

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0290.23
(358 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
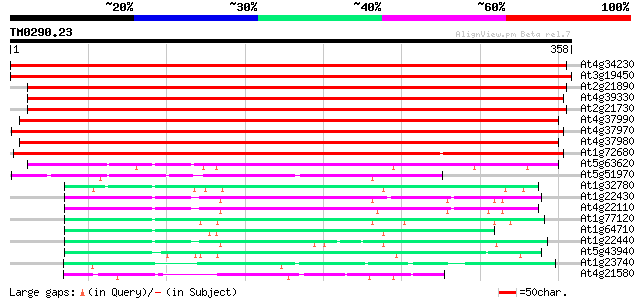
Score E
Sequences producing significant alignments: (bits) Value
At4g34230 cinnamyl alcohol dehydrogenase like protein 585 e-167
At3g19450 putative cinnamyl alcohol dehydrogenase 2 579 e-166
At2g21890 putative cinnamyl-alcohol dehydrogenase 364 e-101
At4g39330 cinnamyl-alcohol dehydrogenase CAD1 364 e-101
At2g21730 putative cinnamyl-alcohol dehydrogenase 359 1e-99
At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2 357 7e-99
At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa 357 7e-99
At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1 339 1e-93
At1g72680 putative cinnamyl-alcohol dehydrogenase 296 2e-80
At5g63620 alcohol dehydrogenase-like protein 97 1e-20
At5g51970 sorbitol dehydrogenase-like protein 78 9e-15
At1g32780 alcohol dehydrogenase like protein 71 1e-12
At1g22430 putative alcohol dehydrogenase 69 3e-12
At4g22110 alcohol dehydrogenase like protein 67 2e-11
At1g77120 alcohol dehydrogenase 64 1e-10
At1g64710 alcohol dehydrogenase (EC 1.1.1.1) like protein 63 2e-10
At1g22440 Very similar to alcohol dehydrogenase 63 2e-10
At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III (pir||S71... 63 3e-10
At1g23740 auxin-induced protein like 57 1e-08
At4g21580 putative NADPH quinone oxidoreductase 54 1e-07
>At4g34230 cinnamyl alcohol dehydrogenase like protein
Length = 357
Score = 585 bits (1507), Expect = e-167
Identities = 277/355 (78%), Positives = 317/355 (89%)
Query: 1 MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM 60
MG +EAER T GWAARDPSGILSPYT+TLR TGP+DV I++ CG+CHTD+HQ KNDLGM
Sbjct: 1 MGIMEAERKTTGWAARDPSGILSPYTYTLRETGPEDVNIRIICCGICHTDLHQTKNDLGM 60
Query: 61 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWN 120
SNYPMVPGHEVVGEV+EVGSDV++FTVG+IVG G LVGCC C+ C+ D+EQYC KKIW+
Sbjct: 61 SNYPMVPGHEVVGEVVEVGSDVSKFTVGDIVGVGCLVGCCGGCSPCERDLEQYCPKKIWS 120
Query: 121 YNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKES 180
YNDVY++G+PTQGGFA+ +V QKFVVKIPEGMA EQ APLLCA VTVYSPLSHFGLK+
Sbjct: 121 YNDVYINGQPTQGGFAKATVVHQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGLKQP 180
Query: 181 GLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQE 240
GLRGGILGLGGVGHMGV IAKAMGHHVTVISSS++K++EA++DLGAD Y++ SD M E
Sbjct: 181 GLRGGILGLGGVGHMGVKIAKAMGHHVTVISSSNKKREEALQDLGADDYVIGSDQAKMSE 240
Query: 241 AADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFI 300
ADSLDY+IDTVPV H LEPYLSLLKLDGKLILMGVIN PLQF+TP++MLGR+ ITGSFI
Sbjct: 241 LADSLDYVIDTVPVHHALEPYLSLLKLDGKLILMGVINNPLQFLTPLLMLGRKVITGSFI 300
Query: 301 GSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKL 355
GS+KETEEMLEF KEKGL+S+IE+V MDY+N AFERLEKNDVRYRFVVDV+GS L
Sbjct: 301 GSMKETEEMLEFCKEKGLSSIIEVVKMDYVNTAFERLEKNDVRYRFVVDVEGSNL 355
>At3g19450 putative cinnamyl alcohol dehydrogenase 2
Length = 365
Score = 579 bits (1493), Expect = e-166
Identities = 279/359 (77%), Positives = 318/359 (87%), Gaps = 1/359 (0%)
Query: 1 MGSLEA-ERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLG 59
MGS+EA E+ +GWAARDPSG+LSPY++TLR+TG DDVYIKV CG+CHTD+HQ+KNDLG
Sbjct: 1 MGSVEAGEKKALGWAARDPSGVLSPYSYTLRSTGADDVYIKVICCGICHTDIHQIKNDLG 60
Query: 60 MSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIW 119
MSNYPMVPGHEVVGEVLEVGSDV++FTVG++VG G++VGCC C C S++EQYC K+IW
Sbjct: 61 MSNYPMVPGHEVVGEVLEVGSDVSKFTVGDVVGVGVVVGCCGSCKPCSSELEQYCNKRIW 120
Query: 120 NYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKE 179
+YNDVY DGKPTQGGFA+T+IV QKFVVKIPEGMA EQ APLLCA VTVYSPLSHFGL
Sbjct: 121 SYNDVYTDGKPTQGGFADTMIVNQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGLMA 180
Query: 180 SGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQ 239
SGL+GGILGLGGVGHMGV IAKAMGHHVTVISSSD+KK+EAIE LGAD Y+VSSD MQ
Sbjct: 181 SGLKGGILGLGGVGHMGVKIAKAMGHHVTVISSSDKKKEEAIEHLGADDYVVSSDPAEMQ 240
Query: 240 EAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSF 299
ADSLDYIIDTVPV HPL+PYL+ LKLDGKLILMGVINTPLQF+TP+V+LGR+ I+GSF
Sbjct: 241 RLADSLDYIIDTVPVFHPLDPYLACLKLDGKLILMGVINTPLQFVTPLVILGRKVISGSF 300
Query: 300 IGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKLIDQ 358
IGSIKETEE+L F KEKGLTS IE V +D +N AFERL KNDVRYRFVVDV GS L+++
Sbjct: 301 IGSIKETEEVLAFCKEKGLTSTIETVKIDELNIAFERLRKNDVRYRFVVDVAGSNLVEE 359
>At2g21890 putative cinnamyl-alcohol dehydrogenase
Length = 375
Score = 364 bits (935), Expect = e-101
Identities = 171/345 (49%), Positives = 244/345 (70%), Gaps = 1/345 (0%)
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAA D SG+LSP+ F+ R G +DV +K+ +CGVCH+D+H +KN G S YP++PGHE+
Sbjct: 9 GWAANDESGVLSPFHFSRRENGENDVTVKILFCGVCHSDLHTIKNHWGFSRYPIIPGHEI 68
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPT 131
VG +VG +VT+F G+ VG G+++G C+ C +C D+E YC K ++ YN DG
Sbjct: 69 VGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFTYNSRSSDGTRN 128
Query: 132 QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGL-KESGLRGGILGLG 190
QGG+++ I+V+ +FV+ IP+G+ + APLLCA +TVYSP+ ++G+ KESG R G+ GLG
Sbjct: 129 QGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGVNGLG 188
Query: 191 GVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIID 250
G+GH+ V I KA G VTVIS S K++EAI+ LGAD++LV++D+ M+EA ++D+IID
Sbjct: 189 GLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTMDFIID 248
Query: 251 TVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEML 310
TV H L P SLLK+ GKL+ +G++ PL ++LGR+ + GS IG +KET+EML
Sbjct: 249 TVSAEHALLPLFSLLKVSGKLVALGLLEKPLDLPIFPLVLGRKMVGGSQIGGMKETQEML 308
Query: 311 EFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKL 355
EF + + S IE++ M IN A +RL K+DVRYRFV+DV S L
Sbjct: 309 EFCAKHKIVSDIELIKMSDINSAMDRLVKSDVRYRFVIDVANSLL 353
>At4g39330 cinnamyl-alcohol dehydrogenase CAD1
Length = 360
Score = 364 bits (934), Expect = e-101
Identities = 170/342 (49%), Positives = 241/342 (69%)
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GW ARD SG+LSP+ F+ R+ G +DV +K+ +CGVCHTD+H +KND G S YP+VPGHE+
Sbjct: 15 GWGARDKSGVLSPFHFSRRDNGENDVTVKILFCGVCHTDLHTIKNDWGYSYYPVVPGHEI 74
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPT 131
VG +VG +VT+F G+ VG G++ G C+ C +C D+E YC + + YN + DG
Sbjct: 75 VGIATKVGKNVTKFKEGDRVGVGVISGSCQSCESCDQDLENYCPQMSFTYNAIGSDGTKN 134
Query: 132 QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGG 191
GG++E I+V+Q+FV++ PE + + APLLCA +TVYSP+ ++G+ E+G G+ GLGG
Sbjct: 135 YGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGMTEAGKHLGVAGLGG 194
Query: 192 VGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDT 251
+GH+ V I KA G VTVISSS K +EAI LGAD++LV++D M+ A ++DYIIDT
Sbjct: 195 LGHVAVKIGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQKMKAAIGTMDYIIDT 254
Query: 252 VPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLE 311
+ H L P L LLK++GKLI +G+ PL+ ++LGR+ + GS +G +KET+EML+
Sbjct: 255 ISAVHALYPLLGLLKVNGKLIALGLPEKPLELPMFPLVLGRKMVGGSDVGGMKETQEMLD 314
Query: 312 FWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
F + +T+ IE++ MD IN A ERL K+DVRYRFV+DV S
Sbjct: 315 FCAKHNITADIELIKMDEINTAMERLAKSDVRYRFVIDVANS 356
>At2g21730 putative cinnamyl-alcohol dehydrogenase
Length = 376
Score = 359 bits (922), Expect = 1e-99
Identities = 171/346 (49%), Positives = 244/346 (70%), Gaps = 2/346 (0%)
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAA D SG+LSP+ F+ R G +DV +K+ +CGVCH+D+H +KN G S YP++PGHE+
Sbjct: 9 GWAANDESGVLSPFHFSRRENGENDVTVKILFCGVCHSDLHTIKNHWGFSRYPIIPGHEI 68
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKP- 130
VG +VG +VT+F G+ VG G+++G C+ C +C D+E YC K ++ YN DG
Sbjct: 69 VGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFTYNSRSSDGTSR 128
Query: 131 TQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGL-KESGLRGGILGL 189
QGG+++ I+V+ +FV+ IP+G+ + APLLCA +TVYSP+ ++G+ KESG R G+ GL
Sbjct: 129 NQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGVNGL 188
Query: 190 GGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYII 249
GG+GH+ V I KA G VTVIS S K++EAI+ LGAD++LV++D+ M+EA ++D+II
Sbjct: 189 GGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTMDFII 248
Query: 250 DTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEM 309
DTV H L P SLLK++GKL+ +G+ PL ++LGR+ + GS IG +KET+EM
Sbjct: 249 DTVSAEHALLPLFSLLKVNGKLVALGLPEKPLDLPIFSLVLGRKMVGGSQIGGMKETQEM 308
Query: 310 LEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKL 355
LEF + + S IE++ M IN A +RL K+DVRYRFV+DV S L
Sbjct: 309 LEFCAKHKIVSDIELIKMSDINSAMDRLAKSDVRYRFVIDVANSLL 354
>At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2
Length = 359
Score = 357 bits (915), Expect = 7e-99
Identities = 171/344 (49%), Positives = 237/344 (68%)
Query: 7 ERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMV 66
++ G AA+D SG+LSP++FT R TG DV KV +CG+CH+D+H VKN+ GMS YP+V
Sbjct: 6 QKEAFGLAAKDNSGVLSPFSFTRRETGEKDVRFKVLFCGICHSDLHMVKNEWGMSTYPLV 65
Query: 67 PGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYV 126
PGHE+VG V EVG+ VT+F GE VG G LV C C +C +E YC K I Y Y
Sbjct: 66 PGHEIVGVVTEVGAKVTKFKTGEKVGVGCLVSSCGSCDSCTEGMENYCPKSIQTYGFPYY 125
Query: 127 DGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGI 186
D T GG+++ ++ E+ FV++IP+ + + APLLCA +TVYSP+ + GL + G+ G+
Sbjct: 126 DNTITYGGYSDHMVCEEGFVIRIPDNLPLDAAAPLLCAGITVYSPMKYHGLDKPGMHIGV 185
Query: 187 LGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLD 246
+GLGG+GH+GV AKAMG VTVIS+S++K+ EAI LGADA+LVS D +++A ++D
Sbjct: 186 VGLGGLGHVGVKFAKAMGTKVTVISTSEKKRDEAINRLGADAFLVSRDPKQIKDAMGTMD 245
Query: 247 YIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKET 306
IIDTV H L P L LLK GKL+++G PL+ ++ R+ + GS IG IKET
Sbjct: 246 GIIDTVSATHSLLPLLGLLKHKGKLVMVGAPEKPLELPVMPLIFERKMVMGSMIGGIKET 305
Query: 307 EEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
+EM++ + +T+ IE+++ DY+N A ERLEK DVRYRFV+DV
Sbjct: 306 QEMIDMAGKHNITADIELISADYVNTAMERLEKADVRYRFVIDV 349
>At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa
Length = 363
Score = 357 bits (915), Expect = 7e-99
Identities = 167/352 (47%), Positives = 240/352 (67%)
Query: 2 GSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMS 61
G E GWAARD SG LSP+ F+ R TG ++V +KV YCG+CH+D+H +KN+ S
Sbjct: 6 GEKEQSVEAFGWAARDSSGHLSPFVFSRRKTGEEEVRVKVLYCGICHSDLHCLKNEWHSS 65
Query: 62 NYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNY 121
YP+VPGHE++GEV E+G+ V++F +G+ VG G +V C+ C +C+ D E YC K I Y
Sbjct: 66 IYPLVPGHEIIGEVSEIGNKVSKFNLGDKVGVGCIVDSCRTCESCREDQENYCTKAIATY 125
Query: 122 NDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESG 181
N V+ DG GG+++ I+V++++ VKIP + APLLCA +++YSP+ +FGL
Sbjct: 126 NGVHHDGTINYGGYSDHIVVDERYAVKIPHTLPLVSAAPLLCAGISMYSPMKYFGLTGPD 185
Query: 182 LRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEA 241
GI+GLGG+GH+GV AKA G VTV+SS+ K K+A++ LGAD +LVS+D M+ A
Sbjct: 186 KHVGIVGLGGLGHIGVRFAKAFGTKVTVVSSTTGKSKDALDTLGADGFLVSTDEDQMKAA 245
Query: 242 ADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIG 301
++D IIDTV H + P + LLK +GKL+L+G P ++LGR+SI GS IG
Sbjct: 246 MGTMDGIIDTVSASHSISPLIGLLKSNGKLVLLGATEKPFDISAFSLILGRKSIAGSGIG 305
Query: 302 SIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
++ET+EM++F E G+ + IEI++MDY+N A +RL K DVRYRFV+D+ +
Sbjct: 306 GMQETQEMIDFAAEHGIKAEIEIISMDYVNTAMDRLAKGDVRYRFVIDISNT 357
>At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1
Length = 357
Score = 339 bits (870), Expect = 1e-93
Identities = 162/344 (47%), Positives = 231/344 (67%)
Query: 7 ERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMV 66
E+ G AA+D SGILSP++F+ R TG DV KV +CG+CHTD+ KN+ G++ YP+V
Sbjct: 6 EKEAFGLAAKDESGILSPFSFSRRATGEKDVRFKVLFCGICHTDLSMAKNEWGLTTYPLV 65
Query: 67 PGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYV 126
PGHE+VG V EVG+ V +F G+ VG G + G C+ C +C E YC K I
Sbjct: 66 PGHEIVGVVTEVGAKVKKFNAGDKVGVGYMAGSCRSCDSCNDGDENYCPKMILTSGAKNF 125
Query: 127 DGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGI 186
D T GG+++ ++ + F+++IP+ + + APLLCA VTVYSP+ + GL + G+ G+
Sbjct: 126 DDTMTHGGYSDHMVCAEDFIIRIPDNLPLDGAAPLLCAGVTVYSPMKYHGLDKPGMHIGV 185
Query: 187 LGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLD 246
+GLGG+GH+ V AKAMG VTVIS+S+RK+ EA+ LGADA+LVS D M++A ++D
Sbjct: 186 VGLGGLGHVAVKFAKAMGTKVTVISTSERKRDEAVTRLGADAFLVSRDPKQMKDAMGTMD 245
Query: 247 YIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKET 306
IIDTV HPL P L LLK GKL+++G PL+ ++ GR+ + GS +G IKET
Sbjct: 246 GIIDTVSATHPLLPLLGLLKNKGKLVMVGAPAEPLELPVFPLIFGRKMVVGSMVGGIKET 305
Query: 307 EEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
+EM++ + +T+ IE+++ DY+N A ERL K DV+YRFV+DV
Sbjct: 306 QEMVDLAGKHNITADIELISADYVNTAMERLAKADVKYRFVIDV 349
>At1g72680 putative cinnamyl-alcohol dehydrogenase
Length = 355
Score = 296 bits (757), Expect = 2e-80
Identities = 147/351 (41%), Positives = 216/351 (60%), Gaps = 1/351 (0%)
Query: 3 SLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSN 62
S E + WAARDPSG+LSP+T T R+ DDV + + +CGVC+ DV +N G S
Sbjct: 4 SESVENECMCWAARDPSGLLSPHTITRRSVTTDDVSLTITHCGVCYADVIWSRNQHGDSK 63
Query: 63 YPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYN 122
YP+VPGHE+ G V +VG +V RF VG+ VG G V C++C C E C K ++ +N
Sbjct: 64 YPLVPGHEIAGIVTKVGPNVQRFKVGDHVGVGTYVNSCRECEYCNEGQEVNCAKGVFTFN 123
Query: 123 DVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGL 182
+ DG T+GG++ I+V +++ KIP E APLLCA +TVY+P+ + + G
Sbjct: 124 GIDHDGSVTKGGYSSHIVVHERYCYKIPVDYPLESAAPLLCAGITVYAPMMRHNMNQPGK 183
Query: 183 RGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAA 242
G++GLGG+GHM V KA G VTV S+S KK+EA+ LGA+ +++SSD M+
Sbjct: 184 SLGVIGLGGLGHMAVKFGKAFGLSVTVFSTSISKKEEALNLLGAENFVISSDHDQMKALE 243
Query: 243 DSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGS 302
SLD+++DT H +PY+SLLK+ G +L+G + ++ + LG R + GS G
Sbjct: 244 KSLDFLVDTASGDHAFDPYMSLLKIAGTYVLVG-FPSEIKISPANLNLGMRMLAGSVTGG 302
Query: 303 IKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
K T++ML+F + IE++ + IN+A ER+ K D++YRFV+D+K S
Sbjct: 303 TKITQQMLDFCAAHKIYPNIEVIPIQKINEALERVVKKDIKYRFVIDIKNS 353
>At5g63620 alcohol dehydrogenase-like protein
Length = 427
Score = 97.4 bits (241), Expect = 1e-20
Identities = 88/372 (23%), Positives = 158/372 (41%), Gaps = 36/372 (9%)
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
G R+P+ L+ F + +++ IK CGVCH+D+H +K ++ ++ P V GHE+
Sbjct: 58 GAVYREPNKPLTIEEFHIPRPKSNEILIKTKACGVCHSDLHVMKGEIPFAS-PCVIGHEI 116
Query: 72 VGEVLEVG-----SDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYN---D 123
GEV+E G + RF +G V G + C C+ C + C+ + YN
Sbjct: 117 TGEVVEHGPLTDHKIINRFPIGSRV-VGAFIMPCGTCSYCAKGHDDLCED-FFAYNRAKG 174
Query: 124 VYVDGKP--------------TQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVY 169
DG+ + GG AE + + +PE + + A L CA T Y
Sbjct: 175 TLYDGETRLFLRHDDSPVYMYSMGGMAEYCVTPAHGLAPLPESLPYSESAILGCAVFTAY 234
Query: 170 SPLSHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAY 229
++H G ++G+GGVG + IA+A G + K + + LGA
Sbjct: 235 GAMAHAAEIRPGDSIAVIGIGGVGSSCLQIARAFGASDIIAVDVQDDKLQKAKTLGATHI 294
Query: 230 LVSSDTTSMQEAAD-----SLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFI 284
+ ++ +++ + +D ++ + +K GK +++G+
Sbjct: 295 VNAAKEDAVERIREITGGMGVDVAVEALGKPQTFMQCTLSVKDGGKAVMIGLSQAGSVGE 354
Query: 285 TPMVMLGRRSI--TGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDY----INKAFERLE 338
+ L RR I GS+ G ++ + E G+ ++ V+ Y KAF+ L
Sbjct: 355 IDINRLVRRKIKVIGSYGGRARQDLPKVVKLAESGIFNLTNAVSSKYKFEDAGKAFQDLN 414
Query: 339 KNDVRYRFVVDV 350
+ + R VV++
Sbjct: 415 EGKIVSRGVVEI 426
>At5g51970 sorbitol dehydrogenase-like protein
Length = 364
Score = 77.8 bits (190), Expect = 9e-15
Identities = 72/289 (24%), Positives = 122/289 (41%), Gaps = 27/289 (9%)
Query: 2 GSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKN----D 57
GS E W + + P F L + GP DV +++ G+C +DVH +K D
Sbjct: 11 GSKVEEENMAAWLVGINTLKIQP--FLLPSVGPHDVRVRMKAVGICGSDVHYLKTMRCAD 68
Query: 58 LGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKK 117
+ PMV GHE G + EVG +V VG+ V + C +C C+ C +
Sbjct: 69 FVVKE-PMVIGHECAGIIEEVGEEVKHLVVGDRVALEPGISCW-RCNLCREGRYNLCPEM 126
Query: 118 IWNYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYS-PLSHFG 176
+ P G A ++ K+PE ++ E+ A +V V++ + G
Sbjct: 127 ------KFFATPPVHGSLANQVVHPADLCFKLPENVSLEEGAMCEPLSVGVHACRRAEVG 180
Query: 177 LKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYL------ 230
+ + L ++G G +G + ++ A+A VI D + + LGAD +
Sbjct: 181 PETNVL---VMGAGPIGLVTMLAARAFSVPRIVIVDVDENRLAVAKQLGADEIVQVTTNL 237
Query: 231 --VSSDTTSMQEAADS-LDYIIDTVPVGHPLEPYLSLLKLDGKLILMGV 276
V S+ +Q+A S +D D + L+ + GK+ L+G+
Sbjct: 238 EDVGSEVEQIQKAMGSNIDVTFDCAGFNKTMSTALAATRCGGKVCLVGM 286
>At1g32780 alcohol dehydrogenase like protein
Length = 394
Score = 70.9 bits (172), Expect = 1e-12
Identities = 84/342 (24%), Positives = 136/342 (39%), Gaps = 42/342 (12%)
Query: 36 DVYIKVHYCGVCHTDVH--QVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGA 93
+V +K+ Y +CHTD+ N+ + +P + GHE VG V VG V G+ V
Sbjct: 37 EVRVKILYSSICHTDLGCWNGTNEAERA-FPRILGHEAVGIVESVGEGVKDVKEGDYV-I 94
Query: 94 GLLVGCCKKCTACQSDIEQYCKK------KIWNYND--------VYVDGKPTQGG----- 134
G C +C C+ + C++ K ND + DG +Q
Sbjct: 95 PTFNGECGECKVCKREESNLCERYHVDPMKRVMVNDGGTRFSTTINKDGGSSQSQPIYHF 154
Query: 135 -----FAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGL 189
F E +++ VVKI +Q++ L C T + + G + GL
Sbjct: 155 LNTSTFTEYTVLDSACVVKIDPNSPLKQMSLLSCGVSTGVGAAWNIANVKEGKSTAVFGL 214
Query: 190 GGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTS------MQEAAD 243
G VG A+A G + ++ K E + +G ++ D T +
Sbjct: 215 GSVGLAVAEGARARGASRIIGVDANASKFEKGKLMGVTDFINPKDLTKPVHQMIREITGG 274
Query: 244 SLDYIID-TVPVGHPLEPYLSLLKLDGKLILMGVINTPLQF-ITPMVMLGRRSITGSFIG 301
+DY + T V E +LS G +L+G+ TP + PM + R ITGS G
Sbjct: 275 GVDYSFECTGNVDVLREAFLSTHVGWGSTVLVGIYPTPRTLPLHPMELFDGRRITGSVFG 334
Query: 302 SIKETEEMLEFWKE--KGLTSMIEIVN----MDYINKAFERL 337
K ++ F ++ KG+ + + + IN AF+ L
Sbjct: 335 GFKPKSQLPNFAQQCMKGVVKLEPFITNELPFEKINDAFQLL 376
>At1g22430 putative alcohol dehydrogenase
Length = 388
Score = 69.3 bits (168), Expect = 3e-12
Identities = 85/343 (24%), Positives = 140/343 (40%), Gaps = 47/343 (13%)
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLG-MSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAG 94
+V IK+ +CHTD+ K G +S +P + GHE VG V +G +V F G++V
Sbjct: 42 EVRIKILCTSLCHTDLTFWKLSFGPISRFPRILGHEAVGVVESIGENVDGFKQGDVV-LP 100
Query: 95 LLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQG--------------------G 134
+ C++C C+S +C + Y + ++ G
Sbjct: 101 VFHPYCEECKDCKSSKTNWCDR----YAEDFISNTRRYGMASRFKDSSGEVIHHFLFVSS 156
Query: 135 FAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGH 194
F+E +V+ +VKI + ++ A L C T E G I GLG VG
Sbjct: 157 FSEYTVVDIAHLVKISPEIPVDKAALLSCGVSTGIGAAWKVANVEEGSTIAIFGLGAVGL 216
Query: 195 MGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYL---------VSSDTTSMQEAADSL 245
A+ G + ++ K E + G ++ +S M E +
Sbjct: 217 AVAEGARLRGAAKIIGIDTNSDKFELGKKFGFTDFINPTLCGEKKISEVIKEMTEG--GV 274
Query: 246 DYIIDTVPVGHPL-EPYLSLLKLDGKLILMGVIN--TPLQFITPMVMLGRRSITGSFIGS 302
DY + V + L E ++S GK +++G+ P+ + +L R I GS G
Sbjct: 275 DYSFECVGLASLLNEAFISTRTGTGKTVMLGMEKHAAPIS-LGSFDLLRGRVICGSLFGG 333
Query: 303 IKETEE---MLEFW--KEKGLTSMI-EIVNMDYINKAFERLEK 339
+K + +++ + KE L S I +N INKAF LE+
Sbjct: 334 LKSKLDIPILVDHYLKKELNLDSFITHELNFKEINKAFALLEE 376
>At4g22110 alcohol dehydrogenase like protein
Length = 389
Score = 66.6 bits (161), Expect = 2e-11
Identities = 80/339 (23%), Positives = 138/339 (40%), Gaps = 43/339 (12%)
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLG-MSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAG 94
+V IK+ +CHTDV K D G ++ +P + GHE VG + +G V F G++V
Sbjct: 43 EVRIKIICTSLCHTDVSFSKIDSGPLARFPRILGHEAVGVIESIGEHVNGFQQGDVV-LP 101
Query: 95 LLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQG--------------------G 134
+ C++C C+S +C + + D ++ G
Sbjct: 102 VFHPHCEECRDCKSSKSNWCAR----FADDFLSNTRRYGMTSRFKDSFGEDIYHFLFVSS 157
Query: 135 FAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGH 194
F+E +V+ +VKI + ++ A L C T E G + GLG VG
Sbjct: 158 FSEYTVVDIAHLVKISPDIPVDKAALLSCGVSTGIGAAWKVANVEKGSTVAVFGLGAVGL 217
Query: 195 MGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSS-------DTTSMQEAADSLDY 247
A+ G + + +K E + G ++ S+ + +DY
Sbjct: 218 AVGEGARLRGAGKIIGVDLNPEKFELGKKFGFTDFINSTLCGENKISEVIKEMTGGGVDY 277
Query: 248 IIDTVPVGHPL-EPYLSLLKLDGKLILMGVIN--TPLQFITPMVMLGRRSITGSFIGSIK 304
+ V + L E + S GK +++G+ TP+ + +L R + GS G +K
Sbjct: 278 SFECVGLPSLLTEAFSSTRTGSGKTVVLGIDKHLTPVS-LGSFDLLRGRHVCGSLFGGLK 336
Query: 305 ---ETEEMLEFW--KEKGLTSMI-EIVNMDYINKAFERL 337
+ +++ + KE L S I + + INKAF+ L
Sbjct: 337 PKLDIPILVDHYLKKELNLDSFITHELKFEEINKAFDLL 375
>At1g77120 alcohol dehydrogenase
Length = 379
Score = 63.9 bits (154), Expect = 1e-10
Identities = 81/335 (24%), Positives = 132/335 (39%), Gaps = 30/335 (8%)
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGL 95
+V IK+ + +CHTDV+ + +P + GHE G V VG VT G+ V +
Sbjct: 36 EVRIKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTDLQPGDHV-LPI 94
Query: 96 LVGCCKKCTACQSDIEQYCKKKIWN----------YNDVYVDGKPT-----QGGFAETII 140
G C +C C S+ C N + ++GKP F+E +
Sbjct: 95 FTGECGECRHCHSEESNMCDLLRINTERGGMIHDGESRFSINGKPIYHFLGTSTFSEYTV 154
Query: 141 VEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVIIA 200
V V KI ++V + C T + + G I GLG VG A
Sbjct: 155 VHSGQVAKINPDAPLDKVCIVSCGLSTGLGATLNVAKPKKGQSVAIFGLGAVGLGAAEGA 214
Query: 201 KAMGHHVTVISSSDRKKKEAIEDLGADAYL--VSSDTTSMQEAADSLDYIID-----TVP 253
+ G + + K+ + ++ G + D Q A+ D +D T
Sbjct: 215 RIAGASRIIGVDFNSKRFDQAKEFGVTECVNPKDHDKPIQQVIAEMTDGGVDRSVECTGS 274
Query: 254 VGHPLEPYLSLLKLDGKLILMGVINTPLQFIT-PMVMLGRRSITGSFIGSIKETEE---M 309
V ++ + + G +L+GV + F T PM L R++ G+F G+ K + +
Sbjct: 275 VQAMIQAFECVHDGWGVAVLVGVPSKDDAFKTHPMNFLNERTLKGTFFGNYKPKTDIPGV 334
Query: 310 LEFWKEKGL---TSMIEIVNMDYINKAFERLEKND 341
+E + K L + V INKAF+ + K +
Sbjct: 335 VEKYMNKELELEKFITHTVPFSEINKAFDYMLKGE 369
>At1g64710 alcohol dehydrogenase (EC 1.1.1.1) like protein
Length = 380
Score = 63.2 bits (152), Expect = 2e-10
Identities = 70/300 (23%), Positives = 117/300 (38%), Gaps = 27/300 (9%)
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSN-YPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAG 94
+V I++ + +CHTD+ K + YP + GHE G V VG V G+ V
Sbjct: 36 EVRIRILFTSICHTDLSAWKGENEAQRAYPRILGHEAAGIVESVGEGVEEMMAGDHV-LP 94
Query: 95 LLVGCCKKCTACQSDIEQYCKK-KIWNYNDVYV-----------DGKP-----TQGGFAE 137
+ G C C C+ D C++ ++ V V D KP F+E
Sbjct: 95 IFTGECGDCRVCKRDGANLCERFRVDPMKKVMVTDGKTRFFTSKDNKPIYHFLNTSTFSE 154
Query: 138 TIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGV 197
+++ V+K+ E+++ L C T + + I GLG VG
Sbjct: 155 YTVIDSACVLKVDPLFPLEKISLLSCGVSTGVGAAWNVADIQPASTVAIFGLGAVGLAVA 214
Query: 198 IIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTS------MQEAADSLDYIIDT 251
A+A G + + K + + G ++ ++ M+ ++Y +
Sbjct: 215 EGARARGASKIIGIDINPDKFQLGREAGISEFINPKESDKAVHERVMEITEGGVEYSFEC 274
Query: 252 VPVGHPL-EPYLSLLKLDGKLILMGVINTP-LQFITPMVMLGRRSITGSFIGSIKETEEM 309
L E +LS G +++GV +P L I PM + RSIT S G K ++
Sbjct: 275 AGSIEALREAFLSTNSGVGVTVMLGVHASPQLLPIHPMELFQGRSITASVFGGFKPKTQL 334
>At1g22440 Very similar to alcohol dehydrogenase
Length = 386
Score = 63.2 bits (152), Expect = 2e-10
Identities = 90/347 (25%), Positives = 142/347 (39%), Gaps = 46/347 (13%)
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLG-MSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAG 94
+V IK+ +CHTDV K D G ++ +P + GHE VG V +G V F G++V
Sbjct: 40 EVRIKILCTSLCHTDVTFWKLDSGPLARFPRILGHEAVGVVESIGEKVDGFKQGDVV-LP 98
Query: 95 LLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQG--------------------G 134
+ C++C C S +C K Y + Y+ G
Sbjct: 99 VFHPQCEECKECISPKSNWCTK----YTNDYLSNTRRYGMTSRFKDSRGEDIHHFIFVSS 154
Query: 135 FAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVG- 193
F E +V+ +VKI + + A L C+ T E G I GLG VG
Sbjct: 155 FTEYTVVDIAHLVKISPEIPVDIAALLSCSVATGLGAAWKVADVEEGSTVVIFGLGAVGL 214
Query: 194 --HMGVII---AKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTS--MQEAAD-SL 245
GV + AK +G + + + K+ I D + L T S ++E D
Sbjct: 215 AVAEGVRLRGAAKIIGVDLNP-AKFEIGKRFGITDF-VNPALCGEKTISEVIREMTDVGA 272
Query: 246 DYIIDTVPVGHPL-EPYLSLLKLDGKLILMGVINTPLQF-ITPMVMLGRRSITGSFIGSI 303
DY + + + + E + S GK I++G+ L + +L R++ G+ G +
Sbjct: 273 DYSFECIGLASLMEEAFKSTRPGSGKTIVLGMEQKALPISLGSYDLLRGRTVCGTLFGGL 332
Query: 304 KETEEM-----LEFWKEKGLTSMI-EIVNMDYINKAFERL-EKNDVR 343
K ++ KE L +I ++ + INKAF L E N +R
Sbjct: 333 KPKLDIPILVDRYLKKELNLEDLITHELSFEEINKAFHLLAEGNSIR 379
>At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III
(pir||S71244)
Length = 379
Score = 62.8 bits (151), Expect = 3e-10
Identities = 83/338 (24%), Positives = 131/338 (38%), Gaps = 40/338 (11%)
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGL 95
+V IK+ Y +CHTD + +P + GHE G V VG VT G+ V
Sbjct: 36 EVRIKILYTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTEVQAGDHV---- 91
Query: 96 LVGC----CKKCTACQSDIEQYCKKK--------IWN--YNDVYVDGKPT-----QGGFA 136
+ C C++C C+S C K + N + V+GKP F+
Sbjct: 92 -IPCYQAECRECKFCKSGKTNLCGKVRSATGVGIMMNDRKSRFSVNGKPIYHFMGTSTFS 150
Query: 137 ETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMG 196
+ +V V KI ++V L C T + + E G I GLG VG
Sbjct: 151 QYTVVHDVSVAKIDPTAPLDKVCLLGCGVPTGLGAVWNTAKVEPGSNVAIFGLGTVGLAV 210
Query: 197 VIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSD-TTSMQEAADSL-----DYIID 250
AK G + D KK E + G + ++ D +QE L DY +
Sbjct: 211 AEGAKTAGASRIIGIDIDSKKYETAKKFGVNEFVNPKDHDKPIQEVIVDLTDGGVDYSFE 270
Query: 251 TVPVGHPLEPYLSLL-KLDGKLILMGVINTPLQFIT-PMVMLGRRSITGSFIGSIKETEE 308
+ + L K G +++GV + + T P ++ R G+ G K +
Sbjct: 271 CIGNVSVMRAALECCHKGWGTSVIVGVAASGQEISTRPFQLVTGRVWKGTAFGGFK-SRT 329
Query: 309 MLEFWKEKGLTSMIEI-------VNMDYINKAFERLEK 339
+ + EK + I++ + + INKAF+ L +
Sbjct: 330 QVPWLVEKYMNKEIKVDEYITHNLTLGEINKAFDLLHE 367
>At1g23740 auxin-induced protein like
Length = 386
Score = 57.4 bits (137), Expect = 1e-08
Identities = 78/319 (24%), Positives = 126/319 (39%), Gaps = 47/319 (14%)
Query: 35 DDVYIKVHYCGVCHTDV--HQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVG 92
D V IKV + D Q K S P VPG++V G V++VGS V G+ V
Sbjct: 106 DQVLIKVVAAALNPVDAKRRQGKFKATDSPLPTVPGYDVAGVVVKVGSAVKDLKEGDEVY 165
Query: 93 AGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEG 152
A N ++ ++G G AE VE+K + P+
Sbjct: 166 A--------------------------NVSEKALEGPKQFGSLAEYTAVEEKLLALKPKN 199
Query: 153 MAPEQVAPLLCAAVTVYSPL--SHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVI 210
+ Q A L A T L + F +S L + G GGVG + + +AK + V
Sbjct: 200 IDFAQAAGLPLAIETADEGLVRTEFSAGKSIL--VLNGAGGVGSLVIQLAKHVYGASKVA 257
Query: 211 SSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHPLEPYLSLLKLDGK 270
+++ +K E + LGAD + ++++ D D + D + + + + ++K GK
Sbjct: 258 ATASTEKLELVRSLGAD-LAIDYTKENIEDLPDKYDVVFDAIGM---CDKAVKVIKEGGK 313
Query: 271 LI-LMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDY 329
++ L G + P G R + S +K+ +E K K +
Sbjct: 314 VVALTGAVTPP----------GFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSR 363
Query: 330 INKAFERLEKNDVRYRFVV 348
+ AF LE N + VV
Sbjct: 364 VADAFSYLETNHATGKVVV 382
>At4g21580 putative NADPH quinone oxidoreductase
Length = 325
Score = 54.3 bits (129), Expect = 1e-07
Identities = 71/255 (27%), Positives = 107/255 (41%), Gaps = 54/255 (21%)
Query: 35 DDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVP----GHEVVGEVLEVGSDVTRFTVGEI 90
D+V I+V + D Q LG+ N P G E G + VG V+R+ VG+
Sbjct: 28 DEVLIRVLATALNRADTLQ---RLGLYNPPPGSSPYLGLECSGTIESVGKGVSRWKVGDQ 84
Query: 91 VGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVEQKFVVKIP 150
V A LL G GG+AE + V + IP
Sbjct: 85 VCA-LLSG----------------------------------GGYAEKVSVPAGQIFPIP 109
Query: 151 EGMAPEQVAPLLCAAVTVYSPLSHFG---LKESGLRGGILGLGGVGHMGVIIAKAMGHHV 207
G++ + A A TV+S + G + ES L G G G+G + IAK +G V
Sbjct: 110 AGISLKDAAAFPEVACTVWSTVFMMGRLSVGESFLIHG--GSSGIGTFAIQIAKHLGVRV 167
Query: 208 TVISSSDRKKKEAIEDLGADA---YLVSSDTTSMQEAAD--SLDYIIDTVPVGHPLEPYL 262
V + SD +K A ++LGAD Y ++ D +D I+D + + L+ L
Sbjct: 168 FVTAGSD-EKLAACKELGADVCINYKTEDFVAKVKAETDGKGVDVILDCIGAPY-LQKNL 225
Query: 263 SLLKLDGKLILMGVI 277
L DG+L ++G++
Sbjct: 226 DSLNFDGRLCIIGLM 240
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,093,715
Number of Sequences: 26719
Number of extensions: 361564
Number of successful extensions: 984
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 930
Number of HSP's gapped (non-prelim): 51
length of query: 358
length of database: 11,318,596
effective HSP length: 100
effective length of query: 258
effective length of database: 8,646,696
effective search space: 2230847568
effective search space used: 2230847568
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0290.23


