

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0288.7
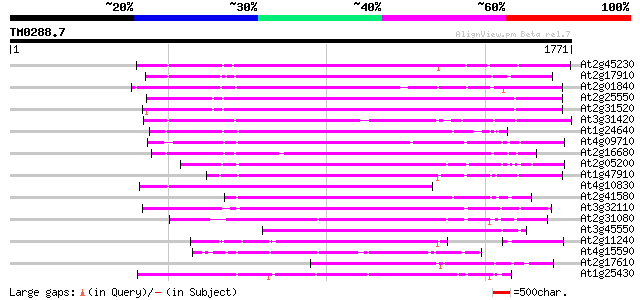
(1771 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g45230 putative non-LTR retroelement reverse transcriptase 798 0.0
At2g17910 putative non-LTR retroelement reverse transcriptase 720 0.0
At2g01840 putative non-LTR retroelement reverse transcriptase 693 0.0
At2g25550 putative non-LTR retroelement reverse transcriptase 679 0.0
At2g31520 putative non-LTR retroelement reverse transcriptase 677 0.0
At3g31420 hypothetical protein 674 0.0
At1g24640 hypothetical protein 652 0.0
At4g09710 RNA-directed DNA polymerase -like protein 645 0.0
At2g16680 putative non-LTR retroelement reverse transcriptase 640 0.0
At2g05200 putative non-LTR retroelement reverse transcriptase 602 e-172
At1g47910 reverse transcriptase, putative 595 e-170
At4g10830 putative protein 588 e-168
At2g41580 putative non-LTR retroelement reverse transcriptase 570 e-162
At3g32110 non-LTR reverse transcriptase, putative 558 e-158
At2g31080 putative non-LTR retroelement reverse transcriptase 503 e-142
At3g45550 putative protein 500 e-141
At2g11240 pseudogene 455 e-128
At4g15590 reverse transcriptase like protein 421 e-117
At2g17610 putative non-LTR retroelement reverse transcriptase 398 e-110
At1g25430 hypothetical protein 360 5e-99
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 798 bits (2060), Expect = 0.0
Identities = 463/1393 (33%), Positives = 716/1393 (51%), Gaps = 49/1393 (3%)
Query: 401 MMILSWNCRGVAATPTGRELRTLCSKNKPTIVFLMETRSKKENLEVIRRSLGFDYLFTVN 460
M ILSWNC+GV TPT R LR + P ++FL ET+ ++ LE + LGF L TV
Sbjct: 1 MRILSWNCQGVGNTPTVRHLREIRGLYFPEVIFLCETKKRRNYLENVVGHLGFFDLHTVE 60
Query: 461 PRGLSGGLALFWKREVHIQILHSSQNYIHTSIFANNSVSDWDCTFVYGDPSSQRRRFLWP 520
P G SGGLAL WK V I++L S + I + + ++ T +YG+P R LW
Sbjct: 61 PIGKSGGLALMWKDSVQIKVLQSDKRLIDALLIWQDK--EFYLTCIYGEPVQAERGELWE 118
Query: 521 QITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVDLKGSR 580
++T L + PW L GDFNE++ EK G + S FR+ +++ GL +V+ G +
Sbjct: 119 RLTRLGLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCLEFRQMLNSCGLWEVNHSGYQ 178
Query: 581 FTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLSPPTWSK- 639
F+W+ N D V + ++DR + N +W +FP A A +I SDHSP++ +L W K
Sbjct: 179 FSWYGNRNDELV-QCRLDRTVANQAWMELFPQAKATYLQKICSDHSPLINNLVGDNWRKW 237
Query: 640 TPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIWSWQRKTFKVAEK 699
FKY+ W + E +++ W + + + +I S +R I W+R + +
Sbjct: 238 AGFKYDKRWVQREGFKDLLCNFWSQQSTKTNAL--MMEKIASCRREISKWKRVSKPSSAV 295
Query: 700 EIGRLKQELQSKQDRPNHLV--DWNEVKNIKLEIDTLWRQEELFWGQRSRVKWA*YGDKN 757
I QELQ K D + D E+ +K E+ + EE FW ++SR+ W GD+N
Sbjct: 296 RI----QELQFKLDAATKQIPFDRRELARLKKELSQEYNNEEQFWQEKSRIMWMRNGDRN 351
Query: 758 SKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGVNQIEDCLRG 817
+K+FHA+T RR +NRI ++ D +G ++ ++++++F SE V + L
Sbjct: 352 TKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYFKKLFASEDVGYTVEELEN 411
Query: 818 FPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAI 877
V+ ++NN L+ P + E+ A + K PGPDG+NG ++ W T+ D + +
Sbjct: 412 LTPLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQITEMV 471
Query: 878 KEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNL 937
+ FF + ++ +N+T + L+PK+ E + FRPIS CN I KVI ++M RLK L
Sbjct: 472 QAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANRLKKILPS 531
Query: 938 LISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEK 997
LIS Q+AFV GRLI DNI ++ E H L ++++AIK D+SK YDR+EW FLEK
Sbjct: 532 LISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEK 591
Query: 998 TLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVL 1057
+ GF W ++ VK Y++ +NG + P RGLRQGDPLSPYLFV+ T++L
Sbjct: 592 AMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEML 651
Query: 1058 SFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASG 1117
ML A++ +I+G + +PP++HL FADDS+ + + + II I+ Y+ ASG
Sbjct: 652 VKMLQSAEQKNQITGLKVARGAPPISHLLFADDSMFYCKVNDEALGQIIRIIEEYSLASG 711
Query: 1118 QRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLGLPAIWGRNKCHSLAWIEER 1177
QR+N KS + FG+ S + + L + G YLGLP + +K +L+++++R
Sbjct: 712 QRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKVATLSYLKDR 771
Query: 1178 VKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQG 1237
+ +K+ GW+ L+ GKE+L+KA+ A+P Y M+ PKT C + S++A FWWK +
Sbjct: 772 LGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESVMAEFWWKNKK 831
Query: 1238 KDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWRVLTNPEALWVCVLKSVYFP 1297
+ RG+HW++W ++ K GG+GF+E N+A+L KQ WR++T ++L V KS YF
Sbjct: 832 EGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKVFKSRYFS 891
Query: 1298 TSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLLIEVWGDNWLSSGEA------ 1351
SD LNA LG PS+ W+S+ + I + IR +G G I VW D W+ + A
Sbjct: 892 KSDPLNAPLGSRPSFAWKSIYEAQVLIKQGIRAVIGNGETINVWTDPWIGAKPAKAAQAV 951
Query: 1352 ----LIQPHNAPDMK-VSELISPHGEGWNSALVRSLFQPELALKVLQTPVSKLNQTDVLF 1406
L+ + A + V +L+ P G WN LV LF +L D
Sbjct: 952 KRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLVSLLFPDNTQENILALRPGGKETRDRFT 1011
Query: 1407 WPHTPKGDYTVSSGF----EIARSNSIKPQALSSRSHNIPELLWRTIWKSQLPQKVKVFL 1462
W ++ G Y+V SG+ EI + PQ + S + +++ IWK +P K+ FL
Sbjct: 1012 WEYSRSGHYSVKSGYWVMTEIINQRN-NPQEVLQPS---LDPIFQQIWKLDVPPKIHHFL 1067
Query: 1463 WKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTRAVWFGSSLQWVVSP 1522
W+ +N L+V NL + + C C ETV H+ F CP+ R L W +SP
Sbjct: 1068 WRCVNNCLSVASNLAYRHLAREKSCVRCPSHGETVNHLLFKCPFAR-------LTWAISP 1120
Query: 1523 QGLGSFDVWL------LQKITALQTISLDFNKDFSTLACILWSIWRGRNKAVFNAIRPDP 1576
W + + ++ + + + + ILW +W+ RN VF
Sbjct: 1121 LPAPPGGEWAESLFRNMHHVLSVHKSQPEESDHHALIPWILWRLWKNRNDLVFKGREFTA 1180
Query: 1577 TFAIQQVASTLSAIDLAKPVTEKVISDGNQNTNQRPTAWRPPFGNLLKINVDASWVSSVP 1636
I + + A + K +V S +T R W+PP +K N D +W +
Sbjct: 1181 PQVILKATEDMDAWNNRKEPQPQVTS----STRDRCVKWQPPSHGWVKCNTDGAWSKDLG 1236
Query: 1637 QSAVGVLIRDKFSTLL-AGSHAKVYSSSPLTAEASAIREGLSLAHNLGCPEIIMESDSKI 1695
VG ++R+ LL G A S L E A+R + +I ESDS+
Sbjct: 1237 NCGVGWVLRNHTGRLLWLGLRALPSQQSVLETEVEALRWAVLSLSRFNYRRVIFESDSQY 1296
Query: 1696 LIDACKSETLVGEASAILQDIFYLKRLFYRCDFAWANRERNNSAHLVAKMCMQNFLLDNW 1755
L+ ++E + + +QDI L R F F + RE NN A A+ + D
Sbjct: 1297 LVSLIQNEMDIPSLAPRIQDIRNLLRHFEEVKFQFTRREGNNVADRTARESLSLMNYDPK 1356
Query: 1756 MFKLPQELRRALV 1768
M+ + + + LV
Sbjct: 1357 MYSITPDWIKNLV 1369
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 720 bits (1859), Expect = 0.0
Identities = 428/1300 (32%), Positives = 671/1300 (50%), Gaps = 36/1300 (2%)
Query: 429 PTIVFLMETRSKKENLEVIRRSLGFDYLFTVNPRGLSGGLALFWKREVHIQILHSSQNYI 488
P I+FLMET++ ++ + + LG+D++ TV P G SGGLA+FWK + I+ L++ +N +
Sbjct: 7 PDILFLMETKNSQDFVYKVFCWLGYDFIHTVEPEGRSGGLAIFWKSHLEIEFLYADKNLM 66
Query: 489 HTSIFANNSVSDWDCTFVYGDPSSQRRRFLWPQITALRFRDNTPWCLLGDFNEILFAHEK 548
+ + N V W + VYG P + R LW + ++ + WCL+GDFN+I EK
Sbjct: 67 DLQVSSRNKV--WFISCVYGLPVTHMRPKLWEHLNSIGLKRAEAWCLIGDFNDIRSNDEK 124
Query: 549 EGMRPQPPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRN 608
G + PS Q F + N + ++ G+ FTW N D +V + K+DR N +W +
Sbjct: 125 LGGPRRSPSSFQCFEHMLLNCSMHELGSTGNSFTWGGNRNDQWV-QCKLDRCFGNPAWFS 183
Query: 609 IFPNASAMAYPQISSDHSPILLDLSPPT-WSKTPFKYEVFWDEHEQCSEVVAEGWLKPLH 667
IFPNA + SDH P+L+ + + F+Y+ D+ C EV+ W +
Sbjct: 184 IFPNAHQWFLEKFGSDHRPVLVKFTNDNELFRGQFRYDKRLDDDPYCIEVIHRSWNSAM- 242
Query: 668 NEDCWKDLENRIKSSKRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNI 727
++ + I+ +R I W+ + A+ I RL+++L + ++ + W ++ I
Sbjct: 243 SQGTHSSFFSLIEC-RRAISVWKHSSDTNAQSRIKRLRKDLDA--EKSIQIPCWPRIEYI 299
Query: 728 KLEIDTLWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEG 787
K ++ + EELFW Q+SR KW GDKN+ FFHA+ R +N + + D
Sbjct: 300 KDQLSLAYGDEELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTR 359
Query: 788 QTEVMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLG 847
++ S +E +F S + + L G KVT+E+N+ LI ++ E+ AV +
Sbjct: 360 NSDKGKIASSFFENLFTSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELEVYNAVFSIN 419
Query: 848 GLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESV 907
APGPDG LFF+ HW ++ + I FF T L + N T + L+PK+ SP+ +
Sbjct: 420 KESAPGPDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRM 479
Query: 908 VQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQ 967
RPIS C+ + K+IS+I+ RLK L ++S QSAFV RLI DNI V+ E H L+
Sbjct: 480 SDLRPISLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLR 539
Query: 968 RGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNG 1027
+ K++MA K DMSK YDR+EW FLE + A GF+ W + ++ V +Y + +NG
Sbjct: 540 TNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLING 599
Query: 1028 FLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFF 1087
+ P RG+RQGDPLSP LFVL T+ L +L +A++ G+I+G +F + HL F
Sbjct: 600 QPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQDKKVSVNHLLF 659
Query: 1088 ADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHM 1147
ADD+LL +AT +E ++ L+ Y + SGQ IN+ KS + FG++ ++K I + +
Sbjct: 660 ADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQIKDWIKSRSGI 719
Query: 1148 AIWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIP 1207
++ G+YLGLP +K +I+E+++ +L GW L+Q GKEVL+K+I A+P
Sbjct: 720 SLEGGTGKYLGLPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALP 779
Query: 1208 LYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQ 1267
+Y M+ PK C L +++ +FWW + R IHW SW+++TL KD+GG GF++ +
Sbjct: 780 VYVMSCFKLPKNLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCF 839
Query: 1268 NLAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRY 1327
N A+LAKQAWRVL +L+ V +S YF SDFL+A+ G PS+ WRS+L GR+ + +
Sbjct: 840 NQALLAKQAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILFGRELLMQG 899
Query: 1328 IRWAVGGGLLIEVWGDNWLSSGEALIQPHNA-----PDMKVSELISPHGEGWNSALVRSL 1382
+R +G G VW D WL G +P N D+KVS+LI P WN ++R L
Sbjct: 900 LRTVIGNGQKTFVWTDKWLHDGSNR-RPLNRRRFINVDLKVSQLIDPTSRNWNLNMLRDL 958
Query: 1383 FQPELALKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIP 1442
F P ++++ + D W H+ G Y+V +G+E ++
Sbjct: 959 F-PWKDVEIILKQRPLFFKEDSFCWLHSHNGLYSVKTGYEFLSKQVHHRLYQEAKVKPSV 1017
Query: 1443 ELLWRTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFF 1502
L+ IW K+++FLWKA H ++ V+ L + I C +C ET+ H+ F
Sbjct: 1018 NSLFDKIWNLHTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDGCLMCDTENETINHILF 1077
Query: 1503 VCPWTRAVWFGSSLQWVVSPQGLGSFDVWLLQKITALQTISLDFNKDFSTLACILWSIWR 1562
CP R VW + L S + S + + I Q L + F + ILW +W+
Sbjct: 1078 ECPLARQVWAITHLSSAGS-EFSNSVYTNMSRLIDLTQQNDLPHHLRFVS-PWILWFLWK 1135
Query: 1563 GRNKAVFNAIRPDPTFAIQQVASTLSAIDLAKPVTEKVISD----GNQNTNQRPTAWRPP 1618
RN +F + + T + +D A + S N + + T W PP
Sbjct: 1136 NRNALLFEG----------KGSITTTLVDKAYEAYHEWFSAQTHMQNDEKHLKITKWCPP 1185
Query: 1619 FGNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAKVYS-SSPLTAEASAIREGLS 1677
LK N+ +W S ++RD +L S SP +A+ + L
Sbjct: 1186 LPGELKCNIGFAWSKQHHFSGASWVVRDSQGKVLLHSRRSFNEVHSPYSAKIRSWEWALE 1245
Query: 1678 LAHNLGCPEIIMESDSKILIDACKS----ETLVGEASAIL 1713
+ +I S + +I A L+G+ S +L
Sbjct: 1246 SMTHHHFDRVIFASSTHEIIQALHKPHEWPLLLGDISELL 1285
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 693 bits (1789), Expect = 0.0
Identities = 417/1381 (30%), Positives = 690/1381 (49%), Gaps = 61/1381 (4%)
Query: 383 HYYQQG*GGGPNHAPPITMMILSWNCRGVAATPTGRELRTLCSKNKPTIVFLMETRSKKE 442
H ++ G G +P M + WNC+G+ T R L + ++FL+ET+ +
Sbjct: 348 HNFESRGGAGTTTSP---MRVGFWNCQGLGQPLTVRRLEEVQRVYFLDMLFLIETKQQDN 404
Query: 443 NLEVIRRSLGFDYLFTVNPRGLSGGLALFWKREVHIQILHSSQNYIHTSIFANNSVSDWD 502
+ +GF+ + ++PRGLSGGL ++WK+ + IQ++ + + N
Sbjct: 405 YTRDLGVKMGFEDMCIISPRGLSGGLVVYWKKHLSIQVISHDVRLVDLYVEYKNFNFYLS 464
Query: 503 CTFVYGDPSSQRRRFLWPQITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRF 562
C +YG P R LW ++ + + PW + GDFNEIL +EK+G R + LQ F
Sbjct: 465 C--IYGHPIPSERHHLWEKLQRVSAHRSGPWMMCGDFNEILNLNEKKGGRRRSIGSLQNF 522
Query: 563 REFVHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQIS 622
++ + D+ KG+ ++W R +DRV N W+ FP P
Sbjct: 523 TNMINCCNMKDLKSKGNPYSWVGK-RQNETIESCLDRVFINSDWQASFPAFETEFLPIAG 581
Query: 623 SDHSPILLDLSPPTWSKT-PFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKS 681
SDH+P+++D++ +K F+Y+ + E + V GW + D ++
Sbjct: 582 SDHAPVIIDIAEEVCTKRGQFRYDRRHFQFEDFVDSVQRGWNRG--RSDSHGGYYEKLHC 639
Query: 682 SKRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEELF 741
++ + W+R+T ++I LK + + + +H + + ++ +++ +R EEL+
Sbjct: 640 CRQELAKWKRRTKTNTAEKIETLKYRVDAAER--DHTLPHQTILRLRQDLNQAYRDEELY 697
Query: 742 WGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEE 801
W +SR +W GD+N+ FF+AST R++RNRI + DAQG + +++ +
Sbjct: 698 WHLKSRNRWMLLGDRNTMFFYASTKLRKSRNRIKAITDAQGIENFRDDTIGKVAENYFAD 757
Query: 802 IFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLF 861
+F + + E+ + G KVT ++N+ L+ +D E+ +AV +G +APG DG F
Sbjct: 758 LFTTTQTSDWEEIISGIAPKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAAF 817
Query: 862 FKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYILK 921
+ + W I + VC ++ FF + + +IN+T + L+PK+ P+ + +RPIS C K
Sbjct: 818 YHHLWDLIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASYK 877
Query: 922 VISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKV 981
+IS+I++ RLK L +IS +Q+AFV G+ I DN+ V+ E H L+ Y+A+K
Sbjct: 878 IISKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKT 937
Query: 982 DMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQ 1041
D+SK YDR+EW FLEK ++ GF P W ++T V +Y++ +NG + P RG+RQ
Sbjct: 938 DISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQ 997
Query: 1042 GDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKE 1101
GDPLSPYLF+ +VLS ML +A+ + +I G + T ++HL FADDSL F A+ +
Sbjct: 998 GDPLSPYLFLFCAEVLSNMLRKAEVNKQIHGMKITKDCLAISHLLFADDSLFFCRASNQN 1057
Query: 1102 ATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLGLPA 1161
+ I Y ASGQ+IN AKS +IFG+ ++ + LL + G+YLGLP
Sbjct: 1058 IEQLALIFKKYEEASGQKINYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGGKYLGLPE 1117
Query: 1162 IWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFC 1221
GR K +I +VKE+ EGW L+ AGKE++IKAI A+P+Y+M P C
Sbjct: 1118 QLGRRKVELFEYIVTKVKERTEGWAYNYLSPAGKEIVIKAIAMALPVYSMNCFLLPTLIC 1177
Query: 1222 NSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWRVLT 1281
N +NSL+ FWW GK+ +EG +GF++ N A+LAKQAWR+LT
Sbjct: 1178 NEINSLITAFWW---GKE---------------NEGDLGFKDLHQFNRALLAKQAWRILT 1219
Query: 1282 NPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLLIEVW 1341
NP++L + K +Y+P + +L A+ G + S+ W S+ G+ + + +R +G G ++W
Sbjct: 1220 NPQSLLARLYKGLYYPNTTYLRANKGGHASYGWNSIQEGKLLLQQGLRVRLGDGQTTKIW 1279
Query: 1342 GDNWLSSGEALIQPHNAP----DMKVSELISPHGEGWNSALVRSLFQPELALKVLQTPVS 1397
D WL + +P P DMKV++L + W+ + + PE +S
Sbjct: 1280 EDPWLPTLPP--RPARGPILDEDMKVADLWRENKREWDPVIFEGVLNPEDQQLAKSLYLS 1337
Query: 1398 KLNQTDVLFWPHTPKGDYTVSSGFEIA-RSNSIKPQALSSRSHNIPELLWRTIWKSQLPQ 1456
D W +T YTV SG+ +A N + + ++ ++P L + IW+ ++
Sbjct: 1338 NYAARDSYKWAYTRNTQYTVRSGYWVATHVNLTEEEIINPLEGDVP--LKQEIWRLKITP 1395
Query: 1457 KVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTRAVWFGSSL 1516
K+K F+W+ +L+ L + I C CC A ET+ H+ F C + + VW ++
Sbjct: 1396 KIKHFIWRCLSGALSTTTQLRNRNIPADPTCQRCCNADETINHIIFTCSYAQVVWRSANF 1455
Query: 1517 QWVVSPQGLGSFDVWLLQKITALQTISLDFNKDFSTLAC-------ILWSIWRGRNKAVF 1569
GS + + + L K+ + I+W +W+ RN+ +F
Sbjct: 1456 S--------GSNRLCFTDNLEENIRLILQGKKNQNLPILNGLMPFWIMWRLWKSRNEYLF 1507
Query: 1570 NAIRPDPTFAIQQVASTLSAIDLAKPVTEKVISDGNQNTNQRPTA----WRPPFGNLLKI 1625
+ P + + Q A + + V + IS +N RP + W P LK
Sbjct: 1508 QQLDRFP-WKVAQKAEQEATEWVETMVNDTAISHNTAQSNDRPLSRSKQWSSPPEGFLKC 1566
Query: 1626 NVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAKVYSS-SPLTAEASAIREGLSLAHNLGC 1684
N D+ +V ++ G ++RD +L AK+ S S L AEA L + G
Sbjct: 1567 NFDSGYVQGRDYTSTGWILRDCNGRVLHSGCAKLQQSYSALQAEALGFLHALQMVWIRGY 1626
Query: 1685 PEIIMESDSKILIDACKSETLVGEASAILQDI-FYLKRLFYRCDFAWANRERNNSAHLVA 1743
+ E D+ L + +L DI F++ +L + + NRERN +A +
Sbjct: 1627 CYVWFEGDNLELTNLINKTEDHHLLETLLYDIRFWMTKLPF-SSIGYVNRERNLAADKLT 1685
Query: 1744 K 1744
K
Sbjct: 1686 K 1686
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 679 bits (1753), Expect = 0.0
Identities = 409/1323 (30%), Positives = 656/1323 (48%), Gaps = 21/1323 (1%)
Query: 431 IVFLMETRSKKENLEVIRRSLGFDYLFTVNPRGLSGGLALFWKREVHIQILHSSQNYIHT 490
I+FL+ET ++ + + + LGF + T P G SGGLAL WK V + ++ + I +
Sbjct: 412 ILFLVETLNQCDKVCKLAYDLGFPNVITQPPNGRSGGLALMWKNNVSLSLISQDERLIDS 471
Query: 491 SIFANNSVSDWDCTFVYGDPSSQRRRFLWPQITALRFRDNTPWCLLGDFNEILFAHEKEG 550
+ NN C VYG P+ R LW + + N W L+GDFNEIL EK G
Sbjct: 472 HVTFNNKSFYLSC--VYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKIG 529
Query: 551 MRPQPPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIF 610
+ + FR V + + D+ KG RF+W R + +DRV N +W F
Sbjct: 530 GPMREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGE-RHTHTVKCCLDRVFINSAWTATF 588
Query: 611 PNASAMAYPQISSDHSPILLDLSP--PTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHN 668
P A SDH P+L+ + P SK F+++ + +V W ++
Sbjct: 589 PYAEIEFLDFTGSDHKPVLVHFNESFPRRSKL-FRFDNRLIDIPTFKRIVQTSWRTNRNS 647
Query: 669 EDCWKDLENRIKSSKRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIK 728
+ RI S ++ + + + +E+ I +L+ L + VD + ++
Sbjct: 648 RST--PITERISSCRQAMARLKHASNLNSEQRIKKLQSSLNRAMESTRR-VDRQLIPQLQ 704
Query: 729 LEIDTLWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQ 788
+ + EE++W Q+SR +W GD+N+ +FHA T R ++NR++ + D QG G
Sbjct: 705 ESLAKAFSDEEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGD 764
Query: 789 TEVMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGG 848
E+ + + IF + G+ F VT +N L SD EI +A+ +G
Sbjct: 765 KEIGNHAQDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGD 824
Query: 849 LKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVV 908
KAPGPDGL F+KN W + V +K+FF T + IN T + ++PK+ +P ++
Sbjct: 825 DKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLS 884
Query: 909 QFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQR 968
+RPI+ CN + KVIS+ +V RLK LN ++S +Q+AF+ GR+I DN+ ++ E H L+
Sbjct: 885 DYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKV 944
Query: 969 GESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGF 1028
+ K YMA+K D+SK YDR+EW FLE T+ +GF W ++ VK Y + +NG
Sbjct: 945 RKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGS 1004
Query: 1029 LSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFA 1088
+TP RG+RQGDPLSPYLF+L D+LS ++ G++ G R +P +THL FA
Sbjct: 1005 PHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFA 1064
Query: 1089 DDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMA 1148
DDSL F +A V+ ++ + ++Y SGQ+INV KS + FG + + +L +
Sbjct: 1065 DDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSRLKQILEIP 1124
Query: 1149 IWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPL 1208
G+YLGLP +GR K +I +RVK++ W L+ AGKE+++K++ A+P+
Sbjct: 1125 NQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPV 1184
Query: 1209 YAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQN 1268
YAM+ PK + + SL+ NFWW+ RGI W +W+++ SK EGG+GFR+ N
Sbjct: 1185 YAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFN 1244
Query: 1269 LAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYI 1328
A+LAKQAWR++ P +L+ V+K+ YF L+A + + S+ W SLL G + +
Sbjct: 1245 DALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLLDGIALLKKGT 1304
Query: 1329 RWAVGGGLLIEVWGDNWLSS--GEALIQPHNAPDMKVSELISPHGE--GWNSALVRSLFQ 1384
R +G G I + DN + S L +M ++ L G W+ + +
Sbjct: 1305 RHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMTINNLFERKGSYYFWDDSKISQFVD 1364
Query: 1385 PELALKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPEL 1444
+ + ++K + D + W + G+YTV SG+ + + + H +L
Sbjct: 1365 QSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRSGYWLLTHDPSTNIPAINPPHGSIDL 1424
Query: 1445 LWRTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVC 1504
R IW + K+K FLW+A +LA L + + CP C E++ H F C
Sbjct: 1425 KTR-IWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFTC 1483
Query: 1505 PWTRAVWFGSSLQWVVSPQGLGSFDVWLLQKITALQ-TISLDFNKDFSTLACILWSIWRG 1563
P+ W S + + F+ + + +Q T DF+K ++W IW+
Sbjct: 1484 PFATMAWRLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHKLLP--VWLIWRIWKA 1541
Query: 1564 RNKAVFNAIRPDPTFAIQQV-ASTLSAIDLAKPVTEKVISDGNQNTNQRPTAWRPPFGNL 1622
RN VFN R P+ + A T ++ + + K + + WR P
Sbjct: 1542 RNNVVFNKFRESPSKTVLSAKAETHDWLNATQ--SHKKTPSPTRQIAENKIEWRNPPATY 1599
Query: 1623 LKINVDASWVSSVPQSAVGVLIRDKFSTLLA-GSHAKVYSSSPLTAEASAIREGLSLAHN 1681
+K N DA + ++ G +IR+ + T ++ GS ++S+PL AE A+ L
Sbjct: 1600 VKCNFDAGFDVQKLEATGGWIIRNHYGTPISWGSMKLAHTSNPLEAETKALLAALQQTWI 1659
Query: 1682 LGCPEIIMESDSKILIDACKSETLVGEASAILQDIFYLKRLFYRCDFAWANRERNNSAHL 1741
G ++ ME D + LI+ + + L+DI + F F + ++ N AH+
Sbjct: 1660 RGYTQVFMEGDCQTLINLINGISFHSSLANHLEDISFWANKFASIQFGFIRKKGNKLAHV 1719
Query: 1742 VAK 1744
+AK
Sbjct: 1720 LAK 1722
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 677 bits (1747), Expect = 0.0
Identities = 413/1341 (30%), Positives = 660/1341 (48%), Gaps = 28/1341 (2%)
Query: 420 LRTLCSKNKPT-------IVFLMETRSKKENLEVIRRSLGFDYLFTVNPRGLSGGLALFW 472
L LC K + T I +ET ++ + + + LGF + T P G SGGLAL W
Sbjct: 168 LSQLCKKGRKTESAGSCSIHDGLETLNQCDKVCKLAYDLGFPNVITQPPNGRSGGLALMW 227
Query: 473 KREVHIQILHSSQNYIHTSIFANNSVSDWDCTFVYGDPSSQRRRFLWPQITALRFRDNTP 532
K V + ++ + I + + NN C VYG P+ R LW + + N
Sbjct: 228 KNNVSLSLISQDERLIDSHVTFNNKSFYLSC--VYGHPTQSERHQLWQTLEHISDNRNAE 285
Query: 533 WCLLGDFNEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPRDGFV 592
W L+GDFNEIL EK G + + FR V + + D+ KG RF+W R
Sbjct: 286 WLLVGDFNEILSNAEKIGGPMREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGE-RHTHT 344
Query: 593 TREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLSP--PTWSKTPFKYEVFWDE 650
+ +DRV N +W FP A SDH P+L+ + P SK F+++ +
Sbjct: 345 VKCCLDRVFINSAWTATFPYAETEFLDFTGSDHKPVLVHFNESFPRRSKL-FRFDNRLID 403
Query: 651 HEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIWSWQRKTFKVAEKEIGRLKQELQS 710
+V W ++ + RI S ++ + + + +E+ I +L+ L
Sbjct: 404 IPTFKRIVQTSWRTNRNSRST--PITERISSCRQAMARLKHASNLNSEQRIKKLQSSLNR 461
Query: 711 KQDRPNHLVDWNEVKNIKLEIDTLWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRN 770
+ VD + ++ + + EE++W Q+SR +W GD+N+ +FHA T R +
Sbjct: 462 AMESTRR-VDRQLIPQLQESLAKAFSDEEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYS 520
Query: 771 RNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRL 830
+NR++ + D QG G E+ + + IF + G+ F VT +N L
Sbjct: 521 QNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDL 580
Query: 831 IMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEI 890
SD EI +A+ +G KAPGPDGL F+KN W + V +K+FF T + I
Sbjct: 581 TKEFSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSI 640
Query: 891 NETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGR 950
N T + ++PK+ +P ++ +RPI+ CN + KVIS+ +V RLK LN ++S +Q+AF+ GR
Sbjct: 641 NHTNICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGR 700
Query: 951 LIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTA 1010
+I DN+ ++ E H L+ + K YMA+K D+SK YDR+EW FLE T+ +GF W
Sbjct: 701 IINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIG 760
Query: 1011 RVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEI 1070
++ VK Y + +NG +TP RG+RQGDPLSPYLF+L D+LS ++ G++
Sbjct: 761 WIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDL 820
Query: 1071 SGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFG 1130
G R +P +THL FADDSL F +A V+ ++ + ++Y SGQ+INV KS + FG
Sbjct: 821 RGVRIGNGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFG 880
Query: 1131 RSASNRVKRDISNLLHMAIWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLL 1190
+ + +L + G+YLGLP +GR K +I +RVK++ W L
Sbjct: 881 SRVYGSTQSKLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFL 940
Query: 1191 NQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKM 1250
+ AGKE+++K++ A+P+YAM+ PK + + SL+ NFWW+ RGI W +W+++
Sbjct: 941 SPAGKEIMLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRL 1000
Query: 1251 TLSKDEGGMGFREFRMQNLAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNP 1310
SK EGG+GFR+ N A+LAKQAWR++ P +L+ V+K+ YF L+A + +
Sbjct: 1001 QYSKKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQ 1060
Query: 1311 SWMWRSLLAGRDFISRYIRWAVGGGLLIEVWGDNWLSS--GEALIQPHNAPDMKVSELIS 1368
S+ W SLL G + + R +G G I + DN + S L +M ++ L
Sbjct: 1061 SYGWASLLDGIALLKKGTRHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMTINNLFE 1120
Query: 1369 PHGE--GWNSALVRSLFQPELALKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARS 1426
G W+ + + + + ++K + D + W + G+YTV SG+ +
Sbjct: 1121 RKGSYYFWDDSKISQFVDQSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRSGYWLLTH 1180
Query: 1427 NSIKPQALSSRSHNIPELLWRTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQ 1486
+ + H +L R IW + K+K FLW+A +LA L + +
Sbjct: 1181 DPSTNIPAINPPHGSIDLKTR-IWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPI 1239
Query: 1487 CPVCCEAQETVEHMFFVCPWTRAVWFGSSLQWVVSPQGLGSFDVWLLQKITALQ-TISLD 1545
CP C E++ H F CP+ W+ S + + F+ + + +Q T D
Sbjct: 1240 CPRCHRENESINHALFTCPFATMAWWLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSD 1299
Query: 1546 FNKDFSTLACILWSIWRGRNKAVFNAIRPDPTFAIQQV-ASTLSAIDLAKPVTEKVISDG 1604
F+K ++W IW+ RN VFN R P+ + A T ++ + + K
Sbjct: 1300 FHKLLP--VWLIWRIWKARNNVVFNKFRESPSKTVLSAKAETHDWLNATQ--SHKKTPSP 1355
Query: 1605 NQNTNQRPTAWRPPFGNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLA-GSHAKVYSSS 1663
+ + WR P +K N DA + ++ G +IR+ + T ++ GS ++S+
Sbjct: 1356 TRQIAENKIEWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNHYGTPISWGSMKLAHTSN 1415
Query: 1664 PLTAEASAIREGLSLAHNLGCPEIIMESDSKILIDACKSETLVGEASAILQDIFYLKRLF 1723
PL AE A+ L G ++ ME D + LI+ + + L+DI + F
Sbjct: 1416 PLEAETKALLAALQQTWIRGYTQVFMEGDCQTLINLINGISFHSSLANHLEDISFWANKF 1475
Query: 1724 YRCDFAWANRERNNSAHLVAK 1744
F + R+ N AH++AK
Sbjct: 1476 ASIQFGFIRRKGNKLAHVLAK 1496
>At3g31420 hypothetical protein
Length = 1491
Score = 674 bits (1738), Expect = 0.0
Identities = 416/1359 (30%), Positives = 669/1359 (48%), Gaps = 83/1359 (6%)
Query: 423 LCSKNKPTIVFLMETRSKKENLEVIRRSLGFDYLFTVNPRGLSGGLALFWKREVHIQILH 482
+C KN I+ L ET+ + + + + LGF TV P GLSGGL +FW V I +
Sbjct: 203 ICRKNLLDIICLSETKQQDDRIRDVGAELGFCNHVTVPPDGLSGGLVVFWNSSVDIFLCF 262
Query: 483 SSQNYIHTSIFANNSVSDWDCTFVYGDPSSQRRRFLWPQITALRF-RDNTPWCLLGDFNE 541
S+ N + + +N + +FVYG P+ R LW ++ L R T W ++GDFNE
Sbjct: 263 SNSNLVDLHVKSNEG--SFYLSFVYGHPNPSHRHHLWERLERLNTTRQGTAWFIMGDFNE 320
Query: 542 ILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVL 601
IL EK G R +P Q FR V D+ G RF+W D VT +DR +
Sbjct: 321 ILSNREKRGGRLRPERTFQDFRNMVRGCNFTDLKSVGDRFSWAGKRGDHHVTCS-LDRTM 379
Query: 602 TNWSWRNIFPNASAMAYPQISSDHSPILLDLSPPTWSKTPF-KYEVFWDEHEQCSEVVAE 660
N W +FP + + SDH P++ ++S + F Y+ E +VV
Sbjct: 380 ANNEWHTLFPESETVFMEYGESDHRPLVTNISAQKEERRGFFSYDSRLTHKEGFKQVVLN 439
Query: 661 GWLKPLHNEDCWKDLENRIKSSKRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVD 720
W + + + L ++ ++ I W++ AE+ I ++ +L +
Sbjct: 440 QWHRRNGSFEGDSSLNRKLVECRQAISRWKKNNRVNAEERIKIIRHKLD--RAIATGTAT 497
Query: 721 WNEVKNIKLEIDTLWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDA 780
E + ++ +++ + EE++W +SR +W GD+N+++FH++T RR RNR+ V D+
Sbjct: 498 PRERRQMRQDLNQAYADEEIYWQTKSRSRWLNAGDRNTRYFHSTTKTRRCRNRLLSVQDS 557
Query: 781 QGTWKEGQTEVMHAFVSHYEEIFQSEGVNQIE--DCLRGFPRKVTAELNNRLIMPASDAE 838
G G + +++++++++S + D +GF +K+T E+N LI P ++ E
Sbjct: 558 DGDICRGDENIAKVAINYFDDLYKSTPNTSLRYADVFQGFQQKITDEINEDLIRPVTELE 617
Query: 839 IAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALV 898
I E+V + + P PDG F++ W I+ V + FF L N T + L+
Sbjct: 618 IEESVFSVAPSRTPDPDGFTADFYQQFWPDIKQKVIDEVTRFFERSELDERHNHTNLCLI 677
Query: 899 PKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITV 958
PKV++P ++ +FRPI+ CN K+IS+I+V RLK L I+ NQ+AFV GRLI +N +
Sbjct: 678 PKVETPTTIAKFRPIALCNVSYKIISKILVNRLKKHLGGAITENQAAFVPGRLITNNAII 737
Query: 959 SQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKG 1018
+ E ++ L+ + + YMA+K D++K YDRLEW FLE+T+ GF+ W R++ V
Sbjct: 738 AHEVYYALKARKRQANSYMALKTDITKAYDRLEWDFLEETMRQMGFNTKWIERIMICVTM 797
Query: 1019 ATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPH 1078
+ + +NG T+ P RG+R GDPLSPYLF+L +VLS M+ QA+ + ++ G R +
Sbjct: 798 VRFSVLINGSPHGTIKPERGIRHGDPLSPYLFILCAEVLSHMIKQAEINKKLKGIRLSTQ 857
Query: 1079 SPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVK 1138
P ++HL FADDS+ F A + T+I K
Sbjct: 858 GPFISHLLFADDSIFFTLANQRSCTAI---------------------------KEPDTK 890
Query: 1139 RDISNLLHMAIWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVL 1198
R + +LL + G+YLGLP + + K +I E+VK+K +GW + L+Q GKEVL
Sbjct: 891 RRMRHLLGIHNEGGEGKYLGLPEQFNKKKKELFNYIIEKVKDKTQGWSKKFLSQGGKEVL 950
Query: 1199 IKAIIQAIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGG 1258
+KA+ A+P+Y+M + K C ++SL+A FWW + +G+HW +W++M++ K EGG
Sbjct: 951 LKAVALAMPVYSMNIFKLTKEVCEEIDSLLARFWWSSGNETKGMHWFTWKRMSIPKKEGG 1010
Query: 1259 MGFREFRMQNLAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLL 1318
+GF+E NLA+L KQ W +L +P L VL+ YFP ++ +NA GR S++W+S+L
Sbjct: 1011 LGFKELENFNLALLGKQTWHLLQHPNCLMARVLRGRYFPETNVMNAVQGRRASFVWKSIL 1070
Query: 1319 AGRDFISRYIRWAVGGGLLIEVWGDNWLSSGEALIQPHNAPDMKVSELISPHGEGWNSAL 1378
GR+ + + +R VG G LI W D WL P ++P + +P
Sbjct: 1071 HGRNLLKKGLRCCVGDGSLINAWLDPWL--------PLHSPRAPYKQEDAPE-------- 1114
Query: 1379 VRSLFQPELALKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRS 1438
Q V + D++ W +T G YTV S + +A P +
Sbjct: 1115 --------------QLLVCSTARDDLIGWHYTKDGMYTVKSAYWLATH---LPGTTGTHP 1157
Query: 1439 HNIPELLWRTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVE 1498
L + +WK++ K+K F WK ++A L + I C CC +ET +
Sbjct: 1158 PPGDIKLKQLLWKTKTAPKIKHFCWKILSGAIATGEMLRYRHINKQSICKRCCRDEETSQ 1217
Query: 1499 HMFFVCPWTRAVWFGSSLQWVVSPQGLGSFDVWLLQKITALQTISLDFNKDFSTLA-CIL 1557
H+FF C + +A W G+ L ++ + V L +K A+ T + + L IL
Sbjct: 1218 HLFFECDYAKATWRGAGLPNLIFQDSI----VTLEEKFRAMFTFNPSTTNYWRQLPFWIL 1273
Query: 1558 WSIWRGRNKAVFNAIRPDPTFAIQQVA--STLSAIDLAKPVTEKVISDGNQNTNQRPTA- 1614
W +WR RN F + P Q+A L D+ V +VI+ ++ + R A
Sbjct: 1274 WRLWRSRNILTFQQ-KHIPWEVTVQLAKQDALEWQDIEDRV--QVINPLSRRHSNRYAAN 1330
Query: 1615 -WRPPFGNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAK-VYSSSPLTAEASAI 1672
W P K N D S+ S++ S G +IRD+ + G AK ++++ L + A+
Sbjct: 1331 RWTRPVCGWKKCNYDGSY-STIINSKAGWVIRDEHGQFIGGGQAKGKHTNNALESALQAL 1389
Query: 1673 REGLSLAHNLGCPEIIMESDSKILIDACKSETLVGEASAILQDIFYLKRLFYRCDFAWAN 1732
+ + G ++ E D+ + + ++DI KR F C F W N
Sbjct: 1390 IIAMQSCWSHGHTKVCFEGDNIEVYQILNEGKARFDVYNWIRDIQAWKRRFQECRFLWIN 1449
Query: 1733 RERNNSAHLVAKMCMQNFLLDNWMFKLPQELRRALVLDA 1771
R N A +AK +Q + +P + AL LD+
Sbjct: 1450 RRNNKPADTLAKGHLQQHEHFKFYSFIPPVIFSALHLDS 1488
>At1g24640 hypothetical protein
Length = 1270
Score = 652 bits (1681), Expect = 0.0
Identities = 380/1146 (33%), Positives = 607/1146 (52%), Gaps = 56/1146 (4%)
Query: 441 KENLEVIRRSLGFDYLFTVNPRGLSGGLALFWKREVHIQILHSSQNYIHTSIFANNSVSD 500
+++L I+ L +D ++TV P G GGLAL WK V + + +N + + +
Sbjct: 4 RDDLVDIQSWLEYDQVYTVEPVGKCGGLALLWKSSVQVDLKFVDKNLMDAQV--QFGAVN 61
Query: 501 WDCTFVYGDPSSQRRRFLWPQITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQ 560
+ + VYGDP +R W +I+ + WC+ GDFN+IL EK G + +
Sbjct: 62 FCVSCVYGDPDRSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGGPRRSDLDCK 121
Query: 561 RFREFVHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQ 620
F E + L+++ G+ FTW D ++ + ++DR N W FP ++
Sbjct: 122 AFNEMIKGCDLVEMPAHGNGFTWAGRRGDHWI-QCRLDRAFGNKEWFCFFPVSNQTFLDF 180
Query: 621 ISSDHSPILLDLSPPTWS-KTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRI 679
SDH P+L+ L S + F+++ + E E + W + H + + +R+
Sbjct: 181 RGSDHRPVLIKLMSSQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNI--SVADRL 238
Query: 680 KSSKRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEE 739
++ ++++ SW+++ + +I +L+ L+ +Q + + V +K ++ +R+EE
Sbjct: 239 RACRKSLSSWKKQNNLNSLDKINQLEAALEKEQSLVWPI--FQRVSVLKKDLAKAYREEE 296
Query: 740 LFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHY 799
+W Q+SR KW G++NSK+FHA+ + R R RI+++ D G + + +++
Sbjct: 297 AYWKQKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYF 356
Query: 800 EEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNG 859
+F+S + D G +V+ +N L+ S EI EAV + APGPDG++
Sbjct: 357 GNLFKSSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSA 416
Query: 860 LFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYI 919
LFF+++W+T+ + V +K+FF+ + AE N T + L+PK P +V RPIS C+ +
Sbjct: 417 LFFQHYWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVL 476
Query: 920 LKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAI 979
K+IS+IM RL+ L ++S QSAFV RLI DNI V+ E H L+ ++MA+
Sbjct: 477 YKIISKIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAV 536
Query: 980 KVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGL 1039
K DMSK YDR+EW++L LL+ GFH W ++ V TY + +N + RGL
Sbjct: 537 KSDMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGL 596
Query: 1040 RQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATV 1099
RQGDPLSP+LFVL T+ L+ +L +AQ +G + G +F+ + P + HL FADDSL +A+
Sbjct: 597 RQGDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASR 656
Query: 1100 KEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLGL 1159
+++ + IL +Y A+GQ IN+ KS + FG ++K I L + G YLGL
Sbjct: 657 EQSLVLQKILKVYGNATGQTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGL 716
Query: 1160 PAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKT 1219
P + +K L ++++R+KEKL+ W L+Q GKEVL+K++ A+P++AM+ P T
Sbjct: 717 PECFSGSKVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPIT 776
Query: 1220 FCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWRV 1279
C +L S +A+FWW R IHW+SWE++ L KD GG+GFR+ + N A+LAKQAWR+
Sbjct: 777 TCENLESAMASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRL 836
Query: 1280 LTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLLIE 1339
L P+ L +LKS YF +DFL+A+L + PS+ WRS+L GR+ +S+ ++ VG G +
Sbjct: 837 LHFPDCLLSRLLKSRYFDATDFLDAALSQRPSFGWRSILFGRELLSKGLQKRVGDGASLF 896
Query: 1340 VWGDNWLSSG--EALIQPHNAPD--MKVSELISPHGEGWNSALVRSLFQPE--LALKVLQ 1393
VW D W+ A + + D +KV L++P W+ ++ LF PE L +K ++
Sbjct: 897 VWIDPWIDDNGFRAPWRKNLIYDVTLKVKALLNPRTGFWDEEVLHDLFLPEDILRIKAIK 956
Query: 1394 TPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLW--RTIWK 1451
+S Q D W GD++V S + +A K Q L S P L +W
Sbjct: 957 PVIS---QADFFVWKLNKSGDFSVKSAYWLAYQT--KSQNLRSEVSMQPSTLGLKTQVWN 1011
Query: 1452 SQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTRAVW 1511
Q K+K+FLWK VC E E+ H F+CP +R +W
Sbjct: 1012 LQTDPKIKIFLWK------------------------VCGELGESTNHTLFLCPLSRQIW 1047
Query: 1512 FGSSLQWVVSPQGLGSFDVWLLQKITALQTISLDFNKDF-----STLACILWSIWRGRNK 1566
S + P G + ++ I L + NK++ ILW IW+ RN
Sbjct: 1048 ALS--DYPFPPDGFSNGSIY--SNINHL--LENKDNKEWPINLRKIFPWILWRIWKNRNS 1101
Query: 1567 AVFNAI 1572
+F I
Sbjct: 1102 FIFEGI 1107
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 645 bits (1664), Expect = 0.0
Identities = 419/1330 (31%), Positives = 660/1330 (49%), Gaps = 76/1330 (5%)
Query: 435 METRSKKENLEVIRRSLGFDYLFTVNPRGLSGGLALFWKREVHIQILHSSQNYIHTSIFA 494
MET+++ E + +G+ + FT+ P GLSGGLAL+WK V ++IL ++ N+I
Sbjct: 1 METKNQDEFISKTFDWMGYAHRFTIPPEGLSGGLALYWKENVEVEILEAAPNFI------ 54
Query: 495 NNSVSDWDCTFVYGDPSSQRRRFLWPQITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQ 554
R W +I++L + ++ W L GDFN+IL EK+G +
Sbjct: 55 ------------------DNRSVFWDKISSLGAQRSSAWLLTGDFNDILDNSEKQGGPLR 96
Query: 555 PPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNAS 614
FR FV +GL D++ G+ +W F+ + ++DR L N SW +FP +
Sbjct: 97 WEGFFLAFRSFVSQNGLWDINHTGNSLSWRGTRYSHFI-KSRLDRALGNCSWSELFPMSK 155
Query: 615 AMAYPQISSDHSPILLDL-SPPTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWK 673
SDH P++ +PP PF+++ E E+ +V E W + +K
Sbjct: 156 CEYLRFEGSDHRPLVTYFGAPPLKRSKPFRFDRRLREKEEIRALVKEVWELARQDSVLYK 215
Query: 674 DLENRIKSSKRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDT 733
I +++I W ++ + K I + +Q L+S + D + + +I E++
Sbjct: 216 -----ISRCRQSIIKWTKEQNSNSAKAIKKAQQALESALSAD--IPDPSLIGSITQELEA 268
Query: 734 LWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMH 793
+RQEELFW Q SRV+W GD+N +FHA+T RR N + + D G + ++
Sbjct: 269 AYRQEELFWKQWSRVQWLNSGDRNKGYFHATTRTRRMLNNLSVIEDGSGQEFHEEEQIAS 328
Query: 794 AFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPG 853
S+++ IF + + ++ +++ N LI +S EI EA+ + KAPG
Sbjct: 329 TISSYFQNIFTTSNNSDLQVVQEALSPIISSHCNEELIKISSLLEIKEALFSISADKAPG 388
Query: 854 PDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPI 913
PDG + FF +W I+ V I+ FF L +NET V L+PK+ +P V +RPI
Sbjct: 389 PDGFSASFFHAYWDIIEADVSRDIRSFFVDSCLSPRLNETHVTLIPKISAPRKVSDYRPI 448
Query: 914 SCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRM 973
+ CN K++++I+ RL+ L+ LIS +QSAFV GR I DN+ ++ E H L+ ++
Sbjct: 449 ALCNVQYKIVAKILTRRLQPWLSELISLHQSAFVPGRAIADNVLITHEILHFLRVSGAKK 508
Query: 974 KDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTL 1033
MAIK DMSK YDR++W FL++ L+ GFH W V+ V +Y +NG ++
Sbjct: 509 YCSMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGSPQGSV 568
Query: 1034 TPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLL 1093
P RGLRQGDPLSPYLF+L T+VLS + +AQ G + G R SP + HL FADD++
Sbjct: 569 VPSRGLRQGDPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVARGSPQVNHLLFADDTMF 628
Query: 1094 FAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSP 1153
F + ++ +IL Y ASGQ IN+AKS + F +KR + L +
Sbjct: 629 FCKTNPTCCGALSNILKKYELASGQSINLAKSAITFSSKTPQDIKRRVKLSLRIDNEGGI 688
Query: 1154 GQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAM 1213
G+YLGLP +GR K + I +R++++ W L+ AGK++L+KA++ ++P YAM
Sbjct: 689 GKYLGLPEHFGRRKRDIFSSIVDRIRQRSHSWSIRFLSSAGKQILLKAVLSSMPSYAMMC 748
Query: 1214 VHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILA 1273
P + C + S++ FWW + R + W SW+K+TL +EGG+GFRE I A
Sbjct: 749 FKLPASLCKQIQSVLTRFWWDSKPDKRKMAWVSWDKLTLPINEGGLGFRE-------IEA 801
Query: 1274 KQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRN-PSWMWRSLLAGRDFISRYIRWAV 1332
K +WR+L P +L VL Y TS F++ S + S WR +LAGRD + + + W++
Sbjct: 802 KLSWRILKEPHSLLSRVLLGKYCNTSSFMDCSASPSFASHGWRGILAGRDLLRKGLGWSI 861
Query: 1333 GGGLLIEVWGDNWL--SSGEALIQP--HNAPDMKVSELISPHGEGWNSALVRSLFQPELA 1388
G G I VW + WL SS + I P D+ V +LI + WN +R P+
Sbjct: 862 GQGDSINVWTEAWLSPSSPQTPIGPPTETNKDLSVHDLICHDVKSWNVEAIRK-HLPQYE 920
Query: 1389 LKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLW-R 1447
++ + ++ L D L W G+YT +G+ +A+ NS L + W +
Sbjct: 921 DQIRKITINALPLQDSLVWLPVKSGEYTTKTGYALAKLNSFPASQL--------DFNWQK 972
Query: 1448 TIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWT 1507
IWK KVK FLWKA +L V L R+ I C C + + ++ H+ +CP+
Sbjct: 973 NIWKIHTSPKVKHFLWKAMKGALPVGEALSRRNIEAEVTCKRCGQTESSL-HLMLLCPYA 1031
Query: 1508 RAVWFGSSLQWVVSPQGLGSFDVWLL--QKITALQTISLDFNKDFSTLACILWSIWRGRN 1565
+ VW + + + S S + L+ +++ AL L + L LW +W+ RN
Sbjct: 1032 KKVWELAPVLFNPSEATHSSVALLLVDAKRMVALPPTGLGSAPLYPWL---LWHLWKARN 1088
Query: 1566 KAVFNAIRPDPTFAIQQVASTLSAIDLAKPVTEKVISDGNQNTNQRPTAWRPPFGNLLKI 1625
+ +F+ + + L AI A+ E + + + P + P LK+
Sbjct: 1089 RLIFD------NHSCSEEGLVLKAILDARAWMEAQLLIHHPS----PISDYPSPTPNLKV 1138
Query: 1626 N---VDASWVSSVPQSAVGVLIRDKFST-LLAGSHAKVYSSSPLTAEASAIREGLSLAHN 1681
VDA+W +S +G ++D + + + + S L AE A+ L A +
Sbjct: 1139 TSCFVDAAWTTS-GYCGMGWFLQDPYKVKIKENQSSSSFVGSALMAETLAVHLALVDALS 1197
Query: 1682 LGCPEIIMESDSKILIDACKSETLVGEASAILQDIFYLKRLFYRCDFAWANRERNNSAHL 1741
G ++ + SD K LI S + E +L DI L F F + R N A
Sbjct: 1198 TGVRQLNVFSDCKELISLLNSGKSIVELRGLLHDIRELSVSFTHLCFFFIPRLSNVVADS 1257
Query: 1742 VAKMCMQNFL 1751
+AK + L
Sbjct: 1258 LAKSALSVIL 1267
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 640 bits (1652), Expect = 0.0
Identities = 393/1226 (32%), Positives = 616/1226 (50%), Gaps = 46/1226 (3%)
Query: 449 RSLGFDYLFTVNPRGLSGGLALFWKREVHIQILHSSQNYIHTSIFANNSVSDWDCTFVYG 508
R LG+DY T+ P G SGGLA+FWK + I L +N + + + W + VYG
Sbjct: 12 RWLGYDYFHTIEPVGKSGGLAIFWKNHLEIDFLFEDKNLLDLKV--SQGKKSWFVSCVYG 69
Query: 509 DPSSQRRRFLWPQITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRFREFVHN 568
+P R L +++++ + N+ WC++GDFN+IL K G + S Q F+ + N
Sbjct: 70 NPVLHLRYLLLDKLSSIGVQRNSAWCMIGDFNDILSNDGKLGGPSRLISSFQPFKNMLLN 129
Query: 569 SGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPI 628
+ + G+ FTW D ++ + K+DR N W +F N+ ++ S H P+
Sbjct: 130 CDMHQMGSSGNSFTWGGTRNDQWI-QCKLDRCFGNSEWFTMFSNSHQWFLEKLGSHHRPV 188
Query: 629 LLD-LSPPTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIW 687
L++ ++ + F Y+ + E QC+ W+ ++ L R+ ++ I
Sbjct: 189 LVNFVNDQEVFRGQFCYDKRFAEDPQCAASTLSSWIGNGISDVSSSML--RMVKCRKAIS 246
Query: 688 SWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEELFWGQRSR 747
W++ + A+ I RL+ EL +++ W+ + I+ ++ +R+EE FW +S+
Sbjct: 247 GWKKNSDFNAQNRILRLRSELD--EEKSKQYPCWSRISVIQTQLGVAFREEESFWRLKSK 304
Query: 748 VKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEG 807
KW GD+NSKFF A R +N + + D G E + ++E +F S
Sbjct: 305 DKWLFGGDRNSKFFQAMVKANRTKNSLRFLVDENGNEHTLNREKGNIASVYFENLFMSSY 364
Query: 808 VNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWA 867
+ L GF +V+ E+N L ++ EI AV + AP
Sbjct: 365 PANSQSALDGFKTRVSEEMNQELTQAVTELEIHSAVFSINVESAP-----------EKLE 413
Query: 868 TIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYILKVISRIM 927
Q S I FF T L E N T + L+PK +P+ + RPIS C+ + K+IS+I+
Sbjct: 414 CCQGSDYIEILGFFETGVLPQEWNHTHLYLIPKFTNPQRMSDIRPISLCSVLYKIISKIL 473
Query: 928 VTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTY 987
+LK L ++SP+QSAF RLI DNI ++ E H L+ + K++M K DMSK Y
Sbjct: 474 SFKLKKHLPSIVSPSQSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMVFKTDMSKAY 533
Query: 988 DRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSP 1047
DR+EW+FL++ L+A GF+ W + ++ V TY + +NG +TP RG+RQGDP+SP
Sbjct: 534 DRVEWSFLQEILVALGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIRQGDPISP 593
Query: 1048 YLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIH 1107
+LFVL T+ L +L QA+ ++SG +F P + HL F DD+ L AT + ++
Sbjct: 594 FLFVLCTEALIHILQQAENSKKVSGIQFNGSGPSVNHLLFVDDTQLVCRATKSDCEQMML 653
Query: 1108 ILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLGLPAIWGRNK 1167
L+ Y SGQ INV KS + FG KR I N + + G+YLGLP +K
Sbjct: 654 CLSQYGHISGQLINVEKSSITFGVKVDEDTKRWIKNRSGIHLEGGTGKYLGLPENLSGSK 713
Query: 1168 CHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSLNSL 1227
+I+E+++ L GW + L+Q GKE+L+K+I A+P+Y M PK C L S+
Sbjct: 714 QDLFGYIKEKLQSHLSGWYDKTLSQGGKEILLKSIALALPVYIMTCFRLPKGLCTKLTSV 773
Query: 1228 VANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWRVLTNPEALW 1287
+ +FWW IHW +K+TL K GG GF++ + N A+LAKQAWR+ ++ +++
Sbjct: 774 MMDFWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAKQAWRLFSDSKSIV 833
Query: 1288 VCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLLIEVWGDNWLS 1347
+ KS YF +DFLNA G PS+ WRS+L GR+ ++ ++ +G G VW D WL
Sbjct: 834 SQIFKSRYFMNTDFLNARQGTRPSYTWRSILYGRELLNGGLKRLIGNGEQTNVWIDKWLF 893
Query: 1348 SGEALIQPHNAPD-----MKVSELISPHGEGWNSALVRSLF-QPELALKVLQTPVSKLNQ 1401
G + +P N MKVS LI P WN + LF + ++ L + Q P+ ++
Sbjct: 894 DGHSR-RPMNLHSLMNIHMKVSHLIDPLTRNWNLKKLTELFHEKDVQLIMHQRPL--ISS 950
Query: 1402 TDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLWRTIWKSQLPQKVKVF 1461
D W T G YTV SG+E + + K + + L+ +W + K+KVF
Sbjct: 951 EDSYCWAGTNNGLYTVKSGYERSSRETFKNLFKEADVYPSVNPLFDKVWSLETVPKIKVF 1010
Query: 1462 LWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTRAVWFGSSLQWVVS 1521
+WKA +LAV+ L + I + C C E ET+ H+ F CP+ R VW S +Q +
Sbjct: 1011 MWKALKGALAVEDRLRSRGIRTADGCLFCKEEIETINHLLFQCPFARQVWALSLIQAPAT 1070
Query: 1522 PQGLGSFDVWLLQKITALQTISLDFN--KDFSTLA-CILWSIWRGRNKAVFNAIRPDPTF 1578
F + I + S +F + T++ +LW IW+ RNK +F
Sbjct: 1071 -----GFGTSIFSNINHVIQNSQNFGIPRHMRTVSPWLLWEIWKNRNKTLFQG---TGLT 1122
Query: 1579 AIQQVASTLSAIDLAKPVTEKVISDGNQNTNQRPTAWRPPFGNLLKINVDASWVSSVPQS 1638
+ + VA +L EK S G + ++ W PP LK N+ +W +
Sbjct: 1123 SSEIVAKAYEECNLWINAQEK--SSGGVSPSEH--KWNPPPAGELKCNIGVAWSRQKQLA 1178
Query: 1639 AVGVLIRDKFSTLL---AGSHAKVYS 1661
V ++RD +L S+++VYS
Sbjct: 1179 GVSWVLRDSMGQVLLHSRRSYSQVYS 1204
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 602 bits (1553), Expect = e-172
Identities = 385/1227 (31%), Positives = 602/1227 (48%), Gaps = 65/1227 (5%)
Query: 540 NEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDR 599
NEIL EK G P+ FR F+ +GL D+ G+ F+W D FV R+++DR
Sbjct: 36 NEILDNSEKRGGPPRDQGSFIDFRSFISKNGLWDLKYSGNPFSWRGMRYDWFV-RQRLDR 94
Query: 600 VLTNWSWRNIFPNASAMAYPQISSDHSPILLDLSPPTWSKT-PFKYEVFWDEHEQCSEVV 658
++N SW FP+ + SDH P+++ + + F+++ +++ + ++
Sbjct: 95 AMSNNSWLESFPSGRSEYLRFEGSDHRPLVVFVDEARVKRRGQFRFDNRLRDNDVVNALI 154
Query: 659 AEGWLKPLHNEDCWKDLENRIKSSKRTIWSWQR----KTFKVAEKEIGRLKQELQSKQDR 714
E W + ++ +R I +W R + ++ EK L++ L +
Sbjct: 155 QETWTNAGD-----ASVLTKMNQCRREIINWTRLQNLNSAELIEKTQKALEEALTADPPN 209
Query: 715 PNHLVDWNEVKNIKLEIDTLWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRI 774
P + + ++ ++ EE FW QRSRV W GD+N+ +FHA T RR +NR+
Sbjct: 210 PT------TIGALTATLEHAYKLEEQFWKQRSRVLWLHSGDRNTGYFHAVTRNRRTQNRL 263
Query: 775 DRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPA 834
+ D G + + ++ ++++IF SE V+ N+ L
Sbjct: 264 TVMEDINGVAQHEEHQISQIISGYFQQIFTSESDGDFSVVDEAIEPMVSQGDNDFLTRIP 323
Query: 835 SDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETI 894
+D E+ +AV + KAPGPDG F+ ++W I V I+ FF++ N +NET
Sbjct: 324 NDEEVKDAVFSINASKAPGPDGFTAGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNETH 383
Query: 895 VALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQD 954
+ L+PK P V +RPI+ CN K++++IM R++ L LIS NQSAFV GR+I D
Sbjct: 384 IRLIPKDLGPRKVADYRPIALCNIFYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISD 443
Query: 955 NITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLT 1014
N+ ++ E H L+ ++ MA+K DMSK YDR+EW FL+K L +GFH IW VL
Sbjct: 444 NVLITHEVLHFLRTSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLE 503
Query: 1015 LVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFR 1074
V +Y +NG + P RGLRQGDPLSP LF+L T+VLS + T+AQR ++ G R
Sbjct: 504 CVTSVSYSFLINGTPQGKVVPTRGLRQGDPLSPCLFILCTEVLSGLCTRAQRLRQLPGVR 563
Query: 1075 FTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSAS 1134
+ + P + HL FADD++ F+++ + + IL+ Y +ASGQ IN KS + F
Sbjct: 564 VSINGPRVNHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQSINFHKSSVTFSSKTP 623
Query: 1135 NRVKRDISNLLHMAIWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAG 1194
VK + +L + G+YLGLP +GR K I +++++K W L+QAG
Sbjct: 624 RSVKGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIRQKSHSWASRFLSQAG 683
Query: 1195 KEVLIKAIIQAIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSK 1254
K+V++KA++ ++PLY+M+ P C + SL+ FWW + R W +W K+T K
Sbjct: 684 KQVMLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRFWWDTKPDVRKTSWVAWSKLTNPK 743
Query: 1255 DEGGMGFREFRMQNLAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMW 1314
+ GG+GFR+ N ++LAK WR+L +PE+L +L Y +S F+ L PS W
Sbjct: 744 NAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSSSFMECKLPSQPSHGW 803
Query: 1315 RSLLAGRDFISRYIRWAVGGGLLIEVWGDNWLSSGEALIQPHNA----PDMKVSELISPH 1370
RS++AGR+ + + W + G + +W D WLS + L+ A D++VS LI+ +
Sbjct: 804 RSIIAGREILKEGLGWLITNGEKVSIWNDPWLSISKPLVPIGPALREHQDLRVSALINQN 863
Query: 1371 GEGWNSALVRSLFQPELALKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIK 1430
W+ + ++ P + Q P D L W G YT SG+ IA SI
Sbjct: 864 TLQWDWNKI-AVILPNYENLIKQLPAPSSRGVDKLAWLPVKSGQYTSRSGYGIASVASIP 922
Query: 1431 -PQALSSRSHNIPELLWRTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPV 1489
PQ + N +WK Q K+K +WKA+ +L V L R+ I PS C
Sbjct: 923 IPQTQFNWQSN--------LWKLQTLPKIKHLMWKAAMEALPVGIQLVRRHISPSAACH- 973
Query: 1490 CCEAQETVEHMFFVCPWTRAVWFGSSLQWVVSPQGLGSFD-VWLLQKITALQTISLDFNK 1548
C A E+ H+FF C + VW + LQ P G D + LL+K L +
Sbjct: 974 RCGAPESTTHLFFHCEFAAQVWELAPLQETTVPPGSSMLDALSLLKKAIILPPTGVTSAA 1033
Query: 1549 DFSTLACILWSIWRGRNKAVFNAIRPDPTFAIQQVASTLSAIDLAKPVTEKVISDGNQNT 1608
F + C ++ +A+ +A+ A Q L P T V+
Sbjct: 1034 LFPWI-CGIYGKLGTMTRAILDAL------AWQSAQRCL-------PKTRNVV------- 1072
Query: 1609 NQRPTAWRPPFGNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAKVYSS------ 1662
P + P + VDA+W++ + G + + S YS+
Sbjct: 1073 --HPISQLPVLRSGYFCFVDAAWIAQSSLAGSGWVFQ---SATALEKETATYSAGCRRLP 1127
Query: 1663 SPLTAEASAIREGLSLAHNLGCPEIIMESDSKILIDACKSETLVGEASAILQDIFYLKRL 1722
S L+AEA AI+ L A LG ++++ SDSK ++DA S + E +L +I L+
Sbjct: 1128 SALSAEAWAIKSALLHALQLGRTDLMVLSDSKSVVDALTSNISINEIYGLLMEIRALRVS 1187
Query: 1723 FYRCDFAWANRERNNSAHLVAKMCMQN 1749
F+ F + +R N A AK+ N
Sbjct: 1188 FHSLCFNFISRSANAIADATAKLSFCN 1214
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 595 bits (1535), Expect = e-170
Identities = 369/1146 (32%), Positives = 562/1146 (48%), Gaps = 48/1146 (4%)
Query: 621 ISSDHSPILLDLSPPT-WSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRI 679
++SDHSP++ ++ K F+++ W + E +++GW L + ++
Sbjct: 2 LASDHSPVIATIADKIPRGKQNFRFDKRWIGKDGLLEAISQGW--NLDSGFREGQFVEKL 59
Query: 680 KSSKRTIWSWQRKTFKVAEKEIGRLKQEL---QSKQDRPNHLVDWNEVKNIKLEIDTLWR 736
+ +R I W++ + I LK EL Q DR E+ + L + +R
Sbjct: 60 TNCRRAISKWRKSLIPFGRQTIEDLKAELDVAQRDDDRSRE-----EITELTLRLKEAYR 114
Query: 737 QEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFV 796
EE +W Q+SR W GD NSKFFHA T +RR RNRI + D G W ++ + V
Sbjct: 115 DEEQYWYQKSRSLWMKLGDNNSKFFHALTKQRRARNRITGLHDENGIWSIEDDDIQNIAV 174
Query: 797 SHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDG 856
S+++ +F + ++ L +T +N+ L A++ E+ A+ ++ KAPGPDG
Sbjct: 175 SYFQNLFTTANPQVFDEALGEVQVLITDRINDLLTADATECEVRAALFMIHPEKAPGPDG 234
Query: 857 LNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCC 916
+ LFF+ WA I+ + + F +N T + L+PK + P + + RPIS C
Sbjct: 235 MTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLC 294
Query: 917 NYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDY 976
N KVIS+I+ RLK L LIS QSAFV GRLI DNI ++QE FH L+ S +
Sbjct: 295 NVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKF 354
Query: 977 MAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPG 1036
MAIK DMSK YD++EW F+E L GF W + ++ + YK+ +NG + P
Sbjct: 355 MAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPE 414
Query: 1037 RGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAE 1096
RGLRQGDPLSPYLF+L T+VL + +A+R I+G + SP ++HL FADDSL F +
Sbjct: 415 RGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVATPSPAVSHLLFADDSLFFCK 474
Query: 1097 ATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQY 1156
A ++ I+ IL Y SGQ+IN +KS + FG + +K DI +L + G Y
Sbjct: 475 ANKEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSY 534
Query: 1157 LGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHF 1216
LGLP G +K +++ +R++ ++ GW L++ GKEV+IK++ +P Y M+
Sbjct: 535 LGLPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRL 594
Query: 1217 PKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQA 1276
PK + L S VA FWW G RG+HW +W+K+ SK +GG+GFR N A+LAKQ
Sbjct: 595 PKAITSKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQL 654
Query: 1277 WRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGL 1336
WR++T P++L+ V K YF S+ L++ +PS+ WRS+++ R + + + VG G
Sbjct: 655 WRLITAPDSLFAKVFKGRYFRKSNPLDSIKSYSPSYGWRSMISARSLVYKGLIKRVGSGA 714
Query: 1337 LIEVWGDNWLSS--------GEALIQPHNAPDMKVSELISPHGEGWNSALVRSLFQPELA 1388
I VW D W+ + G +++ P +KV LI WN L++ LF PE
Sbjct: 715 SISVWNDPWIPAQFPRPAKYGGSIVD----PSLKVKSLIDSRSNFWNIDLLKELFDPEDV 770
Query: 1389 LKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPEL--LW 1446
+ P+ N D L W T G+YTV SG+ AR + + L P+L L
Sbjct: 771 PLISALPIGNPNMEDTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIG-----PDLTTLK 825
Query: 1447 RTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPW 1506
IWK Q P K++ FLW+ + V NL ++ IL C C ++E++ H F C
Sbjct: 826 AYIWKVQCPPKLRHFLWQILSGCVPVSENLRKRGILCDKGCVSCGASEESINHTLFQCHP 885
Query: 1507 TRAVWFGSSLQWVVSPQGLGSFDVWLLQKITALQTI--SLDFNKDFSTLACILWSIWRGR 1564
R +W S + P G F + T L + + D + I+W IW+ R
Sbjct: 886 ARQIWALSQI-----PTAPGIFPSNSI--FTNLDHLFWRIPSGVDSAPYPWIIWYIWKAR 938
Query: 1565 NKAVFNAIRPDP----TFAIQQVASTLSA-IDLAKPVTEKVISDGNQNTNQRPTAWRPPF 1619
N+ VF + DP A+++ S A ++L + D R + F
Sbjct: 939 NEKVFENVDKDPMEILLLAVKEAQSWQEAQVELHSERHGSLSID--SRIRVRDVSQDTTF 996
Query: 1620 GNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAKVYSS-SPLTAEASAIREGLSL 1678
+ +D SW +S S G A V S SPL E A+ +
Sbjct: 997 SG-FRCFIDGSWKASDQFSGTGWFCLSSLGESPTMGAANVRRSLSPLHTEMEALLWAMKC 1055
Query: 1679 AHNLGCPEIIMESDSKILIDACKSETLVGEASAILQDIFYLKRLFYRCDFAWANRERNNS 1738
+ +D L+ S T S L+++ + F + +R N
Sbjct: 1056 MIGADNQNVAFFTDCSDLVKMVSSPTEWPAFSVYLEELQSDREEFTNFSLSLISRSANVK 1115
Query: 1739 AHLVAK 1744
A +A+
Sbjct: 1116 ADKLAR 1121
>At4g10830 putative protein
Length = 1294
Score = 588 bits (1516), Expect = e-168
Identities = 322/928 (34%), Positives = 499/928 (53%), Gaps = 7/928 (0%)
Query: 409 RGVAATPTGRELRTLCSKNKPTIVFLMETRSKKENLEVIRRSLGFDYLFTVNPRGLSGGL 468
+G+ T +L LC K ++FL+ET +K E + + LGF + T P+G SGGL
Sbjct: 370 KGIGVPLTQSQLSNLCKVFKFDVLFLIETLNKCEVISNLASVLGFPNVITQPPQGHSGGL 429
Query: 469 ALFWKREVHIQILHSSQNYIHTSIFANNSVSDWDCTFVYGDPSSQRRRFLWPQITALRFR 528
AL WK V + L+ +I I NN ++ + VYG P R LW L
Sbjct: 430 ALLWKDSVRLSNLYQDDRHIDVHISINNI--NFYLSRVYGHPCQSERHSLWTHFENLSKT 487
Query: 529 DNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPR 588
N PW L+GDFNEIL +EK G + + FR V L D+ G RF+W R
Sbjct: 488 RNDPWILIGDFNEILSNNEKIGGPQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGE-R 546
Query: 589 DGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLSPPTWSKT-PFKYEVF 647
+ +DR N +FP A SDH P+ L L K PF+++
Sbjct: 547 HSHTVKCCLDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFLSLEKTETRKMRPFRFDKR 606
Query: 648 WDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIWSWQRKTFKVAEKEIGRLKQE 707
E V GW K ++ + K L +++++ ++ + + K+ + I +L+
Sbjct: 607 LLEVPHFKTYVKAGWNKAINGQR--KHLPDQVRTCRQAMAKLKHKSNLNSRIRINQLQAA 664
Query: 708 LQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLK 767
L N + + +I+ E+ +R EE +W Q+SR +W GD+N++FFHA T
Sbjct: 665 LDKAMSSVNR-TERRTISHIQRELTVAYRDEERYWQQKSRNQWMKEGDRNTEFFHACTKT 723
Query: 768 RRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELN 827
R + NR+ + D +G G E+ + ++++S G GF VT ++N
Sbjct: 724 RFSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVYESNGRPVSIIDFAGFKPIVTEQIN 783
Query: 828 NRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL* 887
+ L SD EI A+ +G KAPGPDGL F+K+ W + V +K FF T +
Sbjct: 784 DDLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMK 843
Query: 888 AEINETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFV 947
IN T + ++PK+ +PE++ +RPI+ CN + K+IS+ +V RLK L+ ++S +Q+AF+
Sbjct: 844 QSINHTNICMIPKITNPETLSDYRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFI 903
Query: 948 GGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPI 1007
GRL+ DN+ ++ E H L+ + + YMA+K D+SK YDR+EW FLE T+ +GF
Sbjct: 904 PGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSET 963
Query: 1008 WTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRD 1067
W ++ VK Y + VNG T+ P RG+RQGDPLSPYLF+L D+L+ ++ +
Sbjct: 964 WIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAE 1023
Query: 1068 GEISGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGL 1127
G+I G R P +THL FADDSL F ++ V+ ++ + ++Y SGQ+IN++KS +
Sbjct: 1024 GDIRGIRIGNGVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMI 1083
Query: 1128 IFGRSASNRVKRDISNLLHMAIWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKE 1187
FG + + N+L + G+YLGLP +GR K +I ERVK++ W
Sbjct: 1084 TFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVKKRTSSWSA 1143
Query: 1188 TLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSW 1247
L+ AGKE+++K++ ++P+YAM+ P + + +L+ NFWW+ K R I W +W
Sbjct: 1144 KYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNFWWEKNAKKREIPWIAW 1203
Query: 1248 EKMTLSKDEGGMGFREFRMQNLAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLG 1307
+++ SK EGG+GFR+ N A+LAKQ WR++ NP +L+ ++K+ YF L+A
Sbjct: 1204 KRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKARYFREDSILDAKRQ 1263
Query: 1308 RNPSWMWRSLLAGRDFISRYIRWAVGGG 1335
R S+ W S+LAG D I + R+ VG G
Sbjct: 1264 RYQSYGWTSMLAGLDVIKKGSRFIVGDG 1291
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 570 bits (1470), Expect = e-162
Identities = 334/983 (33%), Positives = 513/983 (51%), Gaps = 36/983 (3%)
Query: 678 RIKSSKRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQ 737
R+ ++ I W+R+ A+ I +L++EL ++ W ++ ++ + +R+
Sbjct: 3 RLVECRKAISQWKRENDFNAKSRITKLRRELDKEKSAT--FPSWTQISLLQDVLGDAYRE 60
Query: 738 EELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVS 797
EE FW +SR KW GDKNSKFF A+ R N + + D G + E V+
Sbjct: 61 EEDFWRLKSRDKWMVGGDKNSKFFQATVKANRVSNSLRFLVDENGNEQTVNREKGKIAVT 120
Query: 798 HYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGL 857
+E++F S + ++ L GF ++VT ++N L ++ EI +AV + APGPDG
Sbjct: 121 FFEDLFSSSYPSSMDSVLEGFNKRVTEDMNQDLTKKVNEQEIYKAVFSINAESAPGPDGF 180
Query: 858 NGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCN 917
LFF+ W +++ + I+ FF T L + N T + L+PK+ P + RPIS C+
Sbjct: 181 TALFFQRQWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPISLCS 240
Query: 918 YILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYM 977
+ K+IS+I+ RLK L +++SP QSAFV RL+ DNI ++ E H L+ E KD+M
Sbjct: 241 VMYKIISKILSARLKKYLPVIVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFM 300
Query: 978 AIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGR 1037
K DMSK YDR+EW FL+ LLA GF+ W ++ V +Y + +NG +TP R
Sbjct: 301 VFKTDMSKAYDRVEWPFLKGILLALGFNSTWINWMMACVSSVSYSVLINGQPFGHITPHR 360
Query: 1038 GLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEA 1097
GLRQGDPLSP+LFVL T+ L +L QA++ G+ISG +F P + HL FADD+LL +A
Sbjct: 361 GLRQGDPLSPFLFVLCTEALIHILNQAEKIGKISGIQFNGTGPSVNHLLFADDTLLICKA 420
Query: 1098 TVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYL 1157
+ E I+H L+ Y SGQ IN KS + FG + K+ I N + G+YL
Sbjct: 421 SQLECAEIMHCLSQYGHISGQMINSEKSAITFGAKVNEETKQWIMNRSGIQTEGGTGKYL 480
Query: 1158 GLPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFP 1217
GLP + +K +I+E+++ +L GW L+Q GK++L+K+I A P+YAM
Sbjct: 481 GLPECFQGSKQVLFGFIKEKLQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLS 540
Query: 1218 KTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAW 1277
KT C L S++ +FWW + IHW +K+ L K GG GF++ + N A+LAKQA
Sbjct: 541 KTLCTKLTSVMMDFWWNSVQDKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQAS 600
Query: 1278 RVLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLL 1337
R+ T+ ++L +LKS Y+ SDFL+A+ G PS+ W+S+L GR+ + ++ +G G
Sbjct: 601 RLHTDSDSLLSQILKSRYYMNSDFLSATKGTRPSYAWQSILYGRELLVSGLKKIIGNGEN 660
Query: 1338 IEVWGDNWLSSGE----ALIQPHNAPDMKVSELISPHGEGWNSALVRSLFQ-PELALKVL 1392
VW DNW+ + +Q +KVS+LI P WN ++R LF E+ +
Sbjct: 661 TYVWMDNWIFDDKPRRPESLQIMVDIQLKVSQLIDPFSRNWNLNMLRDLFPWKEIQIICQ 720
Query: 1393 QTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLWRTIWKS 1452
Q P++ ++ D W T G YTV S +++ K + L+ IW
Sbjct: 721 QRPMA--SRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKIWNL 778
Query: 1453 QLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTRAVWF 1512
K+KVFLWK ++AV+ L + +L C +C E ET+ H+ F CP R VW
Sbjct: 779 NSAPKIKVFLWKVLKGAVAVEDRLRTRGVLIEDGCSMCPEKNETLNHILFQCPLARQVWA 838
Query: 1513 GSSLQWVVSP-QGLGSFDVWLLQKITALQTISLDFNKDFS-----TLACILWSIWRGRNK 1566
+ +Q SP G G + + I N + S I+W +W+ RNK
Sbjct: 839 LTPMQ---SPNHGFGDSIFTNVNHV-----IGNCHNTELSPHLRYVSPWIIWILWKNRNK 890
Query: 1567 AVFNAIRPDPTFAIQQVASTLSAIDLAKPVTEKVISDGNQNTNQRPT---AWRPPFGNLL 1623
+F I + +LS + A ++ + ++ PT W PP N L
Sbjct: 891 RLFEGIG----------SVSLSIVGKALEDCKEWLKAHELICSKEPTKDLTWIPPLMNEL 940
Query: 1624 KINVDASWVSSVPQSAVGVLIRD 1646
K N+ +W + V ++R+
Sbjct: 941 KCNIGIAWSKKHQMAGVSWVVRN 963
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 558 bits (1437), Expect = e-158
Identities = 382/1309 (29%), Positives = 630/1309 (47%), Gaps = 75/1309 (5%)
Query: 419 ELR--TLCSKNKPTIVFLMETRSKKENLEVIRRSLGFDYLFTVNPRGLSGGLALFWKREV 476
ELR TL S ++ + ET + + I + LGF+ F V+ G SGGL L W+ +
Sbjct: 582 ELRSITLGSVYITDVLAIFETHAGGDQASRICQGLGFENSFRVDAVGHSGGLWLLWRTGI 641
Query: 477 -HIQILHSSQNYIHTSIFANNSVSDWDCTFVYGDPSSQRRRFLWPQITALRFRDNTPWCL 535
+ ++ S+ +IH N + + VY P++ RR LW ++ + + P +
Sbjct: 642 GEVSVVDSTDQFIHAKDV--NGKDNVNLVVVYAAPTASRRSGLWDRLGDVIRSMDGPVVI 699
Query: 536 LGDFNEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVDLKGSRFTWHSNPRDGFVTRE 595
GDFN I+ E+ G + S F E++++ L+D+ KG++FTW + F +
Sbjct: 700 GGDFNTIVRLDERSGGNGRLSSDSLAFGEWINDHSLIDLGFKGNKFTWKRGREERFFVAK 759
Query: 596 KIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLSPPTWS---KTPFKYEVFWDEHE 652
++DRVL R + AS + P ++SDH+P+ + L+P + PF++E W H
Sbjct: 760 RLDRVLCCAHARLKWQEASVLHLPFLASDHAPLYVQLTPEVSGNRGRRPFRFEAAWLSHP 819
Query: 653 QCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIWSWQRKTFKVAEKEIGRLKQELQSKQ 712
E++ W K + + KV E +L K+
Sbjct: 820 GFKELLLTSWNKDISTPEA----------------------LKVQELLDLHQSDDLLKKE 857
Query: 713 DRPNHLVDWNEVKNIKLEIDTLWRQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRN 772
+ + + D + QEE+ W Q+SR KW +GD+N+KFFH ST+ RR RN
Sbjct: 858 EE------------LLKDFDVVLEQEEVVWMQKSREKWFVHGDRNTKFFHTSTIIRRRRN 905
Query: 773 RIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGVNQIEDCL--RGFPRKVTAELNNRL 830
+I+ + D G W E+ + +Y+ ++ + ++ + + L GF A+ ++ L
Sbjct: 906 QIEMLQDNDGRWLSNAQELETHAIDYYKRLYSLDDLDAVVEQLPQEGFTALSEADFSS-L 964
Query: 831 IMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEI 890
P S E+ A+ +G KAPGPDG +F++ W + +SV + +FFS+ + E
Sbjct: 965 TKPFSPLEVEGAIRSMGKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFFSSGSFPQET 1024
Query: 891 NETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGR 950
N+ +V L+ KV PE + QFRPIS CN + K I+++MV RLK +N LI P Q++F+ GR
Sbjct: 1025 NDVLVVLIAKVLKPEKITQFRPISLCNVLFKTITKVMVGRLKGVINKLIGPAQTSFIPGR 1084
Query: 951 LIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTA 1010
L DNI V QE H ++R + +K +M +K+D+ K YDR+ W LE TL A G W
Sbjct: 1085 LSTDNIVVVQEVVHSMRRKKG-VKGWMLLKLDLEKAYDRIRWDLLEDTLKAAGLPGTWVQ 1143
Query: 1011 RVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEI 1070
++ V+G + +L NG + P RGLRQGDPLSPYLFVL + L ++ + +
Sbjct: 1144 WIMKCVEGPSMRLLWNGEKTDAFKPLRGLRQGDPLSPYLFVLCIERLCHLIESSIAAKKW 1203
Query: 1071 SGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFG 1130
+ + P L+H+ FADD +LFAEA++ + + +L + ASGQ++++ KS + F
Sbjct: 1204 KPIKISQSGPRLSHICFADDLILFAEASIDQIRVLRGVLEKFCGASGQKVSLEKSKIYFS 1263
Query: 1131 RSASNRVKRDISNLLHMAIWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLL 1190
++ + + IS M G+YLG+P + R + + +R +L GWK +L
Sbjct: 1264 KNVLRDLGKRISEESGMKATKDLGKYLGVPILQKRINKETFGEVIKRFSSRLAGWKGRML 1323
Query: 1191 NQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKM 1250
+ AG+ L KA++ +I ++ M+ + P++ + L+ + F W + + H +W ++
Sbjct: 1324 SFAGRLTLTKAVLTSILVHTMSTIKLPQSTLDGLDKVSRAFLWGSTLEKKKQHLVAWTRV 1383
Query: 1251 TLSKDEGGMGFREFRMQNLAILAKQAWRVLTNPEALWVCVLKSVY--FPTSDFLNASLGR 1308
L + EGG+G R N A++AK WRVL + +LW V++S Y D
Sbjct: 1384 CLPRREGGLGIRSATAMNKALIAKVGWRVLNDGSSLWAQVVRSKYKVGDVHDRNWTVAKS 1443
Query: 1309 NPSWMWRSLLAG-RDFISRYIRWAVGGGLLIEVWGDNWLS----SGEALIQPHNAPDMKV 1363
N S WRS+ G RD I R W +G G I W D WLS + ++++ A +
Sbjct: 1444 NWSSTWRSVGVGLRDVIWREQHWVIGDGRQIRFWTDRWLSETPIADDSIVPLSLAQMLCT 1503
Query: 1364 SELISPHGEGWNSALVRSLFQPELALKVLQTPVSKLNQT-DVLFWPHTPKGDYTVSSGFE 1422
+ + G GW+ + + L +L V + D L W T G +TV S F
Sbjct: 1504 ARDLWRDGTGWDMSQIAPFVTDNKRLDLLAVIVDSVTGAHDRLAWGMTSDGRFTVKSAFA 1563
Query: 1423 IARSNSIKPQALSSRSHNIPELLWRTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRIL 1482
+ ++ Q +SS L+ +WK Q P++V+VFLW + ++ R+ +
Sbjct: 1564 MLTNDDSPRQDMSS--------LYGRVWKVQAPERVRVFLWLVVNQAIMTNSERKRRHLC 1615
Query: 1483 PSGQCPVCCEAQETVEHMFFVCPWTRAVWFGSSLQWVVSPQGLGSFDVWLLQKI-TALQT 1541
S C VC E++ H+ CP +W + + S WL + L T
Sbjct: 1616 DSDVCQVCRGGIESILHVLRDCPAMSGIWDRIVPRRLQQSFFTMSLLEWLYSNLRQGLMT 1675
Query: 1542 ISLDFNKDFSTLACILWSIWRGRNKAVF--NAIRPDPTFAIQQVASTLSAIDLAKPVTEK 1599
D++ F A +W W+ R +F N D I+ +A +S I ++ V E
Sbjct: 1676 EGSDWSTMF---AMAVWWGWKWRCSNIFGENKTCRDRVRFIKDLAVEVS-IAYSREV-EL 1730
Query: 1600 VISDGNQNTNQRPTAWRPPFGNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAKV 1659
+S N +P W PP KIN D + + ++ G ++R+ G +
Sbjct: 1731 RLSGLRVN---KPIRWTPPMEGWYKINTDGASRGNPGLASAGGVLRNSAGAWCGGFAVNI 1787
Query: 1660 YSSSPLTAEASAIREGLSLAHNLGCPEIIMESDSKILIDACKSETLVGE 1708
S AE + GL +A + +E DS++++ K T +GE
Sbjct: 1788 GRCSAPLAELWGVYYGLYMAWAKQLTHLELEVDSEVVVGFLK--TGIGE 1834
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 503 bits (1294), Expect = e-142
Identities = 357/1224 (29%), Positives = 577/1224 (46%), Gaps = 116/1224 (9%)
Query: 506 VYGDPSSQRRRFLWPQITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRFREF 565
VY PS RR LW ++ + P + GDFN IL+ E+ G + F ++
Sbjct: 6 VYAAPSVSRRSGLWGELKDVVNGLEGPLLIGGDFNTILWVDERMGGNGRLSPDSLAFGDW 65
Query: 566 VHNSGLMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDH 625
++ L+D+ KG++FTW ++ V +++DRV R + A P ++SDH
Sbjct: 66 INELSLIDLGFKGNKFTWRRGRQESTVVAKRLDRVFVCAHARLKWQEAVVSHLPFMASDH 125
Query: 626 SPILLDLSPPTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRT 685
+P+ + L P +R
Sbjct: 126 APLYVQLEP----------------------------------------------LQQRK 139
Query: 686 IWSWQRKTFKVAEKEIGRLKQELQSKQDR-----PNHLVDWNEVKNIKLEIDTLWRQEEL 740
+ W R+ F +L +++ QD + L+ EV + E+D + QEE
Sbjct: 140 LRKWNREVFGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEV--LLKEMDLVLEQEET 197
Query: 741 FWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYE 800
W Q+SR K+ GD+N+ FFH ST+ RR RNRI+ + W + E+ +++Y+
Sbjct: 198 LWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMALTYYK 257
Query: 801 EIFQSEGVNQIEDCLR--GFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLN 858
++ E V+++ + L GF AE L+ + AE+ AV +G KAPGPDG
Sbjct: 258 RLYSLEDVSEVRNMLPTGGFASISEAE-KAALLQAFTKAEVVSAVKSMGRFKAPGPDGYQ 316
Query: 859 GLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNY 918
+F++ W T+ SV + EFF T L A N+ ++ L+ KV PE + QFRP+S CN
Sbjct: 317 PVFYQQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQFRPVSLCNV 376
Query: 919 ILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMA 978
+ K+I+++MVTRLK+ ++ LI P Q++F+ GRL DNI + QEA H ++R + R K +M
Sbjct: 377 LFKIITKMMVTRLKNVISKLIGPAQASFIPGRLSIDNIVLVQEAVHSMRRKKGR-KGWML 435
Query: 979 IKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRG 1038
+K+D+ K YDR+ W FL++TL A G WT+R++ V + + NG + + P RG
Sbjct: 436 LKLDLEKAYDRVRWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSVLWNGERTDSFVPARG 495
Query: 1039 LRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEAT 1098
LRQGDPLSPYLFVL + L ++ + E + L+H+ FADD +LFAEA+
Sbjct: 496 LRQGDPLSPYLFVLCLERLCHLIEASVGKREWKPIAVSCGGSKLSHVCFADDLILFAEAS 555
Query: 1099 VKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLG 1158
V + I +L + ASGQ++++ KS + F + S +++ IS + G+YLG
Sbjct: 556 VAQIRIIRRVLERFCEASGQKVSLEKSKIFFSHNVSREMEQLISEESGIGCTKELGKYLG 615
Query: 1159 LPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPK 1218
+P + R + + ERV +L GWK L+ AG+ L KA++ +IP++ M+ + P
Sbjct: 616 MPILQKRMNKETFGEVLERVSARLAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPV 675
Query: 1219 TFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWR 1278
+ ++L+ F W + + H SW K+ K EGG+G R R N A++AK WR
Sbjct: 676 STLDTLDRYSRTFLWGSTMEKKKQHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWR 735
Query: 1279 VLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSW--MWRSLLAG-RDFISRYIRWAVGGG 1335
+L + E+LW V++ Y + L P W WRS+ G R+ + + + W G G
Sbjct: 736 LLQDKESLWARVVRKKYKVGGVQDTSWLKPQPRWSSTWRSVAVGLREVVVKGVGWVPGDG 795
Query: 1336 LLIEVWGDNWLSSGEALIQ--PHNAPD---MKVSELISPHGEGWNSALVRSLFQPELALK 1390
I W D WL E L++ P+ +KV+ G GWN ++ L+ PE +
Sbjct: 796 CTIRFWLDRWLLQ-EPLVELGTDMIPEGERIKVAADYWLPGSGWNLEIL-GLYLPETVKR 853
Query: 1391 VLQTPVSK--LNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLWRT 1448
L + V + L D + W T G +TV S + + + + + S +
Sbjct: 854 RLLSVVVQVFLGNGDEISWKGTQDGAFTVRSAYSLLQGDVGDRPNMGS--------FFNR 905
Query: 1449 IWKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTR 1508
IWK P++V+VF+W S N + R+ + + C VC A+ET+ H+ CP
Sbjct: 906 IWKLITPERVRVFIWLVSQNVIMTNVERVRRHLSENAICSVCNGAEETILHVLRDCPAME 965
Query: 1509 AVW------------FGSSLQWVVSPQGLGSFDVWLLQKITALQTISLDFNKDFSTLACI 1556
+W F SL WL + ++ I + TL +
Sbjct: 966 PIWRRLLPLRRHHEFFSQSLL------------EWLFTNMDPVKGI-------WPTLFGM 1006
Query: 1557 -LWSIWRGRNKAVF--NAIRPDPTFAIQQVASTLSAIDLAKPVTEKVISDGNQNTNQRPT 1613
+W W+ R VF I D I+ +A + + + V + N +R
Sbjct: 1007 GIWWAWKWRCCDVFGERKICRDRLKFIKDMAEEVRRVHVG-----AVGNRPNGVRVERMI 1061
Query: 1614 AWRPPFGNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAKVYSSSPLTAEASAIR 1673
W+ P +KI D + + +A G IR+ L G + S + AE
Sbjct: 1062 RWQVPSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGAY 1121
Query: 1674 EGLSLAHNLGCPEIIMESDSKILI 1697
GL +A + G + ++ D K+++
Sbjct: 1122 YGLLIAWDKGFRRVELDLDCKLVV 1145
>At3g45550 putative protein
Length = 851
Score = 500 bits (1288), Expect = e-141
Identities = 277/843 (32%), Positives = 444/843 (51%), Gaps = 19/843 (2%)
Query: 799 YEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLN 858
+ +IF + G+ FP VT +N+ L D+EI EA+ +G KAPGPDGL
Sbjct: 14 FTDIFTTNGIQVSPIDFADFPSSVTNIINSELTQDFRDSEIFEAICQIGDDKAPGPDGLT 73
Query: 859 GLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNY 918
F+K W + + V +K FF + ++ +N T + ++PK+ +P+++ +RPI+ CN
Sbjct: 74 ARFYKQCWDIVGNDVIKEVKLFFESSHMKTSVNHTNICMIPKIQNPQTLSDYRPIALCNV 133
Query: 919 ILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMA 978
+ KVIS+ MV RLK LN ++S +Q+AF+ GR+I DN+ ++ E H L+ + K YMA
Sbjct: 134 LYKVISKCMVNRLKAHLNSIVSDSQAAFIPGRIINDNVMIAHEIMHSLKVRKRVSKTYMA 193
Query: 979 IKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRG 1038
+K D+SK YDR+EW FLE T+ +GF W ++ VK Y + +NG ++P RG
Sbjct: 194 VKTDVSKAYDRVEWDFLETTMRLFGFCDKWIGWIMAAVKSVHYSVLINGSPHGYISPTRG 253
Query: 1039 LRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEAT 1098
+RQGDPLSPYLF+L D+LS ++ G+I G R +P +THL FADDSL F +A
Sbjct: 254 IRQGDPLSPYLFILCGDILSHLIKVKASSGDIRGVRIGNGAPAITHLQFADDSLFFCQAN 313
Query: 1099 VKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLG 1158
V+ ++ + ++Y SGQ+INV KS + FG + + LL++ G+YLG
Sbjct: 314 VRNCQALKDVFDVYEYYSGQKINVQKSLITFGSRVYGSTQTRLKTLLNIPNQGGGGKYLG 373
Query: 1159 LPAIWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPK 1218
LP +GR K +I +RVKE+ W L+ AGKE+L+K++ A+P+YAM+ P+
Sbjct: 374 LPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSPAGKEILLKSVALAMPVYAMSCFKLPQ 433
Query: 1219 TFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWR 1278
+ + SL+ NFWW+ RGI W +W+++ SK EGG+GFR+ N A+LAKQAWR
Sbjct: 434 GIVSEIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWR 493
Query: 1279 VLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLLI 1338
++ P +L+ V+K+ YF + ++A S+ W SLL+G + + R+ +G G I
Sbjct: 494 IIQYPNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLLSGIALLRKGTRYVIGDGKTI 553
Query: 1339 EVWGDNWLSS--GEALIQPHNAPDMKVSELISPHGEG--WNSALVRSLFQPELALKVLQT 1394
+ DN + S L+ + + L G W++A +++ + +
Sbjct: 554 RLGIDNVVDSHPPRPLLTDEQHNGLSLDNLFQHRGHSRCWDNAKLQTFVDQSDHDYIKRI 613
Query: 1395 PVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLWRTIWKSQL 1454
+S ++TD L W + GDYTV SG+ ++ + ++ H +L + IW +
Sbjct: 614 YLSTRSKTDRLIWSYNSTGDYTVRSGYWLSTHDPSNTIPTMAKPHGSVDLKTK-IWNLPI 672
Query: 1455 PQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTRAVWFGS 1514
K+K FLW+ +L L + + CP C E++ H F CP+ W S
Sbjct: 673 MPKLKHFLWRILSKALPTTDRLTTRGMRIDPGCPRCRRENESINHALFTCPFATMAWRLS 732
Query: 1515 SLQWVVSPQGLGSFDVWLLQKITALQTISLDFNKDFSTLACILWSIWRGRNKAVFNAIRP 1574
S + + + + LQ ++ ++ +LW IW+ RN VFN +R
Sbjct: 733 DTPLYRSSILSNNIEDNISNILLLLQNTTITDSQKLIPF-WLLWRIWKARNNVVFNNLRE 791
Query: 1575 DPTFAIQQVAST----LSAIDLAKP--VTEKVISDGNQNTNQRPTAWRPPFGNLLKINVD 1628
P+ + + + L+A P + ++ + GN T W P +K N D
Sbjct: 792 SPSITVVRAKAETNEWLNATQTQGPRRLPKRTTAAGN-------TTWVKPQMPYIKCNFD 844
Query: 1629 ASW 1631
AS+
Sbjct: 845 ASF 847
>At2g11240 pseudogene
Length = 1044
Score = 455 bits (1171), Expect = e-128
Identities = 263/824 (31%), Positives = 432/824 (51%), Gaps = 35/824 (4%)
Query: 571 LMDVDLKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPIL- 629
L D+ G+ +W D V ++DR L+N +W +P + + SDH P+L
Sbjct: 6 LYDLRHSGNFLSWRGKRHD-HVVHCRLDRALSNGAWAEDYPASRCIYLCFEGSDHRPLLT 64
Query: 630 -LDLSPPTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIWS 688
DLS K F+Y+ +++ + +V E W L++ D +E +I + I
Sbjct: 65 HFDLSKKK-KKGVFRYDRRLKNNDEVTALVQEAW--NLYDTDI---VEEKISRCRLEIVK 118
Query: 689 WQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEELFWGQRSRV 748
W R + ++K I +Q+L+ +H + N L + ++ EE +W QRSR
Sbjct: 119 WSRAKQQSSQKLIEENRQKLEEAMSSQDHNQELLSTINTNLLL--AYKAEEEYWKQRSRQ 176
Query: 749 KWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQS-EG 807
W GDKNS +FHA T R N+ + G + + +++ +++++F + EG
Sbjct: 177 LWLALGDKNSGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISEYFQKLFSANEG 236
Query: 808 VNQ--IEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNH 865
I++ ++ F ++ E N EI A + KAPGPDG + FF+++
Sbjct: 237 ARAATIKEAIKPF---ISPEQNPE--------EIKSACFSIHADKAPGPDGFSASFFQSN 285
Query: 866 WATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYILKVISR 925
W T+ ++ I+ FFS+ L IN+T + L+PK+ S + +V +RPI+ C K+IS+
Sbjct: 286 WMTVGPNIVLEIQSFFSSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVFYKIISK 345
Query: 926 IMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSK 985
++ RL+ L +IS NQSAFV R DN+ ++ EA H L+ + + +MA+K +MSK
Sbjct: 346 LLSRRLQPILQEIISENQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMSK 405
Query: 986 TYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPL 1045
YDR+EW F++ + GFH W + +L + +Y +NG +TP RGLRQGDPL
Sbjct: 406 AYDRIEWDFIKLVMQEMGFHQTWISWILQCITTVSYSFLLNGSAQGAVTPERGLRQGDPL 465
Query: 1046 SPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSI 1105
SP+LF++ ++VLS + +AQ DG + G R + +P + HL FADD++ F + +K +
Sbjct: 466 SPFLFIICSEVLSGLCRKAQLDGSLLGLRVSKGNPRVNHLLFADDTIFFCRSDLKSCKTF 525
Query: 1106 IHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLGLPAIWGR 1165
+ IL Y ASGQ IN +KS + F R + +K + +L + + G+YLGLP ++GR
Sbjct: 526 LCILKKYEEASGQMINKSKSAITFSRKTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFGR 585
Query: 1166 NKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSLN 1225
K I +R++++ W L+ AGK ++K+++ ++P Y M+ + C +
Sbjct: 586 KKRDLFNQIVDRIRQRSLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQ 645
Query: 1226 SLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWRVLTNPEA 1285
S + +FWW + + W +W KM +K EGG+GF++ N A+LAK +WR++ +P
Sbjct: 646 SALTHFWWDSSADKKKMCWIAWSKMAKNKKEGGLGFKDITNFNDALLAKLSWRIVQSPSC 705
Query: 1286 LWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLLIEVWGDNW 1345
+ V +L Y TS FL+ S+ S WR + G+D I + +G GL VW + W
Sbjct: 706 VLVRILLGKYCRTSSFLDCSVTAASSHGWRGICTGKDLIKSQLGKVIGSGLDTLVWNEPW 765
Query: 1346 LS-------SGEALIQPHNAPDMKVSELISPHGEGWNSALVRSL 1382
LS G AL Q M V++LI + W+ V L
Sbjct: 766 LSLSTSSTPMGPALEQ---FKSMTVAQLICQTTKSWDREKVWDL 806
Score = 58.2 bits (139), Expect = 4e-08
Identities = 51/200 (25%), Positives = 88/200 (43%), Gaps = 21/200 (10%)
Query: 1556 ILWSIWRGRNKAVFNAIRPDPTFAIQQVASTLSAIDL-AKPVTEKVISDGNQNTNQRPTA 1614
ILW++W RNK +F +S++DL ++ +++ G Q +
Sbjct: 848 ILWNLWNCRNKLIFEQ-------------KHISSMDLISQSISQSTEWLGAQIQASKSKI 894
Query: 1615 WRPPFG------NLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAGSHAKVYS-SSPLTA 1667
P + ++ + DASW Q+ G + D + L + A + SPL A
Sbjct: 895 VIPGISPSEIDLDTIQCSTDASWREETLQAGFGWVFVDHSNHLESHHKAAAMNIRSPLLA 954
Query: 1668 EASAIREGLSLAHNLGCPEIIMESDSKILIDACKSETLVGEASAILQDIFYLKRLFYRCD 1727
+ASA+ + A +LG ++++ SDS+ L+ E E I+ DI L F
Sbjct: 955 KASALSLAIQHAADLGFKKLVVASDSQQLVKVLNGEPHPMELHGIVFDISVLSLNFEENS 1014
Query: 1728 FAWANRERNNSAHLVAKMCM 1747
F++ RE N+ A +AK +
Sbjct: 1015 FSFVKRENNSKADALAKAAL 1034
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 421 bits (1083), Expect = e-117
Identities = 288/932 (30%), Positives = 445/932 (46%), Gaps = 91/932 (9%)
Query: 577 KGSRFTWHSNPRDGFVTREKIDRVL----TNWSWRNIFPNASAMAYPQISSDHSPILLDL 632
KG+RFTW + +++DRVL W+ A+ P + D
Sbjct: 4 KGNRFTWRRGLVESTFVAKRLDRVLFCAHARLKWQE------ALLCPAQNVDAR------ 51
Query: 633 SPPTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIWSWQRK 692
+ PF++E W HE E++ W L L ++K W ++
Sbjct: 52 ------RRPFRFEAAWLSHEGFKELLTASWDTGLSTPVALNRLRWQLKK-------WNKE 98
Query: 693 TFKVAEKEIGRLKQELQSKQDRPNHLVDWNEV----KNIKLEIDTLWRQEELFWGQRSRV 748
F ++ +L++ QD +V +++ + E D L QEE W Q+SR
Sbjct: 99 VFGNIHVRKEKVVSDLKAVQDLLE-VVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSRE 157
Query: 749 KWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGV 808
K GD+N+ FFH ST+ RR RNRI+ + D++ W + + + +Y +++ E V
Sbjct: 158 KLLALGDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYSLEDV 217
Query: 809 NQIEDCL--RGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHW 866
+ + L GFPR +T E N L P + E+ AV +G KAPGPDG +F++ W
Sbjct: 218 SVVRGTLPTEGFPR-LTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCW 276
Query: 867 ATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYILKVISRI 926
T+ +SV + EFF + L N+ ++ L+ KV PE + QFRP+S CN + K+I+++
Sbjct: 277 ETVGESVSKFVMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKM 336
Query: 927 MVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKT 986
MV RLK+ ++ LI P Q++F+ GRL DNI V QEA H ++R + R K +M +K+D+ K
Sbjct: 337 MVIRLKNVISKLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRKKGR-KGWMLLKLDLEKA 395
Query: 987 YDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLS 1046
YDR+ W FL +TL A G W R++ V G L NG + + TP RGLRQGDP+S
Sbjct: 396 YDRIRWDFLAETLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLRQGDPIS 455
Query: 1047 PYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSII 1106
PYLFVL + L + A G+ + P ++H+ FADD +LFAEA+V
Sbjct: 456 PYLFVLCIERLCHQIETAVGRGDWKSISISQGGPKVSHVCFADDLILFAEASV------- 508
Query: 1107 HILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLL----HMAIWDSPGQYLGLPAI 1162
Q++++ KS + F SN V RD+ L+ + G+YLG+P +
Sbjct: 509 ----------AQKVSLEKSKIFF----SNNVSRDLEGLITAETGIGSTRELGKYLGMPVL 554
Query: 1163 WGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCN 1222
R + + ERV +L GWK L+ AG+ L KA++ +IP++ M+ + P +
Sbjct: 555 QKRINKDTFGEVLERVSSRLSGWKSRSLSLAGRITLTKAVLMSIPIHTMSSILLPASLLE 614
Query: 1223 SLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWRVLTN 1282
L+ + NF W + R H SW+K+ K GG+G R + N A+LAK WR+L +
Sbjct: 615 QLDKVSRNFLWGSTVEKRKQHLLSWKKVCRPKAAGGLGLRASKDMNRALLAKVGWRLLND 674
Query: 1283 PEALWVCVLKSVYFPTSDFLNASLGRNPSW--MWRSLLAGRDFISRYIRWAVGGGLLIEV 1340
+LW VL+ Y T ++ L +W WRS+ G +R V G ++
Sbjct: 675 KVSLWARVLRRKYKVTDVHDSSWLVPKATWSSTWRSIGVG-------LREGVAKGWILH- 726
Query: 1341 WGDNWLSSGEALIQPHNAPDMKVSELISPHGEGWNSALVRSLFQPELALKVLQTPVSK-- 1398
+ + L+ P + +V E + G GW+ + P L V K
Sbjct: 727 --EPLCTRATCLLSPEEL-NARVEEFWT-EGVGWDMVKLGQCL-PRSVTDRLHAVVIKGV 781
Query: 1399 LNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLWRTIWKSQLPQKV 1458
L D + W T GD+TV S + + S E ++ IW P++V
Sbjct: 782 LGLRDRISWQGTSDGDFTVGSAYVLLTQ--------EEESKPCMESFFKRIWGVIAPERV 833
Query: 1459 KVFLWKASHNSLAVKYNLWRKRILPSGQCPVC 1490
+VFLW + R+ I G VC
Sbjct: 834 RVFLWLVGQQVIMTNVERVRRHI---GDIEVC 862
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 398 bits (1023), Expect = e-110
Identities = 246/785 (31%), Positives = 399/785 (50%), Gaps = 56/785 (7%)
Query: 951 LIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTA 1010
+I DNI ++ E H L + ++ ++A K+D++K +D++EW F+E + GF W
Sbjct: 1 MITDNILIAHELIHSLHT-KKLVQPFVATKLDITKAFDKIEWGFIEAIMKQMGFSEKWCN 59
Query: 1011 RVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEI 1070
++T + TY + +NG + P RG+RQGDP+SPYL++L T+ LS ++ + + ++
Sbjct: 60 WIMTCITTTTYSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLSALIQASIKAKQL 119
Query: 1071 SGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFG 1130
GF+ + + P ++HL FA DSL+F +AT++E +++++L LY +ASGQ +N KS ++FG
Sbjct: 120 HGFKASRNGPAISHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASGQAVNFQKSAILFG 179
Query: 1131 RSASNRVKRDISNLLHMAIWDSPGQYLGLPAIWGRNKCHSLAWIEERVKEKLEGWKETLL 1190
+ R +S LL + + G+YLGLP GRNK ++ ++I + + +K++ W LL
Sbjct: 180 KGLDFRTSEQLSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNKLL 239
Query: 1191 NQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKM 1250
+ AGKEVLIK+I+ AIP Y+M+ P + + S + FWW I W +W K+
Sbjct: 240 SPAGKEVLIKSIVTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWSKL 299
Query: 1251 TLSKDEGGMGFREFRMQNLAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNP 1310
K GG+ R+ + N+A+LAKQ+WR+L P +L V K+ YFP L+A
Sbjct: 300 NDPKKMGGLAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKATSQS 359
Query: 1311 SWMWRSLLAGRDFISRYIRWAVGGGLLIEVWGDNWLSSGEALIQPHNAP---------DM 1361
S+ W+S+L G ISR +++ G G I++W DNWL + P P +
Sbjct: 360 SYAWKSILHGTKLISRGLKYIAGNGNNIQLWKDNWLP-----LNPPRPPVGTCDSIYSQL 414
Query: 1362 KVSELISPHGEG-WNSALVRSLFQPELALKVLQTPVSKLNQTDVLFWPHTPKGDYTVSSG 1420
KVS+L+ EG WN L+ L + S D + W +T G+Y+V SG
Sbjct: 415 KVSDLLI---EGRWNEDLLCKLIHQNDIPHIRAIRPSITGANDAITWIYTHDGNYSVKSG 471
Query: 1421 FEIARSNSIKPQA-LSSRSHNIPELLWRTIWKSQLPQKVKVFLWKASHNSLAVKYNLWRK 1479
+ + R S + A L S + + ++ IWK P K+K F W+++HN+L NL R+
Sbjct: 472 YHLLRKLSQQQHASLPSPNEVSAQTVFTNIWKQNAPPKIKHFWWRSAHNALPTAGNLKRR 531
Query: 1480 RILPSGQCPVCCEAQETVEHMFFVCPWTRAVWFGSSLQWVVSPQGL-GSFDVWLLQKITA 1538
R++ C C EA E V H+ F C ++ +W + ++ + SF+ Q + +
Sbjct: 532 RLITDDTCQRCGEASEDVNHLLFQCRVSKEIWEQAHIKLCPGDSLMSNSFN----QNLES 587
Query: 1539 LQTISLDFNKDFSTLACILWSIWRGRNKAVFNAIRPDPTFAIQQVASTLSAIDLAKPVTE 1598
+Q ++ KD S I W IW+ RN +FN R +IQ+ L
Sbjct: 588 IQKLNQSARKDVSLFPFIGWRIWKMRNDLIFNNKRWSIPDSIQKA--------LIDQQQW 639
Query: 1599 KVISDGNQNTNQRPTAW----RPPFGNLLKINVDASWVSSVPQSAVGVLIRDKFSTLLAG 1654
K + N+ ++P +G LK + L + +F +L G
Sbjct: 640 KESLNCNEQQQRKPQLHDSHNNKCYGGHLK--------------TLRTLSKAQF--VLRG 683
Query: 1655 SHAKVYSSSPLTAEASAIREGLSLAHNLGCPEIIMESDSKILIDACKSETLVGEASAI-- 1712
+ +S+PL AEA A++ + LG +II D L T E S I
Sbjct: 684 MSSIPPTSTPLEAEAEALKLAMIHLQRLGYEDIIFHGDVAALFQPITRTTHSCEYSPIAT 743
Query: 1713 -LQDI 1716
L+DI
Sbjct: 744 FLRDI 748
>At1g25430 hypothetical protein
Length = 1213
Score = 360 bits (923), Expect = 5e-99
Identities = 317/1228 (25%), Positives = 525/1228 (41%), Gaps = 92/1228 (7%)
Query: 403 ILSWNCRGVAATPTGRELRTLCSKNKPTIVFLMETRSKKENLEVIRRSLGFDYLFTVNPR 462
+ WN RG + NKP ++ET K+ +L + F N
Sbjct: 5 LFCWNIRGFNNVSHRSGFKKWVKANKPIFGGVIETHVKQPKDRKFINALLPGWSFVENYA 64
Query: 463 GLS-GGLALFWKREVHIQILHSSQNYIHTSIFANNSVSDWDCTFVYGDPSSQRRRFLWPQ 521
G + + W V + ++ S I + S S + VY R+ LW +
Sbjct: 65 FSDLGKIWVMWDPSVQVVVVAKSLQMITCEVLLPGSPSWIIVSVVYAANEVASRKELWIE 124
Query: 522 ITALRFRD---NTPWCLLGDFNEILFAHEKEGMRPQPPSV---LQRFREFVHNSGLMDVD 575
I + + PW +LGDFN++L + +E P +V ++ FR+ + + L D+
Sbjct: 125 IVNMVVSGIIGDRPWLVLGDFNQVL--NPQEHSNPVSLNVDINMRDFRDCLLAAELSDLR 182
Query: 576 LKGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLSPP 635
KG+ FTW + V + KIDR+L N SW +FP++ + SDH + L
Sbjct: 183 YKGNTFTWWNKSHTTPVAK-KIDRILVNDSWNALFPSSLGIFGSLDFSDHVSCGVVLEET 241
Query: 636 TW-SKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDCWKDLENRIKSSKRTIWSWQRKTF 694
+ +K PFK+ + ++ +V + W + ++K+ K+ I + R +
Sbjct: 242 SIKAKRPFKFFNYLLKNLDFLNLVRDNWFTLNVVGSSMFRVSKKLKALKKPIKDFSRLNY 301
Query: 695 KVAEKEIGRLKQELQSKQDRPNHLVDWNEVK-NIKLEIDTLWR----QEELFWGQRSRVK 749
EK L QDR L D + + +LE + W EE F+ Q+SR+
Sbjct: 302 SELEKRTKEAHDFLIGCQDRT--LADPTPINASFELEAERKWHILTAAEESFFRQKSRIS 359
Query: 750 WA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGVN 809
W GD N+K+FH R + N I + D G + Q ++ S++ + E
Sbjct: 360 WFAEGDGNTKYFHRMADARNSSNSISALYDGNGKLVDSQEGILDLCASYFGSLLGDEVDP 419
Query: 810 QIED--------CLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLF 861
+ + R P +V EL + S+ +I A+ L K+ GPDG F
Sbjct: 420 YLMEQNDMNLLLSYRCSPAQV-CELESTF----SNEDIRAALFSLPRNKSCGPDGFTAEF 474
Query: 862 FKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYILK 921
F + W+ + V AIKEFFS+ L + N T + L+PK+ +P FRPISC N + K
Sbjct: 475 FIDSWSIVGAEVTDAIKEFFSSGCLLKQWNATTIVLIPKIVNPTCTSDFRPISCLNTLYK 534
Query: 922 VISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKV 981
VI+R++ RL+ L+ +IS QSAF+ GR + +N+ ++ + H S + +KV
Sbjct: 535 VIARLLTDRLQRLLSGVISSAQSAFLPGRSLAENVLLATDLVH--GYNWSNISPRGMLKV 592
Query: 982 DMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQ 1041
D+ K +D + W F+ L A + + + T+ + +NG +GLRQ
Sbjct: 593 DLKKAFDSVRWEFVIAALRALAIPEKFINWISQCISTPTFTVSINGGNGGFFKSTKGLRQ 652
Query: 1042 GDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKE 1101
GDPLSPYLFVLA + S +L G I + + ++HL FADD ++F +
Sbjct: 653 GDPLSPYLFVLAMEAFSNLLHSRYESGLIH-YHPKASNLSISHLMFADDVMIFFDGGSFS 711
Query: 1102 ATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQYLGLPA 1161
I L+ + SG ++N KS L + N+++ + + I P +YLGLP
Sbjct: 712 LHGICETLDDFASWSGLKVNKDKSHLYL--AGLNQLESNANAAYGFPIGTLPIRYLGLPL 769
Query: 1162 IWGRNKCHSLAWIEERVKEKLEGWKETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFC 1221
+ + + + E++ + W L+ AG+ LI ++I + M+ PK
Sbjct: 770 MNRKLRIAEYEPLLEKITARFRSWVNKCLSFAGRIQLISSVIFGSINFWMSTFLLPKGCI 829
Query: 1222 NSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLAILAKQAWRVLT 1281
+ SL + F W G + SW + L K EGG+G R N + + WR+
Sbjct: 830 KRIESLCSRFLWSGNIEQAKGIKVSWAALCLPKSEGGLGLRRLLEWNKTLSMRLIWRLFV 889
Query: 1282 NPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDFISRYIRWAVGGGLLIEVW 1341
++LW + F G++ SW W+ LL+ R +++ VG GL + W
Sbjct: 890 AKDSLWADWQHLHHLSRGSFWAVEGGQSDSWTWKRLLSLRPLAHQFLVCKVGNGLKADYW 949
Query: 1342 GDNWLSSGEAL-----IQPHNAPDMKVSELISPHGE-GWNSALVRSLFQPELALKVLQTP 1395
DNW S G I P + ++++ S E GW + RS + + P
Sbjct: 950 YDNWTSLGPLFRIIGDIGPSSLRVPLLAKVASAFSEDGWRLPVSRSAPAKGIHDHLCTVP 1009
Query: 1396 VSKLNQTDVLFWPHTPKGDYTVSSGFEIARS-NSIKPQALSSRSHNIPELLW-RTIWKSQ 1453
V Q DV + + G + GF A++ +I+P+A W +IW
Sbjct: 1010 VPSTAQEDVDRYEWSVNG--FLCQGFSAAKTWEAIRPKATVKS--------WASSIWFKG 1059
Query: 1454 LPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVEHMFFVCPWTRAVW-- 1511
K +W + N L + L + S C +C A E+ +H+ +C ++ VW
Sbjct: 1060 AVPKYAFNMWVSHLNRLLTRQRLASWGHIQSDACVLCSFASESRDHLLLICEFSAQVWRL 1119
Query: 1512 --------------FGSSLQWV--VSPQGLGSFDVWLLQKITALQTISLDFNKDFSTLAC 1555
+ L WV SP+ LL+KI +
Sbjct: 1120 VFRRICPRQRLFSSWSELLSWVRQSSPEAPP-----LLRKIVS---------------QV 1159
Query: 1556 ILWSIWRGRNKAVFNAIRPDPTFAIQQV 1583
+++++WR RN + N++R P + V
Sbjct: 1160 VVYNLWRQRNNLLHNSLRLAPAVIFKLV 1187
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.334 0.144 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,834,341
Number of Sequences: 26719
Number of extensions: 1704560
Number of successful extensions: 6669
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 110
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 5983
Number of HSP's gapped (non-prelim): 266
length of query: 1771
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1658
effective length of database: 8,299,349
effective search space: 13760320642
effective search space used: 13760320642
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0288.7


