

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0290.16
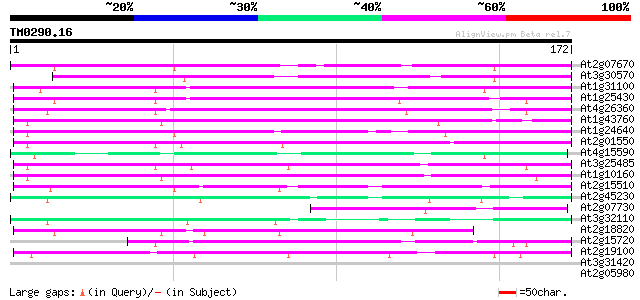
(172 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g07670 putative non-LTR retrolelement reverse transcriptase 71 3e-13
At3g30570 putative reverse transcriptase 70 6e-13
At1g31100 hypothetical protein 66 8e-12
At1g25430 hypothetical protein 62 2e-10
At4g26360 putative protein 61 3e-10
At1g43760 reverse transcriptase, putative 54 3e-08
At1g24640 hypothetical protein 53 9e-08
At2g01550 putative non-LTR retroelement reverse transcriptase 52 2e-07
At4g15590 reverse transcriptase like protein 52 2e-07
At3g25485 unknown protein 52 2e-07
At1g10160 putative reverse transcriptase 51 4e-07
At2g15510 putative non-LTR retroelement reverse transcriptase 49 1e-06
At2g45230 putative non-LTR retroelement reverse transcriptase 47 4e-06
At2g07730 putative non-LTR retroelement reverse transcriptase 47 5e-06
At3g32110 non-LTR reverse transcriptase, putative 45 3e-05
At2g18820 putative non-LTR retroelement reverse transcriptase 43 7e-05
At2g15720 putative non-LTR retroelement reverse transcriptase 43 1e-04
At2g19100 putative non-LTR retroelement reverse transcriptase 40 8e-04
At3g31420 hypothetical protein 39 0.001
At2g05980 putative non-LTR retroelement reverse transcriptase 39 0.001
>At2g07670 putative non-LTR retrolelement reverse transcriptase
Length = 913
Score = 70.9 bits (172), Expect = 3e-13
Identities = 57/180 (31%), Positives = 84/180 (46%), Gaps = 18/180 (10%)
Query: 1 KFTWHRPNNQAQ---SRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLR---QTFQD 54
KFTW R ++ RLDRVL + W S L SDH P+ ++ + +
Sbjct: 650 KFTWKRGRVESTFVAKRLDRVLCRPQTRLKWQEASVTHLPFFASDHAPIYIQLEPEVRSN 709
Query: 55 WGPKPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGD 114
+PFR WL L++ SW G V L L+S LK+WNREVFGD
Sbjct: 710 PLRRPFRFEAAWLTHSGFKDLLQASWNTE-----GETPVA--LAALKSKLKKWNREVFGD 762
Query: 115 LRSRREVVVTQINNLDLKEEEVGLDQAEVDLRK--ALFNDFWAILRLHESFLCQKARSKW 172
+ R+E + +N + + +E + ++Q + L K L +F +L E QK+R KW
Sbjct: 763 VNRRKE---SLMNEIKVVQELLEINQTDNLLSKEEELIKEFDVVLEQEEVLWFQKSREKW 819
>At3g30570 putative reverse transcriptase
Length = 1099
Score = 70.1 bits (170), Expect = 6e-13
Identities = 48/164 (29%), Positives = 80/164 (48%), Gaps = 15/164 (9%)
Query: 14 RLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTF---QDWGPKPFRVLNCWLGDH 70
RLDRVL + W + L SDH P+ L+ ++ +PFR WL
Sbjct: 244 RLDRVLCCSHARLKWQEATLTHLPFLASDHAPLYLQLALGVGKNRFRRPFRFEAAWLSHP 303
Query: 71 RLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRSRREVVVTQINNLD 130
L+ SWK G E LKG+++ L++WN+EVFGD++ ++E ++ +I +
Sbjct: 304 GFKELLLASWK-------GNIPTPEALKGIQTTLRKWNKEVFGDIQEKKEKLMVEIKTV- 355
Query: 131 LKEEEVGLDQAEVDLRK--ALFNDFWAILRLHESFLCQKARSKW 172
++ + +Q + LR+ L +F +L E+ K+R KW
Sbjct: 356 --QDSLYSNQTDDLLRRESELIIEFDVVLEQEETLWLHKSREKW 397
>At1g31100 hypothetical protein
Length = 1090
Score = 66.2 bits (160), Expect = 8e-12
Identities = 48/178 (26%), Positives = 81/178 (44%), Gaps = 12/178 (6%)
Query: 2 FTWHRPN--NQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDH--CPVLLRQTFQDWGP 57
FTW + +LDR+LV+ W + +P+ + D SDH C V++
Sbjct: 99 FTWWNKSATRPVAKKLDRILVNESWCSRFPSAYAVFGEPDFSDHASCGVIINPLMHR-EK 157
Query: 58 KPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRS 117
+PFR N L + +LV W + V G F + +KLK L++ ++ ++ E F +L
Sbjct: 158 RPFRFYNFLLQNPDFISLVGELWYSINVVGSSMFKMSKKLKALKNPIRTFSMENFSNLEK 217
Query: 118 RREVVVTQINNLDLKEEEVGLDQAEVD---LRKALFNDFWAILRLHESFLCQKARSKW 172
R V + +NL L + L + L + +++ ESF CQ++R W
Sbjct: 218 R----VKEAHNLVLYRQNKTLSDPTIPNAALEMEAQRKWLILVKAEESFFCQRSRVTW 271
>At1g25430 hypothetical protein
Length = 1213
Score = 62.0 bits (149), Expect = 2e-10
Identities = 51/177 (28%), Positives = 82/177 (45%), Gaps = 10/177 (5%)
Query: 2 FTWHRPNNQAQ--SRLDRVLVSNEWGTFWPNCSQLVLNRDVSDH--CPVLLRQTFQDWGP 57
FTW ++ ++DR+LV++ W +P+ + + D SDH C V+L +T
Sbjct: 188 FTWWNKSHTTPVAKKIDRILVNDSWNALFPSSLGIFGSLDFSDHVSCGVVLEETSIK-AK 246
Query: 58 KPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRS 117
+PF+ N L + LV ++W + V G F V +KLK L+ +K+++R + +L
Sbjct: 247 RPFKFFNYLLKNLDFLNLVRDNWFTLNVVGSSMFRVSKKLKALKKPIKDFSRLNYSELEK 306
Query: 118 R-REVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAIL-RLHESFLCQKARSKW 172
R +E I D + A +L W IL ESF QK+R W
Sbjct: 307 RTKEAHDFLIGCQDRTLADPTPINASFELEA---ERKWHILTAAEESFFRQKSRISW 360
>At4g26360 putative protein
Length = 1141
Score = 61.2 bits (147), Expect = 3e-10
Identities = 50/179 (27%), Positives = 79/179 (43%), Gaps = 14/179 (7%)
Query: 2 FTWHRPNNQ--AQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDH--CPVLLRQTFQDWGP 57
FTW + ++DR+LV+ W +P+ L D SDH C V+L +
Sbjct: 180 FTWWNKSKTRPVAKKIDRILVNESWSNLFPSSFGLFGPPDFSDHASCGVVL-ELDPIKAK 238
Query: 58 KPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRS 117
+PF+ N L + LV + W V G F V +KLK L+ +K+++R + +L
Sbjct: 239 RPFKFFNFLLKNPEFLNLVWDVWYSTNVVGSSMFRVSKKLKALKKPIKDFSRLNYSNLEK 298
Query: 118 RRE---VVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAIL-RLHESFLCQKARSKW 172
R E + NL L + E++ ++ W IL ESF Q++R W
Sbjct: 299 RTEEAHETLLSFQNLTLDNPSLENAAHELEAQRK-----WQILATAEESFFRQRSRVTW 352
>At1g43760 reverse transcriptase, putative
Length = 622
Score = 54.3 bits (129), Expect = 3e-08
Identities = 45/177 (25%), Positives = 80/177 (44%), Gaps = 10/177 (5%)
Query: 2 FTW--HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCP-VLLRQTFQDWGPK 58
+TW H+ +N +LDR + + +W + +P+ + VSDH P +++ + K
Sbjct: 267 YTWSNHQDDNPIIRKLDRAIANGDWFSSFPSAIAVFELSGVSDHSPCIIILENLPKRSKK 326
Query: 59 PFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRSR 118
FR + + +W++ G F + E LK + K NR+ FG+++ +
Sbjct: 327 CFRYFSFLSTHPTFLVSLTVAWEEQIPVGSHMFSLGEHLKAAKKCCKLLNRQGFGNIQHK 386
Query: 119 REVVVTQINNLD---LKEEEVGLDQAEVDLRKALFNDFWAILRLHESFLCQKARSKW 172
+ + + ++ L L + E RK +N F A L ESF QK+R KW
Sbjct: 387 TKEALDSLESIQSQLLTNPSDSLFRVEHVARKK-WNFFAAAL---ESFYRQKSRIKW 439
>At1g24640 hypothetical protein
Length = 1270
Score = 52.8 bits (125), Expect = 9e-08
Identities = 50/175 (28%), Positives = 77/175 (43%), Gaps = 13/175 (7%)
Query: 2 FTW--HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLR-QTFQDWGPK 58
FTW R ++ Q RLDR + EW F+P +Q L+ SDH PVL++ + QD
Sbjct: 142 FTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSDHRPVLIKLMSSQDSYRG 201
Query: 59 PFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRSR 118
FR +L + + +W + K V ++L+ R L W ++ +L S
Sbjct: 202 QFRFDKRFLFKEDVKEAIIRTWS--RGKHGTNISVADRLRACRKSLSSWKKQ--NNLNS- 256
Query: 119 REVVVTQINNLDLK-EEEVGLDQAEVDLRKALFNDFWAILRLHESFLCQKARSKW 172
+ +IN L+ E+E L L D R E++ QK+R KW
Sbjct: 257 ----LDKINQLEAALEKEQSLVWPIFQRVSVLKKDLAKAYREEEAYWKQKSRQKW 307
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 52.0 bits (123), Expect = 2e-07
Identities = 44/180 (24%), Positives = 73/180 (40%), Gaps = 10/180 (5%)
Query: 2 FTW--HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDH--CPVLLRQT--FQDW 55
FTW R N+ +LDRV+V+ W +P + SDH C + L Q
Sbjct: 602 FTWCNKRDNDPIWKKLDRVMVNEAWKMVYPQSYNVFEAGGCSDHLRCRINLNMNSGAQVR 661
Query: 56 GPKPFRVLNCWLGDHRLPALVENSWKD---MKVKGWGAFMVKEKLKGLRSVLKEWNREVF 112
G KPF+ +N LVEN W++ + + F +KLK L+ L+ +E
Sbjct: 662 GNKPFKFVNAVADMEEFKPLVENFWRETEPIHMSTSSLFRFTKKLKALKPKLRGLAKEKM 721
Query: 113 GDLRSRREVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAILRLHESFLCQKARSKW 172
G+L R + + Q +++ + + I + E +L Q ++ W
Sbjct: 722 GNLVKRTREAYLSLCQAQQSNSQ-NPSQRAMEIESEAYVRWDRIASIEEKYLKQVSKLHW 780
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 51.6 bits (122), Expect = 2e-07
Identities = 51/178 (28%), Positives = 70/178 (38%), Gaps = 33/178 (18%)
Query: 1 KFTWHR---PNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTFQDWGP 57
+FTW R + RLDRVL C+ L + CP D
Sbjct: 7 RFTWRRGLVESTFVAKRLDRVLF----------CAHARLKWQEALLCPA----QNVDARR 52
Query: 58 KPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRS 117
+PFR WL L+ SW L LR LK+WN+EVFG++
Sbjct: 53 RPFRFEAAWLSHEGFKELLTASWDT-------GLSTPVALNRLRWQLKKWNKEVFGNIHV 105
Query: 118 RREVVVTQINNLDLKEEEVGLDQAEVD----LRKALFNDFWAILRLHESFLCQKARSK 171
R+E VV+ DLK + L+ + D L +F +L E+ QK+R K
Sbjct: 106 RKEKVVS-----DLKAVQDLLEVVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREK 158
>At3g25485 unknown protein
Length = 979
Score = 51.6 bits (122), Expect = 2e-07
Identities = 44/181 (24%), Positives = 80/181 (43%), Gaps = 12/181 (6%)
Query: 2 FTW--HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDH--CPVLLRQTFQD--W 55
+TW + N+ +LDRVLV+N+W +P + SDH C + L +
Sbjct: 143 YTWSNRQENDLILKKLDRVLVNNKWKMSFPQSYNVFEAGGCSDHLRCRINLNIDVGERVK 202
Query: 56 GPKPFRVLNCWLGDHRLPALVENSWKDMK---VKGWGAFMVKEKLKGLRSVLKEWNREVF 112
G KPF+ +N +VE WK + + F +KLK L+ +++ +E
Sbjct: 203 GKKPFKFINAIAEMQEFRPMVEAYWKTTEPVYMSTSTMFRFTKKLKALKPKIRQLAKEKM 262
Query: 113 GDLRSRREVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAIL-RLHESFLCQKARSK 171
G+L R + + +EE + + + + ++ W +L + E +L QK++
Sbjct: 263 GNLVKRTKEAHDTL--CQKQEENLKSPSPQAMIEETEAHEKWELLAEIEEKYLKQKSKLH 320
Query: 172 W 172
W
Sbjct: 321 W 321
>At1g10160 putative reverse transcriptase
Length = 1118
Score = 50.8 bits (120), Expect = 4e-07
Identities = 41/175 (23%), Positives = 76/175 (43%), Gaps = 6/175 (3%)
Query: 2 FTW--HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCP-VLLRQTFQDWGPK 58
FTW H+ +N +LDR L + EW +P+ + SDH P ++L K
Sbjct: 187 FTWSNHQQDNPILRKLDRALANGEWFAVFPSALAVFDPPGDSDHAPCIILIDNQPPPSKK 246
Query: 59 PFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRSR 118
F+ + A + +W++ + G F +++ LK + + NR F +++ R
Sbjct: 247 SFKYFSFLSSHPSYLAALSTAWEENTLVGSHMFSLRQHLKVAKLCCRTLNRLRFSNIQQR 306
Query: 119 REVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAILRLH-ESFLCQKARSKW 172
+T++ D++ E + + R+ + W ESF QK+R +W
Sbjct: 307 TAQSLTRLE--DIQVELLTSPSDTLFRREHVARKQWIFFAAALESFFRQKSRIRW 359
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 48.9 bits (115), Expect = 1e-06
Identities = 48/176 (27%), Positives = 77/176 (43%), Gaps = 15/176 (8%)
Query: 2 FTWHRPNNQA--QSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLR--QTFQDWGP 57
F+W N Q RLDR + W +P +Q L++ SD PVL+R +T +++
Sbjct: 145 FSWGGKRNSLWIQCRLDRCFGNKNWFRHFPVSNQEFLDKRGSDRRPVLVRLSKTKEEYKG 204
Query: 58 KPFRVLNCWLGDHRLPALVENSWK-DMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLR 116
FR + ++ +W + +V G V E+LK RS L W +E
Sbjct: 205 N-FRFDKRLFNQPLVKEAIQQAWNGNQRV---GGMQVLERLKKCRSALSRWKQE----NN 256
Query: 117 SRREVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAILRLHESFLCQKARSKW 172
S ++Q E+ G +++V L ND + E+F QK+R+KW
Sbjct: 257 SNSLTCISQARAALELEQSSGFPRSKVVF--TLKNDLCKANQDEETFWSQKSRAKW 310
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 47.4 bits (111), Expect = 4e-06
Identities = 41/177 (23%), Positives = 71/177 (39%), Gaps = 15/177 (8%)
Query: 1 KFTWHRPNNQ--AQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTFQDWGP- 57
+F+W+ N Q RLDR + + W +P L + SDH P++ +W
Sbjct: 178 QFSWYGNRNDELVQCRLDRTVANQAWMELFPQAKATYLQKICSDHSPLINNLVGDNWRKW 237
Query: 58 KPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLRS 117
F+ W+ L+ N W K M EK+ R + +W R +
Sbjct: 238 AGFKYDKRWVQREGFKDLLCNFWSQQSTKTNALMM--EKIASCRREISKWKRV----SKP 291
Query: 118 RREVVVTQIN-NLDLKEEEVGLDQAEV-DLRKALFNDFWAILRLHESFLCQKARSKW 172
V + ++ LD +++ D+ E+ L+K L ++ E F +K+R W
Sbjct: 292 SSAVRIQELQFKLDAATKQIPFDRRELARLKKELSQEY----NNEEQFWQEKSRIMW 344
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 47.0 bits (110), Expect = 5e-06
Identities = 29/83 (34%), Positives = 45/83 (53%), Gaps = 9/83 (10%)
Query: 93 VKEKLKGLRSVLKEWNREVFGDLRSRREVVVTQI----NNLDLKEEEVGLDQAEVDLRKA 148
+++ L GL+ LK+WNREVFGD+ ++E +V I + LD+ + L Q E+
Sbjct: 31 IRKALLGLQRKLKKWNREVFGDIHVKKEKLVADIKAVQDLLDVMHSDELLAQEEI----- 85
Query: 149 LFNDFWAILRLHESFLCQKARSK 171
L D +L E+ QK+R K
Sbjct: 86 LLKDLDLVLEQEETLWFQKSREK 108
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 44.7 bits (104), Expect = 3e-05
Identities = 53/179 (29%), Positives = 67/179 (36%), Gaps = 46/179 (25%)
Query: 1 KFTWHRPNNQ---AQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTFQ---D 54
KFTW R + RLDRVL W S L L SDH P+ ++ T + +
Sbjct: 744 KFTWKRGREERFFVAKRLDRVLCCAHARLKWQEASVLHLPFLASDHAPLYVQLTPEVSGN 803
Query: 55 WGPKPFRVLNCWLGDHRLPALVENSW-KDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFG 113
G +PFR WL L+ SW KD+ A V+E L
Sbjct: 804 RGRRPFRFEAAWLSHPGFKELLLTSWNKDISTP--EALKVQELL---------------- 845
Query: 114 DLRSRREVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAILRLHESFLCQKARSKW 172
DL Q ++L KEEE L DF +L E QK+R KW
Sbjct: 846 DLH--------QSDDLLKKEEE-------------LLKDFDVVLEQEEVVWMQKSREKW 883
>At2g18820 putative non-LTR retroelement reverse transcriptase
Length = 1412
Score = 43.1 bits (100), Expect = 7e-05
Identities = 42/160 (26%), Positives = 69/160 (42%), Gaps = 21/160 (13%)
Query: 2 FTWHRPNNQAQ--SRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCP--VLLRQTFQDWGP 57
FTW N+ +LDRVL++ E+ + +P+ ++ + SDH LR Q P
Sbjct: 489 FTWGNKRNEGLICKKLDRVLLNPEYNSAYPHSYCIMDSGGCSDHLRGRFHLRSAIQK--P 546
Query: 58 K-PFRVLNCWLGDHRLPALVENSWK---DMKVKGWGAFMVKEKLKGLRSVLKEWNREVFG 113
K PF+ N VE+ WK ++ F +KLK L+ +LK+ +R
Sbjct: 547 KGPFKFTNVIAAHPEFMPKVEDFWKNTTELFPSTSTLFRFSKKLKELKPILKDLSRNNLS 606
Query: 114 DLRSRREVV-----------VTQINNLDLKEEEVGLDQAE 142
DL R +T +N D+ +E + ++ E
Sbjct: 607 DLTRRATYAYEELCRCQTKSLTTLNPHDIVDESLAFERWE 646
>At2g15720 putative non-LTR retroelement reverse transcriptase
Length = 444
Score = 42.7 bits (99), Expect = 1e-04
Identities = 40/142 (28%), Positives = 65/142 (45%), Gaps = 12/142 (8%)
Query: 37 NRDVSDH--CPVLLRQTFQDWGPKPFRVLNCWLGDHRLPALVENSWKDMKVKGWGAFMVK 94
N D SDH C V+L +PF+ N L + +V +W V G F V
Sbjct: 51 NLDFSDHVSCGVVLEADGISV-KRPFKFYNFLLNNEEFLNVVIENWYSTNVVGSHMFRVS 109
Query: 95 EKLKGLRSVLKEWNREVFGDLRSRREVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDF- 153
+KLK L+ +K+++R + + R + + + L+ +++ L A DL AL +
Sbjct: 110 KKLKALKKPIKDFSRLNYSGIELRTK----EAHEWFLRRQDLTLANAS-DLNAALELEAQ 164
Query: 154 --WAIL-RLHESFLCQKARSKW 172
W +L ESF Q++R W
Sbjct: 165 RKWLLLSSAEESFFHQRSRVSW 186
>At2g19100 putative non-LTR retroelement reverse transcriptase
Length = 1447
Score = 39.7 bits (91), Expect = 8e-04
Identities = 45/186 (24%), Positives = 79/186 (42%), Gaps = 22/186 (11%)
Query: 2 FTWH--RPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTFQDW---- 55
+TW R ++ +LDRV+V++ W +P + DH + R D
Sbjct: 595 YTWSNKREHDLIAKKLDRVMVNDVWTQSFPQSYSVFEAGGCLDH--LRGRINLNDGPGSI 652
Query: 56 --GPKPFRVLNCWLGDHRLPALVENSWKDMK---VKGWGAFMVKEKLKGLRSVLKEWNRE 110
G +PF+ +N V++ WK+ + + F +KLK L+ +L+ +E
Sbjct: 653 VRGKRPFKFVNVLTEMEDFKPTVDSYWKETEPIFLSTSSLFRFSKKLKSLKPLLRNLAKE 712
Query: 111 VFGDL-RSRREVVVTQINNLDLKEEEVGLDQAEVDLRK--ALFNDFWA-ILRLHESFLCQ 166
G+L + RE T K++E L+ + K +D W + L E FL +
Sbjct: 713 RLGNLVKKTREAYDTL-----CKKQESTLNNPTPNAMKEEVEAHDRWEHVAGLEEKFLKK 767
Query: 167 KARSKW 172
K++ W
Sbjct: 768 KSKLHW 773
>At3g31420 hypothetical protein
Length = 1491
Score = 39.3 bits (90), Expect = 0.001
Identities = 36/176 (20%), Positives = 68/176 (38%), Gaps = 11/176 (6%)
Query: 1 KFTW--HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTFQDWGPK 58
+F+W R ++ LDR + +NEW T +P + + SDH P++ + Q +
Sbjct: 359 RFSWAGKRGDHHVTCSLDRTMANNEWHTLFPESETVFMEYGESDHRPLVTNISAQKEERR 418
Query: 59 PFRVLNCWLGDHR--LPALVENSWKDMKVKGWGAFMVKEKLKGLRSVLKEWNREVFGDLR 116
F + L H+ +V N W G + KL R + W + +
Sbjct: 419 GFFSYDSRL-THKEGFKQVVLNQWHRRNGSFEGDSSLNRKLVECRQAISRWKKNNRVNAE 477
Query: 117 SRREVVVTQINNLDLKEEEVGLDQAEVDLRKALFNDFWAILRLHESFLCQKARSKW 172
R +++ ++ + + A R+ + D E + K+RS+W
Sbjct: 478 ERIKIIRHKL------DRAIATGTATPRERRQMRQDLNQAYADEEIYWQTKSRSRW 527
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 39.3 bits (90), Expect = 0.001
Identities = 33/123 (26%), Positives = 49/123 (39%), Gaps = 6/123 (4%)
Query: 2 FTW--HRPNNQAQSRLDRVLVSNEWGTFWPNCSQLVLNRDVSDHCPVLLRQTFQDWGPK- 58
FTW R +LDRVLV+ W +P + +SDH ++ + PK
Sbjct: 424 FTWCNKREEGLVNKKLDRVLVNEVWTRKFPAVYSIFEAGGISDHLRCRIKFASGNTQPKG 483
Query: 59 PFRVLNCWLGDHRLPALVENSWKDMKV---KGWGAFMVKEKLKGLRSVLKEWNREVFGDL 115
PF+ N +V++ W F +KLKGL+ +LK R +L
Sbjct: 484 PFKFNNVLTTLPEFLQIVKDFWDGTLALFHPTSAMFRFSKKLKGLKPILKSLGRSTLHNL 543
Query: 116 RSR 118
R
Sbjct: 544 SVR 546
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,997,342
Number of Sequences: 26719
Number of extensions: 163046
Number of successful extensions: 486
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 423
Number of HSP's gapped (non-prelim): 57
length of query: 172
length of database: 11,318,596
effective HSP length: 92
effective length of query: 80
effective length of database: 8,860,448
effective search space: 708835840
effective search space used: 708835840
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0290.16


