/77 (36%), Positives = 31/77 (39%), Gaps = 6/77 (7%)
Query: 182 IRVREYDRDYSRSPSRDSRRSYSRSISRSPYLSRSRSRSRSYSG------RSISLSPTAK 235
+R RE R R R YSRS S P RSRSRS Y S+SP +
Sbjct: 122 MRARERGGGRFRDRRRTPPRYYSRSRSPPPRRGRSRSRSGDYYSPPPRRHHPRSISPREE 181
Query: 236 RSRRLLSLSRCGLLRGS 252
R S SR GS
Sbjct: 182 RYDGRRSYSRSPASDGS 198
>At2g37220 putative RNA-binding protein
Length = 289
Score = 52.8 bits (125), Expect = 2e-07
Identities = 55/210 (26%), Positives = 85/210 (40%), Gaps = 28/210 (13%)
Query: 9 IYVGNLPGDIRLREVEDIFYKFGPIVDIDL---KIPPRPPGYAFVQFEDARDAEDAIYYR 65
++VGNLP ++ ++ +F G + +++ KI R G+ FV + E A
Sbjct: 93 LFVGNLPFNVDSAQLAQLFESAGNVEMVEVIYDKITGRSRGFGFVTMSSVSEVEAAAQQF 152
Query: 66 DGYDFDGFRLRVELAHG--------GRGSSSSVDRYSSYSGGSSSRGASRRSDYRVLVTG 117
+GY+ DG LRV RG SS S GG GA S RV V
Sbjct: 153 NGYELDGRPLRVNAGPPPPKREDGFSRGPRSSFGSSGSGYGGGGGSGAG--SGNRVYVGN 210
Query: 118 LPPSASWQDLKDHMRKAGDVCFSQVFRERGGMT----GIVDYTNYDDMKYALRKLDDSEF 173
L L+ + G V ++V +R G V Y + +++ A++ LD ++
Sbjct: 211 LSWGVDDMALESLFSEQGKVVEARVIYDRDSGRSKGFGFVTYDSSQEVQNAIKSLDGAD- 269
Query: 174 RNAFSRAYIRVREYDRDYSRSPSRDSRRSY 203
IRV S + +R RR Y
Sbjct: 270 ---LDGRQIRV-------SEAEARPPRRQY 289
Score = 43.5 bits (101), Expect = 1e-04
Identities = 24/79 (30%), Positives = 43/79 (54%), Gaps = 3/79 (3%)
Query: 5 SSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDL---KIPPRPPGYAFVQFEDARDAEDA 61
S +YVGNL + +E +F + G +V+ + + R G+ FV ++ +++ ++A
Sbjct: 202 SGNRVYVGNLSWGVDDMALESLFSEQGKVVEARVIYDRDSGRSKGFGFVTYDSSQEVQNA 261
Query: 62 IYYRDGYDFDGFRLRVELA 80
I DG D DG ++RV A
Sbjct: 262 IKSLDGADLDGRQIRVSEA 280
>At1g71800 cleavage stimulation factor like protein
Length = 461
Score = 52.8 bits (125), Expect = 2e-07
Identities = 28/94 (29%), Positives = 49/94 (51%), Gaps = 3/94 (3%)
Query: 2 SSRSSRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDL---KIPPRPPGYAFVQFEDARDA 58
SS R ++VGN+P D ++ +I + GP+V L + +P GY F +++D A
Sbjct: 4 SSSQRRCVFVGNIPYDATEEQLREICGEVGPVVSFRLVTDRETGKPKGYGFCEYKDEETA 63
Query: 59 EDAIYYRDGYDFDGFRLRVELAHGGRGSSSSVDR 92
A Y+ +G +LRV+ A +G+ + D+
Sbjct: 64 LSARRNLQSYEINGRQLRVDFAENDKGTDKTRDQ 97
>At4g14300 ribonucleoprotein like protein
Length = 406
Score = 50.1 bits (118), Expect = 1e-06
Identities = 38/134 (28%), Positives = 62/134 (45%), Gaps = 17/134 (12%)
Query: 6 SRTIYVGNLPGDIRLREVEDIFYKFGPIVDIDL---KIPPRPPGYAFVQFEDARDAEDAI 62
++ I+VG LP + E F +GP+ D+ + + RP G+ FV F D+ DA D++
Sbjct: 104 TKKIFVGGLPPTLTDEEFRQYFEVYGPVTDVAIMYDQATNRPRGFGFVSF-DSEDAVDSV 162
Query: 63 YYRDGYDFDGFRLRVELA------HGGRGSS---SSVDRYSSYSGGSSS----RGASRRS 109
++ +D G ++ V+ A GG G S Y Y G SS ++R
Sbjct: 163 LHKTFHDLSGKQVEVKRALPKDANPGGGGRSMGGGGSGGYQGYGGNESSYDGRMDSNRFL 222
Query: 110 DYRVLVTGLPPSAS 123
++ + GLP S
Sbjct: 223 QHQSVGNGLPSYGS 236
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,160,733
Number of Sequences: 26719
Number of extensions: 277671
Number of successful extensions: 1651
Number of sequences better than 10.0: 219
Number of HSP's better than 10.0 without gapping: 100
Number of HSP's successfully gapped in prelim test: 121
Number of HSP's that attempted gapping in prelim test: 1026
Number of HSP's gapped (non-prelim): 468
length of query: 259
length of database: 11,318,596
effective HSP length: 97
effective length of query: 162
effective length of database: 8,726,853
effective search space: 1413750186
effective search space used: 1413750186
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0299.13


