

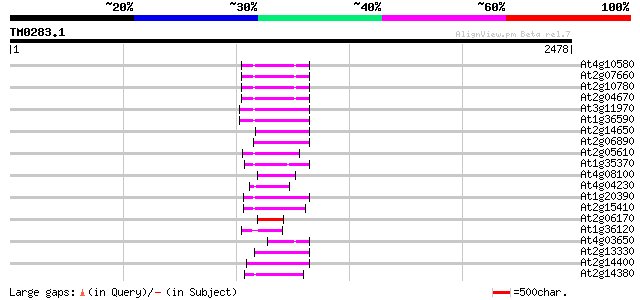
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0283.1
(2478 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g10580 putative reverse-transcriptase -like protein 170 1e-41
At2g07660 putative retroelement pol polyprotein 166 1e-40
At2g10780 pseudogene 165 4e-40
At2g04670 putative retroelement pol polyprotein 164 8e-40
At3g11970 hypothetical protein 150 7e-36
At1g36590 hypothetical protein 150 7e-36
At2g14650 putative retroelement pol polyprotein 144 6e-34
At2g06890 putative retroelement integrase 140 7e-33
At2g05610 putative retroelement pol polyprotein 130 1e-29
At1g35370 hypothetical protein 130 1e-29
At4g08100 putative polyprotein 120 8e-27
At4g04230 putative transposon protein 120 1e-26
At1g20390 hypothetical protein 116 2e-25
At2g15410 putative retroelement pol polyprotein 109 2e-23
At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein 107 1e-22
At1g36120 putative reverse transcriptase gb|AAD22339.1 105 3e-22
At4g03650 putative reverse transcriptase 105 4e-22
At2g13330 F14O4.9 103 1e-21
At2g14400 putative retroelement pol polyprotein 102 3e-21
At2g14380 putative retroelement pol polyprotein 101 6e-21
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 170 bits (430), Expect = 1e-41
Identities = 94/304 (30%), Positives = 161/304 (52%), Gaps = 7/304 (2%)
Query: 1022 PYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASE 1081
P+ E P P +M + ++++ + L KG IR S SPW +V +
Sbjct: 480 PFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFV----K 535
Query: 1082 LERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREED 1141
+ G+ RL I+Y+ LN+V RYPLP +L+ +L T FSK D+ SGY+QI + E D
Sbjct: 536 KKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEAD 595
Query: 1142 RYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSKGID 1200
KTAF +GH+E+ VMP GL NAP+ + +MN +F +++ F I+++DD+LV+SK +
Sbjct: 596 VRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPE 655
Query: 1201 QHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDE 1260
+ HL + +E +++ L K ++ + FLG+ + ++ +E + +P
Sbjct: 656 EQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWP-R 714
Query: 1261 LKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQS 1319
T+++ FLG Y F+ + P+ K K+ P WS+E +K+++ S
Sbjct: 715 PTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTS 774
Query: 1320 LPIV 1323
P++
Sbjct: 775 TPVL 778
Score = 32.3 bits (72), Expect = 3.6
Identities = 19/66 (28%), Positives = 34/66 (50%), Gaps = 6/66 (9%)
Query: 2410 QQPSVKSRIENILGNIQSSICSCKSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCW 2469
+ +K +++++LG + I S W +V K+ G+ RL I+Y+ LN+
Sbjct: 503 EMAELKKQLKDLLG--KGFIRPSTSPWGAPVLFVKKKD----GSFRLCIDYRELNRVTVK 556
Query: 2470 IRYPIP 2475
RYP+P
Sbjct: 557 NRYPLP 562
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 166 bits (421), Expect = 1e-40
Identities = 95/304 (31%), Positives = 161/304 (52%), Gaps = 7/304 (2%)
Query: 1022 PYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASE 1081
P+ E P P +M + ++++ E L KG IR S SPW +V +
Sbjct: 143 PFTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFV----K 198
Query: 1082 LERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREED 1141
+ G+ RL I+Y+ LNKV +YPLP +L+ +L FSK D+ SGY+QI + D
Sbjct: 199 KKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTD 258
Query: 1142 RYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSKGID 1200
KTAF +GH+E+ VMP GL NAP+ + +MN +F +++ F I+++DD+LV SK +
Sbjct: 259 VRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWE 318
Query: 1201 QHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDE 1260
H +HL +E ++++ L K ++ FLG+ I ++ + K++P
Sbjct: 319 AHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-R 377
Query: 1261 LKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQS 1319
+ T+++ FLG Y F+ + + PL + K++ WS+E S E+K ++ +
Sbjct: 378 PRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLIN 437
Query: 1320 LPIV 1323
P++
Sbjct: 438 APVL 441
>At2g10780 pseudogene
Length = 1611
Score = 165 bits (417), Expect = 4e-40
Identities = 93/304 (30%), Positives = 162/304 (52%), Gaps = 7/304 (2%)
Query: 1022 PYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASE 1081
P+ E P P +M + ++++ E L+KG IR S SPW +V +
Sbjct: 648 PFTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFV----K 703
Query: 1082 LERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREED 1141
+ G+ RL I+Y+ LNKV +YPLP +L+ +L FSK D+ SGY+QI + D
Sbjct: 704 KKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTD 763
Query: 1142 RYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSKGID 1200
KTAF + H+E+ VMP GL NAP+ + +MN +F +++ F I++++D+LV+SK +
Sbjct: 764 VRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWE 823
Query: 1201 QHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDE 1260
H +HL +E ++++ L K ++ FLG+ I ++ + K++P
Sbjct: 824 AHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-R 882
Query: 1261 LKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQS 1319
+ T+++ FLG Y F+ + + PL + K++ WS+E S E+K ++ +
Sbjct: 883 PRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTN 942
Query: 1320 LPIV 1323
P++
Sbjct: 943 APVL 946
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 164 bits (414), Expect = 8e-40
Identities = 92/304 (30%), Positives = 158/304 (51%), Gaps = 7/304 (2%)
Query: 1022 PYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASE 1081
P+ E P P +M + ++++ + L KG IR S SPW +V +
Sbjct: 506 PFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFV----K 561
Query: 1082 LERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREED 1141
+ G+ RL I+Y+ LN V +YPLP +L+ +L T FSK D+ SGY+QI + E D
Sbjct: 562 KKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEAD 621
Query: 1142 RYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSKGID 1200
KTAF +GH+E+ VMP L NAP+ + +MN +F +++ F I+++DD+LV+SK +
Sbjct: 622 VRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPE 681
Query: 1201 QHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDE 1260
+H HL + +E +++ L K ++ + FLG+ + ++ +E + +P
Sbjct: 682 EHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWP-R 740
Query: 1261 LKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQS 1319
T+++ FL Y F+ + P+ K K+ P WS E +K+++ S
Sbjct: 741 PTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTS 800
Query: 1320 LPIV 1323
P++
Sbjct: 801 TPVL 804
Score = 31.2 bits (69), Expect = 7.9
Identities = 19/66 (28%), Positives = 33/66 (49%), Gaps = 6/66 (9%)
Query: 2410 QQPSVKSRIENILGNIQSSICSCKSSWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCW 2469
+ +K ++E++LG + I S W +V K+ G+ RL I+Y+ LN
Sbjct: 529 EMTELKKQLEDLLG--KGFIRPSTSPWGAPVLFVKKKD----GSFRLCIDYRGLNWVTVK 582
Query: 2470 IRYPIP 2475
+YP+P
Sbjct: 583 NKYPLP 588
>At3g11970 hypothetical protein
Length = 1499
Score = 150 bits (380), Expect = 7e-36
Identities = 91/322 (28%), Positives = 161/322 (49%), Gaps = 15/322 (4%)
Query: 1013 YPKVH---TIALPYIDEFNEK------DIPTKARPIQMNEQYLKYCREEIGEYLNKGLIR 1063
YP + T P+ ++ N K P RP + + + + + L G ++
Sbjct: 572 YPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQ 631
Query: 1064 HSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIF 1123
S SP++ V + + G RL ++Y+ LN + +P+P DL+ L IF
Sbjct: 632 ASSSPYASPVVLV----KKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIF 687
Query: 1124 SKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYM- 1182
SK D+++GY+Q+ + +D KTAF GH+E+ VMP GL NAP+ +Q +MN IF P++
Sbjct: 688 SKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLR 747
Query: 1183 NFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQG 1242
F +V+ DD+LV+S +++H QHL++ E+++ N L K K +LG+ I
Sbjct: 748 KFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQ 807
Query: 1243 QITPIQRSLEFAKKFPDELKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSPPW 1302
I ++ K++P K QL+ FLG Y F+ + ++ PL + ++ W
Sbjct: 808 GIETDPAKIKAVKEWPQPTTLK-QLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEW 866
Query: 1303 SEEMSHSVREVKKLVQSLPIVT 1324
+ + ++K + P+++
Sbjct: 867 TAVAQQAFEDLKAALCQAPVLS 888
>At1g36590 hypothetical protein
Length = 1499
Score = 150 bits (380), Expect = 7e-36
Identities = 91/322 (28%), Positives = 161/322 (49%), Gaps = 15/322 (4%)
Query: 1013 YPKVH---TIALPYIDEFNEK------DIPTKARPIQMNEQYLKYCREEIGEYLNKGLIR 1063
YP + T P+ ++ N K P RP + + + + + L G ++
Sbjct: 572 YPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQ 631
Query: 1064 HSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIF 1123
S SP++ V + + G RL ++Y+ LN + +P+P DL+ L IF
Sbjct: 632 ASSSPYASPVVLV----KKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIF 687
Query: 1124 SKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYM- 1182
SK D+++GY+Q+ + +D KTAF GH+E+ VMP GL NAP+ +Q +MN IF P++
Sbjct: 688 SKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLR 747
Query: 1183 NFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQG 1242
F +V+ DD+LV+S +++H QHL++ E+++ N L K K +LG+ I
Sbjct: 748 KFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQ 807
Query: 1243 QITPIQRSLEFAKKFPDELKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSPPW 1302
I ++ K++P K QL+ FLG Y F+ + ++ PL + ++ W
Sbjct: 808 GIETDPAKIKAVKEWPQPTTLK-QLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEW 866
Query: 1303 SEEMSHSVREVKKLVQSLPIVT 1324
+ + ++K + P+++
Sbjct: 867 TAVAQQAFEDLKAALCQAPVLS 888
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 144 bits (363), Expect = 6e-34
Identities = 77/241 (31%), Positives = 132/241 (53%), Gaps = 3/241 (1%)
Query: 1085 GALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYK 1144
G+ RL I+Y+ LN V +YPLP +L+ +L T FSK D+ SGY+ I + E D K
Sbjct: 514 GSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRK 573
Query: 1145 TAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSKGIDQHV 1203
TAF +GH+E+ VMP GL NAP+ + +MN +F ++ F I+++DD+LV+SK +++H
Sbjct: 574 TAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHE 633
Query: 1204 QHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKE 1263
HL + +E +++ L K ++ + FLG+ + ++ +E + +
Sbjct: 634 VHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTP-TN 692
Query: 1264 KTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLPI 1322
T+++ FLG Y F+ + P+ K K+ P WS E +K+++ S P+
Sbjct: 693 ATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPV 752
Query: 1323 V 1323
+
Sbjct: 753 L 753
>At2g06890 putative retroelement integrase
Length = 1215
Score = 140 bits (354), Expect = 7e-33
Identities = 74/252 (29%), Positives = 135/252 (53%), Gaps = 6/252 (2%)
Query: 1077 LNASELER---GALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYY 1133
+ EL+R G+ R+ + + +N V +P+P D++ LH +IFSK D+KSGY+
Sbjct: 456 VETKELQRQKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYH 515
Query: 1134 QISVREEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDV 1192
QI + E D +KTAF G YEW VMP GL +APS + +MN + ++ F IVY DD+
Sbjct: 516 QIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDI 575
Query: 1193 LVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLE 1252
LV+S+ + +H++HL+ + +++K L + KK FLG+ + + + ++
Sbjct: 576 LVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKVDEEKVK 635
Query: 1253 FAKKFPDELKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKN-SPPWSEEMSHSVR 1311
+ +P K +++ F G + F + + APL + ++K+ W + + +
Sbjct: 636 AIRDWPSP-KTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQEEAFQ 694
Query: 1312 EVKKLVQSLPIV 1323
+K + + P++
Sbjct: 695 SLKDKLTNAPVL 706
Score = 35.0 bits (79), Expect = 0.55
Identities = 32/125 (25%), Positives = 58/125 (45%), Gaps = 16/125 (12%)
Query: 600 DMQISRKEVGSFCEQYGVEPLQAPS-SKARRINRSKTKPQGYKKRKPFQKKPYEQNPSTQ 658
+ Q+ RK S YG + P+ + R + KP + + KPY + Q
Sbjct: 90 EQQVKRKS--SSRSSYGSGTIAKPTYQREERTSSYHNKPIVSPRA---ESKPYA---AVQ 141
Query: 659 QPTNKKNVNPKK-KNV-CWKCGKPGHYANKCKTQQKI-----NELDLDQKLKDSLISVLI 711
K ++ + ++V C+KC GHYAN+C ++ + E++ ++++ DS S+
Sbjct: 142 DHKGKAEISTSRVRDVRCYKCQGKGHYANECPNKRVMILLDNGEIEPEEEIPDSPSSLKE 201
Query: 712 NSEDP 716
N E P
Sbjct: 202 NEELP 206
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 130 bits (327), Expect = 1e-29
Identities = 78/256 (30%), Positives = 131/256 (50%), Gaps = 6/256 (2%)
Query: 1026 EFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERG 1085
E E P RP + + + + L G I+ S SP++ V + + G
Sbjct: 36 ELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLASGTIQASSSPYASPVVLV----KKKDG 91
Query: 1086 ALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKT 1145
RL ++Y+ LN + R+P+P DL+ L ++SK D+++GY+Q+ + D +KT
Sbjct: 92 TWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVYSKIDLRAGYHQVRMDPLDIHKT 151
Query: 1146 AFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYM-NFTIVYLDDVLVFSKGIDQHVQ 1204
AF GHYE+ VMP GL NAP+ +Q++MN F P++ F +V+ DD+L++S +++H +
Sbjct: 152 AFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFLRKFVLVFFDDILIYSTSMEEHKK 211
Query: 1205 HLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEK 1264
HLE E+++ + L K + +LG+ I I ++ + +P + K
Sbjct: 212 HLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISGEGIATDPAKIKAVQDWPVPVNLK 271
Query: 1265 TQLQRFLGCVNYVADF 1280
QL FLG Y F
Sbjct: 272 -QLCGFLGLTGYYRRF 286
>At1g35370 hypothetical protein
Length = 1447
Score = 130 bits (327), Expect = 1e-29
Identities = 83/294 (28%), Positives = 151/294 (51%), Gaps = 19/294 (6%)
Query: 1036 ARPI-QMNEQYLKYCREEIG----EYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLV 1090
A P+ Q +Y+ + ++EI + + G I+ S SP++ V + + G RL
Sbjct: 578 ANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLV----KKKDGTWRLC 633
Query: 1091 INYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVP 1150
++Y LN + R+ +P DL+ L +FSK D+++GY+Q+ + +D KTAF
Sbjct: 634 VDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTH 693
Query: 1151 FGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYM-NFTIVYLDDVLVFSKGIDQHVQHLEKF 1209
GH+E+ VM GL NAP+ +Q++MN +F ++ F +V+ DD+L++S I++H +HL
Sbjct: 694 NGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLV 753
Query: 1210 IEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQR 1269
E+++ + L K LG+ I +I ++ K++P K Q++
Sbjct: 754 FEVMRLHKLFAKGSK--------EHLGHFISAREIETDPAKIQAVKEWPTPTTVK-QVRG 804
Query: 1270 FLGCVNYVADFIPNIRIVCAPLFKRLRKNSPPWSEEMSHSVREVKKLVQSLPIV 1323
FLG Y F+ N ++ PL + + WS E + +K ++ + P++
Sbjct: 805 FLGFAGYYRRFVRNFGVIAGPLHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVL 858
>At4g08100 putative polyprotein
Length = 1054
Score = 120 bits (302), Expect = 8e-27
Identities = 58/167 (34%), Positives = 97/167 (57%), Gaps = 1/167 (0%)
Query: 1094 KPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGH 1153
+ +N + R+P+P D + +LH +IFSK D+KSGY+Q ++E D +KTA
Sbjct: 529 RAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQRL 588
Query: 1154 YEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSKGIDQHVQHLEKFIEI 1212
YEW VMP GL NAP+ + +MN + ++ F IVY DD+LV+SK ++ HV HL+ +++
Sbjct: 589 YEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVLDL 648
Query: 1213 IKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPD 1259
++K L + KK FLG+ + I + ++ +++P+
Sbjct: 649 LRKEKLYANLKKCTFCTDNLVFLGFVVSADGIKVDEEKVKAIREWPN 695
>At4g04230 putative transposon protein
Length = 315
Score = 120 bits (301), Expect = 1e-26
Identities = 64/179 (35%), Positives = 99/179 (54%), Gaps = 5/179 (2%)
Query: 1057 LNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKR 1116
++KG IR S SP + V G R+ ++ + +N + R+P+P D++
Sbjct: 125 IHKGYIRESLSPCAVPVLLVPKKD----GTWRMCLDCRAINNITIKYRHPIPRLYDMLDE 180
Query: 1117 LHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMND 1176
L IFSK D++SGY+Q+ +RE D +KTAF G YE VMP GL NAPS + +MN
Sbjct: 181 LSGAIIFSKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQ 240
Query: 1177 IFYPYM-NFTIVYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRF 1234
+ ++ F +VY DD+L+++K H+Q LE ++ ++K GL + KK K F
Sbjct: 241 VLRSFIGKFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTFCSDKFFF 299
>At1g20390 hypothetical protein
Length = 1791
Score = 116 bits (290), Expect = 2e-25
Identities = 80/294 (27%), Positives = 139/294 (47%), Gaps = 8/294 (2%)
Query: 1033 PTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSP-WSCIGFYVLNASELERGALRLVI 1091
P K + ++ + + EE+ + L G I K P W V + + G R+ +
Sbjct: 777 PVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVV----KKKNGKWRVCV 832
Query: 1092 NYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPF 1151
+Y LNK YPLP+ L++ + S D SGY QI + ++D+ KT+FV
Sbjct: 833 DYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDR 892
Query: 1152 GHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQHVQHLEKFI 1210
G Y + VM GLKNA + YQ +N + + T+ VY+DD+LV S + HV+HL K
Sbjct: 893 GTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCF 952
Query: 1211 EIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRF 1270
+++ G+ ++ K T FLGY + + I + + + P + ++QR
Sbjct: 953 DVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSP-RNAREVQRL 1011
Query: 1271 LGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLPIV 1323
G + + FI C P + L++ + W ++ + ++K + + PI+
Sbjct: 1012 TGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPIL 1065
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 109 bits (273), Expect = 2e-23
Identities = 76/276 (27%), Positives = 131/276 (46%), Gaps = 8/276 (2%)
Query: 1033 PTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSP-WSCIGFYVLNASELERGALRLVI 1091
P K + ++ K EE+ + L+ G I + P W V + + G R+ I
Sbjct: 782 PLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVV----KKKNGKWRVCI 837
Query: 1092 NYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPF 1151
++ LNK +PLP+ L++ + S D SGY QI + + DR KT F+
Sbjct: 838 DFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQ 897
Query: 1152 GHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQHVQHLEKFI 1210
G Y + VMP GLKNA + Y ++N +F ++ ++ VY+DD+LV S ++H+ HL +
Sbjct: 898 GTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCF 957
Query: 1211 EIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRF 1270
+++ + + ++ K T FLGY + + I + + P + ++QR
Sbjct: 958 QVLNRYNMKLNPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSP-RNTREVQRL 1016
Query: 1271 LGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEE 1305
+G + + FI C P ++ LR N W E+
Sbjct: 1017 IGRIAALNRFISRSTDKCLPFYQLLRANKRFEWDEK 1052
>At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 587
Score = 107 bits (266), Expect = 1e-22
Identities = 48/116 (41%), Positives = 74/116 (63%), Gaps = 1/116 (0%)
Query: 1094 KPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGH 1153
K +NK+ R+P+P D++ L +FSK D++SGY+QI +R D +KT F G
Sbjct: 401 KAINKITIKYRFPIPQLDDMLDELSGSKVFSKIDLRSGYHQIRIRPGDEWKTDFKSKDGL 460
Query: 1154 YEWNVMPQGLKNAPSEYQNIMNDIFYPYM-NFTIVYLDDVLVFSKGIDQHVQHLEK 1208
YEW VMP G+ NAPS + +MN I P+ +F +VY DD+L++SK +++++H K
Sbjct: 461 YEWQVMPFGMSNAPSTFMRLMNQILRPFTGSFVVVYFDDILIYSKTKEEYLEHCLK 516
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 105 bits (262), Expect = 3e-22
Identities = 59/182 (32%), Positives = 91/182 (49%), Gaps = 23/182 (12%)
Query: 1022 PYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASE 1081
P+ E P P +M + ++++ + L KG IR S S W G
Sbjct: 524 PFTIELELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGAPG-------- 575
Query: 1082 LERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREED 1141
LN+V +YPLP +L+ +L T FSK D+ GY+Q + E D
Sbjct: 576 --------------LNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEAD 621
Query: 1142 RYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSKGID 1200
KTAF +GH+E+ VMP GL NAP+ +MN +F +++ F I+++DD+LV+ K +
Sbjct: 622 VRKTAFRTRYGHFEFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVYFKSPE 681
Query: 1201 QH 1202
+H
Sbjct: 682 EH 683
>At4g03650 putative reverse transcriptase
Length = 839
Score = 105 bits (261), Expect = 4e-22
Identities = 56/187 (29%), Positives = 100/187 (52%), Gaps = 3/187 (1%)
Query: 1139 EEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMN-FTIVYLDDVLVFSK 1197
E D KTAF +GH+E+ VMP GL NAP+ + +MN +F +++ F I+++DD+LV+SK
Sbjct: 518 EADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSK 577
Query: 1198 GIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKF 1257
++H HL + +E +++ L K ++ + FLG+ + ++ +E + +
Sbjct: 578 SPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW 637
Query: 1258 PDELKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKL 1316
P T+++ FLG Y FI + P+ K K+ P WS E +K++
Sbjct: 638 P-RPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEM 696
Query: 1317 VQSLPIV 1323
+ S P++
Sbjct: 697 LTSTPVL 703
>At2g13330 F14O4.9
Length = 889
Score = 103 bits (257), Expect = 1e-21
Identities = 73/244 (29%), Positives = 124/244 (49%), Gaps = 5/244 (2%)
Query: 1083 ERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDR 1142
+ G R+ I+++ LNK +PLP+ L++ + S D GY QI +R + +
Sbjct: 290 KNGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQ 349
Query: 1143 YKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQ 1201
KTAF+ G Y + VMP GLKNA + YQ ++N +F + TI VY+DD+LV S
Sbjct: 350 EKTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKD 409
Query: 1202 HVQHLEKFIEIIKKNGLVVSAKKMKIFETKTR-FLGYEIYQGQITPIQRSLEFAKKFPDE 1260
HV L + +I+ K + ++ +K FE ++R FL Y + + I + + + P
Sbjct: 410 HVPQLRECFKILIKFEMKLNPEKCS-FEVQSREFLEYLVTERGIEANPKQIAAFIEMPSP 468
Query: 1261 LKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQS 1319
K ++QR G + + FI C P ++ LRK W+++ + +++K +
Sbjct: 469 -KMAREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTE 527
Query: 1320 LPIV 1323
PI+
Sbjct: 528 PPIL 531
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 102 bits (254), Expect = 3e-21
Identities = 76/279 (27%), Positives = 130/279 (46%), Gaps = 6/279 (2%)
Query: 1047 KYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVINYKPLNKVLKWIRYP 1106
K +E+ + L G IR + P V+ + G R+ I++ LNK +P
Sbjct: 478 KAVNDEVDKLLKIGSIREVQYPDWLANTVVVKK---KNGKDRVCIDFTDLNKACPKDSFP 534
Query: 1107 LPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMPQGLKNA 1166
LP+ L++ + S D SGY QI + ED+ KT F+ G Y + VMP GL+NA
Sbjct: 535 LPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNA 594
Query: 1167 PSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKM 1225
+ Y ++N +F ++ T+ VY+DD+L+ S + HV+HLE+ I+ + + ++ K
Sbjct: 595 GATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKC 654
Query: 1226 KIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLGCVNYVADFIPNIR 1285
FLGY + + I + P K ++QR G + + FI
Sbjct: 655 TFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSP-KNFKEVQRLTGRIAALNRFISRST 713
Query: 1286 IVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQSLPIV 1323
P ++ L+ N W E+ + ++K + S P++
Sbjct: 714 DKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVL 752
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 101 bits (251), Expect = 6e-21
Identities = 72/264 (27%), Positives = 124/264 (46%), Gaps = 7/264 (2%)
Query: 1035 KARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSP-WSCIGFYVLNASELERGALRLVINY 1093
K + ++ + K +E+ + L+ G I K P W V ++ R+ I++
Sbjct: 499 KQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWLANPVVVKKKND----KWRVCIDF 554
Query: 1094 KPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFGH 1153
LNK +PLP+ +++ + S D SGY QI + ++D+ KT+F++ G
Sbjct: 555 TDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQIPMHKDDQEKTSFIIDRGT 614
Query: 1154 YEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQHVQHLEKFIEI 1212
Y + VMP GLKN + YQ ++N +F P + T+ VY+DD+LV S H+ HL+ E
Sbjct: 615 YCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDMLVKSTRSADHIDHLKACFET 674
Query: 1213 IKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQLQRFLG 1272
+ K + ++ K T FLGY + + I + + + K ++QR G
Sbjct: 675 LNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRAILDLQSP-RNKKEVQRLTG 733
Query: 1273 CVNYVADFIPNIRIVCAPLFKRLR 1296
+ + FI P ++ LR
Sbjct: 734 RIAGLNRFIARSTDKSLPFYQLLR 757
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 54,706,165
Number of Sequences: 26719
Number of extensions: 2443309
Number of successful extensions: 7487
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 7261
Number of HSP's gapped (non-prelim): 223
length of query: 2478
length of database: 11,318,596
effective HSP length: 116
effective length of query: 2362
effective length of database: 8,219,192
effective search space: 19413731504
effective search space used: 19413731504
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0283.1


