

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281a.3
(1430 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
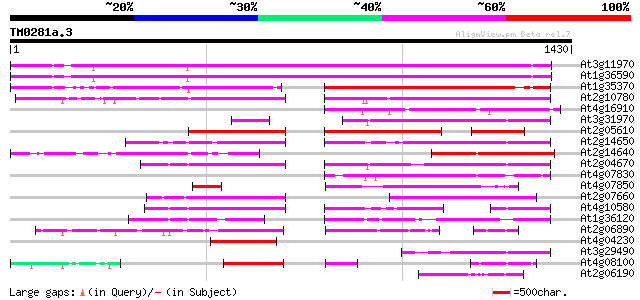
Score E
Sequences producing significant alignments: (bits) Value
At3g11970 hypothetical protein 921 0.0
At1g36590 hypothetical protein 916 0.0
At1g35370 hypothetical protein 466 e-131
At2g10780 pseudogene 311 2e-84
At4g16910 retrotransposon like protein 303 6e-82
At3g31970 hypothetical protein 299 9e-81
At2g05610 putative retroelement pol polyprotein 298 1e-80
At2g14650 putative retroelement pol polyprotein 295 1e-79
At2g14640 putative retroelement pol polyprotein 289 7e-78
At2g04670 putative retroelement pol polyprotein 286 5e-77
At4g07830 putative reverse transcriptase 274 2e-73
At4g07850 putative polyprotein 268 2e-71
At2g07660 putative retroelement pol polyprotein 258 2e-68
At4g10580 putative reverse-transcriptase -like protein 255 1e-67
At1g36120 putative reverse transcriptase gb|AAD22339.1 234 3e-61
At2g06890 putative retroelement integrase 211 2e-54
At4g04230 putative transposon protein 194 3e-49
At3g29490 hypothetical protein 170 5e-42
At4g08100 putative polyprotein 167 3e-41
At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein 161 2e-39
>At3g11970 hypothetical protein
Length = 1499
Score = 921 bits (2380), Expect = 0.0
Identities = 521/1409 (36%), Positives = 785/1409 (54%), Gaps = 52/1409 (3%)
Query: 2 KLEIPIFSGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWERCNPSPT 61
K++ P F G W K+E +F + ED +++ + + A +W Q + +
Sbjct: 111 KIDFPRFDGTRLKEWLFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLE 170
Query: 62 ----WEAFKLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALKEIDHDFVR 117
W+ + + RF+ +P L L++ + +Y ++FE + + +++
Sbjct: 171 VLYDWKGYVKLLKERFEDDC-DDPMAELKHLQETDGIIDYHQKFELIKTRVN-LSEEYLV 228
Query: 118 GIFLNGLKEEIKAEVRLIQKSLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVVTVEPQN 177
++L GL+ + + VR+ Q T F Y K +P N
Sbjct: 229 SVYLAGLRTDTQMHVRMFQPQ-----------------TVRHCLFLGKTYEKAHPKKPAN 271
Query: 178 NTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFK-------HLTGAQMREKRDKNLCFR 230
T S+ G + G FK ++ +M ++R K LC+
Sbjct: 272 TTWSTNRSAPTGGYNKYQKEGESKTDHYGNKGNFKPVSQQPKKMSQQEMSDRRSKGLCYF 331
Query: 231 CDEPYSREHKCRNKQLRMIIMEDDDDADCGEFEDALSGEFNSLQLSLCSMAGLTSTKSWK 290
CDE Y+ EH +K+ ++ M+ D++ + E + + Q+S+ +++G+ K+ +
Sbjct: 332 CDEKYTPEHYLVHKKTQLFRMDVDEEFEDAREELVNDDDEHMPQISVNAVSGIAGYKTMR 391
Query: 291 MVGQIGNQSIVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKCSQL 350
+ G + I +LID G++HNF+ +L V V V DG K+R +GK +
Sbjct: 392 VKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKVTDF 451
Query: 351 GIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLGDIKANFGKMELTLKRGKEVYTLKGDP 410
+Q + L L G+D+VLG++WL +LG I F K+E+ K + L G
Sbjct: 452 SWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLT 511
Query: 411 SLTKSELSYGALMQILLEEGEGLLVHC--DTGEKVNKKECEI---------PEIITGVLL 459
S + E+ L Q L E+ L + C + E + C I ++ VL
Sbjct: 512 SGSVREVKAQKL-QKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLN 570
Query: 460 QFDTVFQDPQGLPP-RRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVI 518
++ +F +P LPP R +H+H I L EG+ N RPY+ +QKNE++KLV ++L G +
Sbjct: 571 EYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTV 630
Query: 519 RPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKL 578
+ S SP++SP++LVKKKDG WRLCVDYR LN +T+ + FPIP+IE+L+DELGGA +FSK+
Sbjct: 631 QASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKI 690
Query: 579 DLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFA 638
DL++GYHQ+RM DI+KTAF+TH GH+E+LVMPFGLTNAP+TFQ LMN + KP+LRKF
Sbjct: 691 DLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFV 750
Query: 639 LVFFDDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEGGG 698
LVFFDDILVYS L H HL+QV ++ + L A KC+F +EYLGH +S +G
Sbjct: 751 LVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIE 810
Query: 699 C*PIKGGSYEGLANP*RYQGNEGVPRFDWLL*TLCQGLWENCKTLD*PSEERWVQLDRIS 758
P K + + P + G + L ++ + ++
Sbjct: 811 TDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAVA 870
Query: 759 NCSF*AFKNSHG*LASVSCARFYKTICG*D*CLQQRAWSCLDARGQTLAFWSQGLSPRAQ 818
+F K + +S F K Q + L G LA+ S+ L +
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQL 930
Query: 819 HKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQRLFS*DQFKWAVKLVGL 878
H S+YE+EL+A++ A++KWRHYL+ HFI++TDQ+SLK+L +QRL + Q +W KL+
Sbjct: 931 HLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEF 990
Query: 879 DFEIQYKPGSENTVADALSR----KMMYSALSILTSAEKEDWNSELQQDSKLVKVIQDLL 934
D+EIQY+ G EN VADALSR ++++ A++++ +D + DS+L +I L
Sbjct: 991 DYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLLKDIQAGYANDSQLQDIITALQ 1050
Query: 935 ISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTGGHSGHFRTYKRIASFL 994
P S F + + +L K+++V+ N I+ H + GGHSG T++R+
Sbjct: 1051 RDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKGLF 1110
Query: 995 YWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGK 1054
YW+GM DI+ ++ C CQ+ K A GLLQPLPIP +W ++SMDF+ GLP GK
Sbjct: 1111 YWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVSGGK 1170
Query: 1055 DTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWK 1114
++VVVDRL+K AHF AL HPYSA VA ++ + KLHG PT+IVSDRD +F S FW+
Sbjct: 1171 TVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPTSIVSDRDVVFTSEFWR 1230
Query: 1115 ELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTN 1174
E F + G LKL+++YHPQ+DGQTEV NRCLETYLRC +P+ W WL AE+W+NTN
Sbjct: 1231 EFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYNTN 1290
Query: 1175 YNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMK 1234
Y+SS+ M+PF+ +YG+ PP+ L S+V V + Q+R+ +L LK +L++AQH+MK
Sbjct: 1291 YHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRMK 1350
Query: 1235 VQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLE 1294
AD+HR EF GD+VY+KLQPY+ +S+ R KL+P+++GPYKI+DR G VAYKL
Sbjct: 1351 QFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLA 1410
Query: 1295 LPAHSKIHPVFHVSLLKQALKPTQQPQPLPPMLNEELELEVVPEDVIDHR---EDAQGNL 1351
LP++S++HPVFHVS LK + LP ++ + E VPE V++ + +
Sbjct: 1411 LPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVMQD--VFEKVPEKVVERKMVNRQGKAVT 1468
Query: 1352 EVLIKWEQLPTCDNTWELAAVIQDKFPLF 1380
+VL+KW P + TWE +Q FP F
Sbjct: 1469 KVLVKWSNEPLEEATWEFLFDLQKTFPEF 1497
>At1g36590 hypothetical protein
Length = 1499
Score = 916 bits (2367), Expect = 0.0
Identities = 520/1409 (36%), Positives = 784/1409 (54%), Gaps = 52/1409 (3%)
Query: 2 KLEIPIFSGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWERCNPSPT 61
K++ P F G W K+E +F + ED +++ + + A +W Q + +
Sbjct: 111 KIDFPRFDGTRLKEWLFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLE 170
Query: 62 ----WEAFKLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALKEIDHDFVR 117
W+ + + RF+ +P L L++ + +Y ++FE + + +++
Sbjct: 171 VLYDWKGYVKLLKERFEDDC-DDPMAELKHLQETDGIIDYHQKFELIKTRVN-LSEEYLV 228
Query: 118 GIFLNGLKEEIKAEVRLIQKSLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVVTVEPQN 177
++L GL+ + + VR+ Q T F Y K +P N
Sbjct: 229 SVYLAGLRTDTQMHVRMFQPQ-----------------TVRHCLFLGKTYEKAHLKKPAN 271
Query: 178 NTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFK-------HLTGAQMREKRDKNLCFR 230
T S+ G + G FK ++ +M ++R K LC+
Sbjct: 272 TTWSTNRSAPTGGYNKYQKEGESKTDHYGNKGNFKPVSQQPKKMSQQEMSDRRSKGLCYF 331
Query: 231 CDEPYSREHKCRNKQLRMIIMEDDDDADCGEFEDALSGEFNSLQLSLCSMAGLTSTKSWK 290
CDE Y+ EH +K+ ++ M+ D++ + E + + Q+S+ +++G+ K+ +
Sbjct: 332 CDEKYTPEHYLVHKKTQLFRMDVDEEFEDAREELVNDDDEHMPQISVNAVSGIAGYKTMR 391
Query: 291 MVGQIGNQSIVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKCSQL 350
+ G + I +LID G++HNF+ +L V V V DG K+R +GK +
Sbjct: 392 VKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKVTDF 451
Query: 351 GIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLGDIKANFGKMELTLKRGKEVYTLKGDP 410
+Q + L L G+D+VLG++WL +LG I F K+E+ K + L G
Sbjct: 452 SWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLT 511
Query: 411 SLTKSELSYGALMQILLEEGEGLLVHC--DTGEKVNKKECEI---------PEIITGVLL 459
S + E+ L Q L E+ L + C + E + C I ++ VL
Sbjct: 512 SGSVREVKAQKL-QKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLN 570
Query: 460 QFDTVFQDPQGLPP-RRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVI 518
++ +F +P LPP R +H+H I L EG+ N RPY+ +QKNE++KLV ++L G +
Sbjct: 571 EYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTV 630
Query: 519 RPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKL 578
+ S SP++SP++LVKKKDG WRLCVDYR LN +T+ + FPIP+IE+L+DELGGA +FSK+
Sbjct: 631 QASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKI 690
Query: 579 DLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFA 638
DL++GYHQ+RM DI+KTAF+TH GH+E+LVMPFGLTNAP+TFQ LMN + KP+LRKF
Sbjct: 691 DLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFV 750
Query: 639 LVFFDDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEGGG 698
LVFFDDILVYS L H HL+QV ++ + L A KC+F +EYLGH +S +G
Sbjct: 751 LVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIE 810
Query: 699 C*PIKGGSYEGLANP*RYQGNEGVPRFDWLL*TLCQGLWENCKTLD*PSEERWVQLDRIS 758
P K + + P + G + L ++ + ++
Sbjct: 811 TDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAVA 870
Query: 759 NCSF*AFKNSHG*LASVSCARFYKTICG*D*CLQQRAWSCLDARGQTLAFWSQGLSPRAQ 818
+F K + +S F K Q + L G LA+ S+ L +
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQL 930
Query: 819 HKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQRLFS*DQFKWAVKLVGL 878
H S+YE+EL+A++ A++KWRHYL+ HFI++TDQ+SLK+L +QRL + Q +W KL+
Sbjct: 931 HLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEF 990
Query: 879 DFEIQYKPGSENTVADALSR----KMMYSALSILTSAEKEDWNSELQQDSKLVKVIQDLL 934
D+EIQY+ G EN VADALSR ++++ A++++ +D + DS+L +I L
Sbjct: 991 DYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLLKDIQAGYANDSQLQDIITALQ 1050
Query: 935 ISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTGGHSGHFRTYKRIASFL 994
P S F + + +L K+++V+ N I+ H + GGHSG T++R+
Sbjct: 1051 RDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKGLF 1110
Query: 995 YWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGK 1054
Y +GM DI+ ++ C CQ+ K A GLLQPLPIP +W ++SMDF+ GLP GK
Sbjct: 1111 YSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVSGGK 1170
Query: 1055 DTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWK 1114
++VVVDRL+K AHF AL HPYSA VA ++ + KLHG PT+IVSDRD +F S FW+
Sbjct: 1171 TVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKLHGCPTSIVSDRDVVFTSEFWR 1230
Query: 1115 ELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTN 1174
E F + G LKL+++YHPQ+DGQTEV NRCLETYLRC +P+ W WL AE+W+NTN
Sbjct: 1231 EFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYNTN 1290
Query: 1175 YNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMK 1234
Y+SS+ M+PF+ +YG+ PP+ L S+V V + Q+R+ +L LK +L++AQH+MK
Sbjct: 1291 YHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRMK 1350
Query: 1235 VQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLE 1294
AD+HR EF GD+VY+KLQPY+ +S+ R KL+P+++GPYKI+DR G VAYKL
Sbjct: 1351 QFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLA 1410
Query: 1295 LPAHSKIHPVFHVSLLKQALKPTQQPQPLPPMLNEELELEVVPEDVIDHR---EDAQGNL 1351
LP++S++HPVFHVS LK + LP ++ + E VPE V++ + +
Sbjct: 1411 LPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVMQD--VFEKVPEKVVERKMVNRQGKAVT 1468
Query: 1352 EVLIKWEQLPTCDNTWELAAVIQDKFPLF 1380
+VL+KW P + TWE +Q FP F
Sbjct: 1469 KVLVKWSNEPLEEATWEFLFDLQKTFPEF 1497
>At1g35370 hypothetical protein
Length = 1447
Score = 466 bits (1200), Expect = e-131
Identities = 244/583 (41%), Positives = 356/583 (60%), Gaps = 32/583 (5%)
Query: 802 RGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQ 861
+G LA+ S+ L + H S+YE+EL+A + A++KWRHYL+ HFI++TDQ+SLK+L +Q
Sbjct: 885 KGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQ 944
Query: 862 RLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSR----KMMYSALSILTSAEKEDWN 917
RL + Q +W KL+ D+EIQY+ G EN VADALSR ++++ ALSI+ ++
Sbjct: 945 RLNTPVQQQWLPKLLEFDYEIQYRQGKENLVADALSRVEGSEVLHMALSIVECDFLKEIQ 1004
Query: 918 SELQQDSKLVKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPT 977
+ D L +I L P + + + + +L K+++V+ ++ H +
Sbjct: 1005 VAYESDGVLKDIISALQQHPDAKKHYSWSQDILRRKSKIVVPNDVEITNKLLQWLHCSGM 1064
Query: 978 GGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVW 1037
GG SG +++R+ S YW+GM DI+ F+ C CQ+ K A GLLQPLPIP ++W
Sbjct: 1065 GGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIW 1124
Query: 1038 MDISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFP 1097
D+SMDF+ GLP GK ++VVVDRL+K AHF AL HPYSA VA F+ + K HG P
Sbjct: 1125 CDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHGCP 1184
Query: 1098 TTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKP 1157
T+IVSDRD LF S+FWKE FK+ G +L++S++YHPQ+DGQTEV NRCLE YLRC +P
Sbjct: 1185 TSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHARP 1244
Query: 1158 KQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDG 1217
W WLP AE+W+NTNY+SS+ M+PF+ +YG+ PP+ L S+V V + Q+R+
Sbjct: 1245 HLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQEREN 1304
Query: 1218 VLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFY 1277
+L LK +L++AQH+MK AD+HR F+ GD+VY+KLQPY+ +S+ R KL+P+++
Sbjct: 1305 MLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKYF 1364
Query: 1278 GPYKILDRVGPVAYKLELPAHSKIHPVFHVSLLKQALKPTQQPQPLPPMLNEELELEVVP 1337
GPYKI+++ G V ++ TQ P LP + E P
Sbjct: 1365 GPYKIIEKCGEV-------------------MVGNVTTSTQLPSVLPDI------FEKAP 1399
Query: 1338 EDVIDH---REDAQGNLEVLIKWEQLPTCDNTWELAAVIQDKF 1377
E +++ + + VL+KW P + TW+ Q KF
Sbjct: 1400 EYILERKLVKRQGRAATMVLVKWIGEPVEEATWKFLFDRQQKF 1442
Score = 363 bits (932), Expect = e-100
Identities = 232/717 (32%), Positives = 376/717 (52%), Gaps = 81/717 (11%)
Query: 2 KLEIPIFSGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWERCNPSP- 60
K++ P F G W K+E +F + E+ +++ + + A +W + +
Sbjct: 116 KIDFPRFDGSRINEWLFKVEEFFGVDFTPEEMKVKMVAIHFDSHAATWHHSFIQSGIGLD 175
Query: 61 ---TWEAFKLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALKEIDHDFVR 117
W + + RF+ + +P L L++ + EY ++FE L + +++
Sbjct: 176 VFFNWPEYVKLLKDRFEDAC-DDPMAELKKLQETDGIVEYHQQFELIKVRLN-LSEEYLV 233
Query: 118 GIFLNGLKEEIKAEVRLIQKSLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVVTVEPQN 177
++L GL+ + + VR+ + + + R Y +
Sbjct: 234 SVYLAGLRTDTQMHVRMFEPKTVRD------------------CLRLGKYYE-------- 267
Query: 178 NTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFKHLTGAQMREKRDK-NLCFRCDEPYS 236
K+ SS S GG ++ + ++ +K D LC+ CDE ++
Sbjct: 268 RAHPKKTVSSTWS-----------QKGTRSGGSYRPVK--EVEQKSDHLGLCYFCDEKFT 314
Query: 237 REHKCRNKQLRMIIMEDDDDADCGEFEDA---LSGEFNSL----QLSLCSMAGLTSTKSW 289
EH +K+ ++ M+ D+ EFEDA LS + + Q+S+ +++G++ K+
Sbjct: 315 PEHYLVHKKTQLFRMDVDE-----EFEDAVEVLSDDDHEQKPMPQISVNAVSGISGYKTM 369
Query: 290 KMVGQIGNQSIVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKCSQ 349
+ G + + + +LID G++HNFI + +L V V V DG K+ G+
Sbjct: 370 GVKGTVDKRDLFILIDSGSTHNFIDSTVAAKLGCHVESAGLTKVAVADGRKLNVDGQIKG 429
Query: 350 LGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLGDIKANFGKMELTLKRGKEVYTLKGD 409
+Q+ + L L G+D+VLG++WL +LG I F K+E+ + L G
Sbjct: 430 FTWKLQSTTFQSDILLIPLQGVDMVLGVQWLETLGRISWEFKKLEMQFFYKNQRVWLHGI 489
Query: 410 PSLTKSELSYGALMQILLEEGEGLLVHCDTGEKVNKKECEIPEI------------ITGV 457
+ + ++ L + ++ + +V C E V+ +E EI I + +
Sbjct: 490 ITGSVRDIKAHKLQKTQADQIQLAMV-C-VREVVSDEEQEIGSISALTSDVVEESVVQNI 547
Query: 458 LLQFDTVFQDPQGLPP-RRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAG 516
+ +F VF +P LPP R +HDH I L EGA N RPY+ +QK+E++K+V +M+++G
Sbjct: 548 VEEFPDVFAEPTDLPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSG 607
Query: 517 VIRPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFS 576
I+ S SPF+SP++LVKKKDG WRLCVDY LN +T+ ++F IP+IE+L+DELGG+ VFS
Sbjct: 608 TIQVSSSPFASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFS 667
Query: 577 KLDLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRK 636
K+DL++GYHQ+RM DI+KTAF+TH GH+E+LVM FGLTNAP+TFQSLMN V + +LRK
Sbjct: 668 KIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRK 727
Query: 637 FALVFFDDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVS 693
F LVFFDDIL+YS + H HL+ V ++ H L F + S E+LGH +S
Sbjct: 728 FVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHKL--------FAKGSKEHLGHFIS 776
>At2g10780 pseudogene
Length = 1611
Score = 311 bits (796), Expect = 2e-84
Identities = 214/614 (34%), Positives = 312/614 (49%), Gaps = 44/614 (7%)
Query: 802 RGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQ 861
+G +A+ S+ L ++ ++ E+ A+V ++ WR YL G + TD KSLK++ Q
Sbjct: 973 KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQ 1032
Query: 862 RLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKM--------------MYSALSI 907
+ Q +W + + +I Y PG N VADALSR+ M L +
Sbjct: 1033 PELNLRQRRWMELVADYNLDIAYHPGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHV 1092
Query: 908 -----------LTSAEKEDWNSEL----QQDSKLVKVIQDLLIS-PTSHPGFEFKKGLLY 951
L +A++ D S + ++D ++ Q+ TS+ G G +
Sbjct: 1093 NALSKEVEPLGLGAADQADLLSRIRLAQERDEEIKGWAQNNKTEYQTSNNGTIVVNGRVC 1152
Query: 952 YKNRLVLSKASNRIPMIISECHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCE 1011
N L + I+ E H + H G + Y+ + + +W GMK D+ +V C
Sbjct: 1153 VPNDRALKEE------ILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCP 1206
Query: 1012 ICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPK-VRGK-DTVLVVVDRLTKFAH 1069
CQ K P GLLQ LPIP W I+MDFV GLP ++ K + V VVVDRLTK AH
Sbjct: 1207 TCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAH 1266
Query: 1070 FYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTS 1129
F A+ A+ +A +I E+V+LHG P +IVSDRD F S FWK K GT++ LST+
Sbjct: 1267 FMAISDKDGAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTA 1326
Query: 1130 YHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYG 1189
YHPQTD Q+E T + LE LR V W +L EF +N ++ +S GMSP++ALYG
Sbjct: 1327 YHPQTDEQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYG 1386
Query: 1190 RDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEG 1249
R L + R + + + LK L +AQ + K A+K R+ +EF G
Sbjct: 1387 RACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELEFQVG 1446
Query: 1250 DWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAH-SKIHPVFHVS 1308
D VYLK YK T KL+PR+ GPYK+++RVG VAYKL+LP + H VFHVS
Sbjct: 1447 DLVYLKAMTYKGAGRFTSR-KKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVFHVS 1505
Query: 1309 LLKQALKPTQQP-QPLPPMLNEELELEVVPEDVIDH-REDAQGNLEVLIK--WEQLPTCD 1364
L++ L ++ + +PP L E + +E P ++D + +G L+K W +
Sbjct: 1506 QLRKCLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMTKGTRGKARDLLKVLWNCRGREE 1565
Query: 1365 NTWELAAVIQDKFP 1378
TWE ++ FP
Sbjct: 1566 YTWETENKMKANFP 1579
Score = 263 bits (671), Expect = 7e-70
Identities = 213/719 (29%), Positives = 337/719 (46%), Gaps = 70/719 (9%)
Query: 16 WTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWE--RCNPSPTWEAFKLAVVRRF 73
W +L+R F ED + + LEG A +W+ E R + ++ F+ +++
Sbjct: 194 WRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGDEVRSFADFEDEFNKKY 253
Query: 74 QPSMIQNPFELL-LSLKQVG-TVDEYVEEFEK---YAGALKEIDHDFVRGIFLNGLKEEI 128
P + E L L Q TV EY EEF + Y G E + +R F+ GL+ EI
Sbjct: 254 FPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRYVGRELEEEQAQLRR-FIRGLRIEI 312
Query: 129 KAEV---------RLIQKSLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVVTVEPQNNT 179
+ L++++ +IE+ ++ R + RNN +K
Sbjct: 313 RNHCLVRTFNSVSELVERAAMIEEG----IEEERYLNREKAPIRNNQSTKPA-------- 360
Query: 180 DQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFKHLTGAQMREKRDKNLCFRCDEPYSREH 239
D+K+ V + ++ + +N G GA C RC S++H
Sbjct: 361 DKKRKFDKVDNTKSDAKTGECVTCGKNHSGTCWKAIGA----------CGRCG---SKDH 407
Query: 240 ------KCRNKQLRMIIMEDDDDADCGEFEDA------LSGEFNSLQLSLCSMAGLTSTK 287
K Q +++ E CG+ L+ E + Q GL
Sbjct: 408 AIQSCPKMEPGQSKVLGEETRTCFYCGKTGHLKRECPKLTAEKQAGQRDNRGGNGLPPPP 467
Query: 288 SWKMVGQIGNQSIVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKC 347
+ V + D G + I+ ++ N T V G + G
Sbjct: 468 KRQAVASRVYELSEEANDAG-NFRAITGGFRKEPN-----TDYGMVRAAGGQAMYPTGLV 521
Query: 348 SQLGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLGDIKANFG--KMELTLKRGKEVYT 405
+ +++ + + + + L DV+LG++WL G KA+ + + +R + +
Sbjct: 522 RGISVVVNGVNMPADLIIVPLKKHDVILGMDWL---GKYKAHIDCHRGRIQFERDEGMLK 578
Query: 406 LKGDPSLTKSELSYGALMQILLEEG-EGLLVHCDTGEKVNKKECEIPEIITGVLLQFDTV 464
+G + + S + + +L +G E L T E E + I+ +F V
Sbjct: 579 FQGIRTTSGSLVISAIQAERMLGKGCEAYLATITTKEVGASAELKDIPIVN----EFSDV 634
Query: 465 FQDPQGLPPRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISP 524
F G+PP R I L+ G + PY+ + +++K + E+L+ G IRPS SP
Sbjct: 635 FAAVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSP 694
Query: 525 FSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGY 584
+ +P++ VKKKDG +RLC+DYR LNKVT+ NK+P+P I+EL+D+LGGA+ FSK+DL SGY
Sbjct: 695 WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGY 754
Query: 585 HQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDD 644
HQI ++ D+ KTAFRT H+EF+VMPFGLTNAP+ F +MN V + +L +F ++F +D
Sbjct: 755 HQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIND 814
Query: 645 ILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEGGGC*PIK 703
ILVYS H HL+ VL L +H L A KCSF Q S+ +LGH++S +G P K
Sbjct: 815 ILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEK 873
>At4g16910 retrotransposon like protein
Length = 687
Score = 303 bits (775), Expect = 6e-82
Identities = 218/639 (34%), Positives = 311/639 (48%), Gaps = 59/639 (9%)
Query: 802 RGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQ 861
+G +A+ S+ L ++ + E+ A+V A++ WR YL G + TD KSLK++ Q
Sbjct: 65 KGSVIAYASRQLRKHEKNYPTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKYIFTQ 124
Query: 862 RLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSR---------------KMMYSALS 906
+ Q +W + D I Y PG N V DALSR MM +
Sbjct: 125 PELNLRQRRWIKLVADYDLNIAYHPGKANQVVDALSRLRTVVEAERNQVNLVNMMGTLHL 184
Query: 907 ILTSAEKEDWNSELQQDSKLVKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRI- 965
S E E + L+ I+ G+ Y + + R+
Sbjct: 185 NALSKEVEPLGLRAANQADLLSRIRSAQERDEEIKGWAQNNKTEYQSSNNGTIVVNGRVC 244
Query: 966 --------PMIISECHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSK 1017
I+ E H + H G + Y+ + + +W GMK D+ +V +K
Sbjct: 245 GPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV--------AK 296
Query: 1018 YSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPK-VRGK-DTVLVVVDRLTKFAHFYALGH 1075
P G+LQ LPIP W I MDFV GLP ++ K + V VVVDRLTK AHF A+
Sbjct: 297 EEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISD 356
Query: 1076 PYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTD 1135
+A+ +A +I E+V+LHG P +IVSDRD F S FWK K+ GT++ LST+YHPQTD
Sbjct: 357 KDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNLSTAYHPQTD 416
Query: 1136 GQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLL 1195
GQ+E T + LE LR V W +L EF +N ++ +S GMSP++ALYGR
Sbjct: 417 GQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGR----- 471
Query: 1196 LKAGAIPSRVEEVNQLFQQRDGVLGE-------LKQNLLKAQHQMKVQADKHRRLVEFNE 1248
AG P V + V+ E LK L +AQ + K A+K R+ +EF
Sbjct: 472 --AGRTPLCWTPVGERRLFGPAVVDETTKKMKFLKIKLKEAQDRQKSYANKRRKELEFQV 529
Query: 1249 GDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAH-SKIHPVFHV 1307
GD VYLK YK T KL PR+ GPYK+++RVG VAYKL+LP H VFHV
Sbjct: 530 GDLVYLKAMTYKGAGRFTSR-KKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNVFHV 588
Query: 1308 SLLKQALKPTQQP-QPLPPMLNEELELEVVPEDVIDH-REDAQGNLEVLIK--WEQLPTC 1363
S L++ L ++ + +PP L E + +E P ++D ++ +G L+K W
Sbjct: 589 SQLRKCLSEQEESMEDVPPGLKENMTVEAWPVRIMDQMKKGTRGKSMDLLKILWNCGGRE 648
Query: 1364 DNTWELAAVIQDKFPLFPLEDKVKLLGGVLIGLGLSQPL 1402
+ TWE ++ FP + K +G +G LS P+
Sbjct: 649 EYTWETETKMKANFP-----EWFKEMGKDQLGFDLSNPI 682
>At3g31970 hypothetical protein
Length = 1329
Score = 299 bits (765), Expect = 9e-81
Identities = 202/556 (36%), Positives = 290/556 (51%), Gaps = 30/556 (5%)
Query: 848 VRTDQKSLKFLTDQRLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSI 907
V TD KSLK++ Q + Q +W + D EI Y G N VADALSRK + ++
Sbjct: 768 VFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAYHSGKANVVADALSRKRVGGSVEA 827
Query: 908 LTSA----------------EKEDWNSELQQDSKLVKVIQDLLISPTSHPGFEFK---KG 948
L S E D ++L +L + + LI+ + G E++ G
Sbjct: 828 LVSEIGALRLCVMAQEPLGLEAVD-RADLLTRVRLAQEKDEGLIAASKADGSEYQFAANG 886
Query: 949 LLYYKNRLVLSKASNRIPMIISECHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVE 1008
+ R+ + K I+SE H + H + Y+ + + W GMK D+ N+V
Sbjct: 887 TILVHGRVCVPKDEELRREILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWVT 946
Query: 1009 DCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGKDTVLVVVDRLTKFA 1068
+C++CQ K PGGLLQ LPI W I+MDFV GLP R KD + V+VDRLTK A
Sbjct: 947 ECDVCQLVKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLPVSRTKDAIWVIVDRLTKSA 1006
Query: 1069 HFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLST 1128
HF A+ A +A ++ E+V+LHG P +IVSDRD F S FW+ GT++++ST
Sbjct: 1007 HFLAIRKTDGAVLLAKKYVSEIVELHGVPVSIVSDRDSKFTSAFWRAFQGEMGTKVQMST 1066
Query: 1129 SYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALY 1188
+YHPQTDGQ+E T + LE LR V + W D L EF +N +Y +S M+PF+ALY
Sbjct: 1067 AYHPQTDGQSERTIQTLEDMLRMCVLDRGGHWADHLSLVEFAYNNSYQASIRMAPFEALY 1126
Query: 1189 GRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNE 1248
GR L + R + + LK N+ +AQ + + ADK RR +EF
Sbjct: 1127 GRPCRTPLCWTQVGERSIYGADYVLETTERIRVLKLNMKEAQDRQRSYADKRRRELEFEV 1186
Query: 1249 GDWVYLKLQPYK--LKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELP-AHSKIHPVF 1305
GD VYLK+ + +SI+ KL PR+ GP++I++RVGPVAY+LELP H VF
Sbjct: 1187 GDRVYLKMAMLRGPNRSISE---TKLTPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVF 1243
Query: 1306 HVSLLKQAL-KPTQQPQPLPPMLNEELELEVVPEDVIDHR-EDAQGNLEVLIK--WEQLP 1361
HVS+L++ L K + + L + LE P +++ R ++ + LIK W
Sbjct: 1244 HVSMLRKCLHKDDEVLAKILEDLQPNMTLEARPVRILERRIKELRRKKIPLIKVLWNCDG 1303
Query: 1362 TCDNTWELAAVIQDKF 1377
+ TWE A ++ F
Sbjct: 1304 VTEETWEPEARMKASF 1319
Score = 84.3 bits (207), Expect = 4e-16
Identities = 40/96 (41%), Positives = 58/96 (59%)
Query: 566 LDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSL 625
+ E G S + + + I + +A++ KTAFRT GH+EF+VMPFGLTN P+ F L
Sbjct: 552 MPESVGQVAVSDIRVVHEFEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRL 611
Query: 626 MNEVLKPYLRKFALVFFDDILVYSPDLLTHTTHLQQ 661
MN V + +L +F ++F DDILVYS H ++
Sbjct: 612 MNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVQSEE 647
Score = 37.4 bits (85), Expect = 0.063
Identities = 38/149 (25%), Positives = 65/149 (43%), Gaps = 11/149 (7%)
Query: 9 SGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQ--WWERCNPSPTWEAFK 66
S ++A W ++E F + R+ + LEG A W++ R +W F
Sbjct: 163 SPEEADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAHMSWADFV 222
Query: 67 LAVVRRFQPSMIQNPFEL-LLSLKQ-VGTVDEYVEEFEK---YAGALKEIDHDFVRGIFL 121
++ P + E L L Q +V EY EF + YAG + D +R FL
Sbjct: 223 AEFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNRLLVYAGRGMQDDQAQMRR-FL 281
Query: 122 NGLKEEIKAEVRLIQ---KSLLIEQKNQI 147
GL+ +++ R++Q K+ L+E ++
Sbjct: 282 RGLRPDLRVRCRVLQYATKAALVETAAEV 310
Score = 34.7 bits (78), Expect = 0.41
Identities = 31/133 (23%), Positives = 57/133 (42%), Gaps = 5/133 (3%)
Query: 277 LCSMAGLTSTKSWKMVGQIGNQSIV---VLIDCGASHNFISHKLVEQLNLTVAE-TQTYT 332
L +A + + +G ++I VL D GASH FI+ + + N+ Q
Sbjct: 386 LAHIAAAPQGYTTREIGSTSKRAITEAHVLFDSGASHCFITPESASRGNIRGDPGEQLGA 445
Query: 333 VEVGDGYKIRCKGKCSQLGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLG-DIKANFG 391
V+V G + G+ + I I + + + + DV+LG++WL + + G
Sbjct: 446 VKVAGGQFLAVLGRAKGVDIQIAGESMPVDLIISPVELYDVILGMDWLDHYRVHLDCHRG 505
Query: 392 KMELTLKRGKEVY 404
++ G+ VY
Sbjct: 506 RVSFERPEGRLVY 518
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 298 bits (764), Expect = 1e-80
Identities = 140/248 (56%), Positives = 183/248 (73%), Gaps = 1/248 (0%)
Query: 457 VLLQFDTVFQDPQGLPP-RRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEA 515
V+ +F +F +P LPP R HDH I L E + N RPY+ +QKNE++K+V +ML +
Sbjct: 10 VVTEFPDIFVEPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLAS 69
Query: 516 GVIRPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVF 575
G I+ S SP++SP++LVKKKDG WRLCVDYR LN +T+ ++FPIP+IE+L+DELGG+ V+
Sbjct: 70 GTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVY 129
Query: 576 SKLDLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLR 635
SK+DL++GYHQ+RM DI KTAF+TH GHYE+LVMPFGLTNAP++FQSLMN KP+LR
Sbjct: 130 SKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFLR 189
Query: 636 KFALVFFDDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGE 695
KF LVFFDDIL+YS + H HL+ V ++ H L A KC+F +EYLGH +SGE
Sbjct: 190 KFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISGE 249
Query: 696 GGGC*PIK 703
G P K
Sbjct: 250 GIATDPAK 257
Score = 254 bits (649), Expect = 2e-67
Identities = 131/303 (43%), Positives = 189/303 (62%), Gaps = 4/303 (1%)
Query: 803 GQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQR 862
G LAF S+ L + H S+YE+EL+A++ ++KWRHYL+ HFI++TDQ+SLK+L +QR
Sbjct: 306 GHPLAFISRQLKGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQR 365
Query: 863 LFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSR----KMMYSALSILTSAEKEDWNS 918
L + Q +W KL+ D+EIQYK G EN VADALSR ++++ ALS++ ++ +
Sbjct: 366 LNTPIQQQWLPKLLEFDYEIQYKQGKENLVADALSRVEGSEVLHMALSVVECDLLKEIQA 425
Query: 919 ELQQDSKLVKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTG 978
D + +I L S + + +G+L KN++V+ S I+ H + G
Sbjct: 426 GYVTDGDIQGIITILQQQADSKKHYTWSQGVLRRKNKIVVPNNSGIKDTILRWLHCSGMG 485
Query: 979 GHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWM 1038
GHSG T++R+ YW+ M DI+ F+ C CQ+ K A GLLQPLPIP ++W
Sbjct: 486 GHSGKEVTHQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWS 545
Query: 1039 DISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPT 1098
D+SMDF+ GLP GK ++VVVDRL+K AHF AL HPYSA VA ++ + KLHG P+
Sbjct: 546 DVSMDFIDGLPLSNGKTVIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVFKLHGCPS 605
Query: 1099 TIV 1101
+IV
Sbjct: 606 SIV 608
Score = 136 bits (342), Expect = 1e-31
Identities = 65/135 (48%), Positives = 99/135 (73%)
Query: 1177 SSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQ 1236
S + M+P++A+YG+ PPL L S+V V + Q+R+ ++ LK +L++AQH+MK
Sbjct: 621 SHSTMTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQL 680
Query: 1237 ADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELP 1296
AD+H EF GD+V++KLQPY+ +S+ R KL+P+++GPYK++DR G VAYKL+LP
Sbjct: 681 ADQHITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLP 740
Query: 1297 AHSKIHPVFHVSLLK 1311
A+S++HPVFHVS L+
Sbjct: 741 ANSQVHPVFHVSQLR 755
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 295 bits (755), Expect = 1e-79
Identities = 210/586 (35%), Positives = 297/586 (49%), Gaps = 54/586 (9%)
Query: 802 RGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQ 861
RG+ +A+ S+ L + ++ E+ A++ A++ WR YL G + V TD KSLK++ Q
Sbjct: 780 RGKVIAYASRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQ 839
Query: 862 RLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSILTSAEKEDWNSELQ 921
+ Q +W + D EI Y PG N VADALS K + +A Q
Sbjct: 840 PELNLRQRQWMELVADYDLEIAYHPGKANVVADALSHKRVGAAPG--------------Q 885
Query: 922 QDSKLVKVIQDLLISPTSHP--GFE-FKKGLLYYKNRLVLSKASNRIPMIISECHDTPTG 978
LV I L + + G E + L + RL K
Sbjct: 886 SVEALVSEIGALRLCAVAREPLGLEAVDRADLLTRVRLAQEK------------------ 927
Query: 979 GHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWM 1038
G YK S D+ N+V +C++CQ K PGG+LQ LPIP W
Sbjct: 928 -DEGLIAAYKAEGS--------EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWD 978
Query: 1039 DISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPT 1098
I++DFV GLP R KD + V+VDRLTK AHF A+ A +A ++ E+VKLHG P
Sbjct: 979 FITIDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPV 1038
Query: 1099 TIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPK 1158
+IVSDRD F S FW+ GT++++ST+YHPQT GQ+E T + LE LR V
Sbjct: 1039 SIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGG 1098
Query: 1159 QWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGV 1218
W D L EF +N +Y +S GM+PF+ALY R L + R Q+
Sbjct: 1099 HWADHLSLVEFAYNNSYPASIGMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTER 1158
Query: 1219 LGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYK--LKSIATRPCAKLAPRF 1276
+ LK N+ +AQ + + ADK RR +EF GD VYLK+ + +SI+ KL+PR+
Sbjct: 1159 IRVLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISE---TKLSPRY 1215
Query: 1277 YGPYKILDRVGPVAYKLELP-AHSKIHPVFHVSLLKQAL-KPTQQPQPLPPMLNEELELE 1334
GP++I++RVGPVAY+LELP H VFHVS+L++ L K + +P L + LE
Sbjct: 1216 MGPFRIVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLE 1275
Query: 1335 VVPEDVIDHR-EDAQGNLEVLIK--WEQLPTCDNTWELAAVIQDKF 1377
P V++ R ++ + LIK W+ TWE A ++ +F
Sbjct: 1276 ARPVRVLERRIKELRRKKIPLIKVLWDCDGVTKETWEPEARMKARF 1321
Score = 253 bits (646), Expect = 6e-67
Identities = 156/411 (37%), Positives = 221/411 (52%), Gaps = 40/411 (9%)
Query: 295 IGNQSIVVLIDCGASHNFISHKLVEQLNLTVAE-TQTYTVEVGDGYKIRCKGKCSQLGIM 353
+G VL D GASH FI+ + + N+ Q V+V G + G+ + I
Sbjct: 308 VGGVEAHVLFDSGASHCFITPESASRGNIRGDPGEQLGAVKVAGGQFVAVLGRTKGVDIQ 367
Query: 354 IQALEIVQNFYLFGLNGMDVVLGIEWLASLGDIKANFGKMELTLKRGKEVYTLKGDPSLT 413
I + + + + DV+LG++WL + ++ L RG+ +
Sbjct: 368 IAGESMPADLIISPVELYDVILGMDWL--------DHYRVHLDCHRGRVSF--------- 410
Query: 414 KSELSYGALMQILLEEGEG-LLVHCDTGEKVNKKECEIPEIITGVLLQFDTVFQDPQGLP 472
E G+L+ + G L++ EK+ +K CE L+F+ VFQ QGLP
Sbjct: 411 --ERPEGSLVYQGVRPTSGSLVISAVQAEKMIEKGCE-------AYLEFEDVFQSLQGLP 461
Query: 473 PRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISPFSSPIILV 532
P R I L+ G + PY+ + E++K + ++L A P++ V
Sbjct: 462 PSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGA------------PVLFV 509
Query: 533 KKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDA 592
KKKDG +RLC+DYR LN VT+ NK+P+P I+ELLD+L GA FSK+DL SGYH I + +A
Sbjct: 510 KKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEA 569
Query: 593 DIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPDL 652
D+ KTAFRT GH+EF+VMPFGLTNAP+ F LMN V + L +F ++F DDILVYS L
Sbjct: 570 DVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSL 629
Query: 653 LTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEGGGC*PIK 703
H HL++V+ L + L A KCSF Q + +LGHIVS EG P K
Sbjct: 630 EEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEK 680
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 289 bits (740), Expect = 7e-78
Identities = 151/319 (47%), Positives = 211/319 (65%), Gaps = 7/319 (2%)
Query: 1076 PYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTD 1135
P ++ VA F+ +VKLHG P +IVSD DP+F+S FW+E +K++ T+L +ST+YHPQTD
Sbjct: 628 PLLSRSVAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTD 687
Query: 1136 GQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLL 1195
GQTEV NRC+E +LRCFV PKQW ++PWAE+W+NT +++S GM+PF+ALYGR PP
Sbjct: 688 GQTEVVNRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGR-PPSP 746
Query: 1196 LKAGAIPSRV-EEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYL 1254
+ A + S V E+N+ RD +L ELKQ+L+ A + MK QAD R V F GDWV L
Sbjct: 747 IPAYELGSVVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLL 806
Query: 1255 KLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAHSKIHPVFHVSLLKQAL 1314
++QPY+ K++ R KL+ RFYGP+++ + G VAY+L LP ++IHPVFHVSLLK +
Sbjct: 807 RIQPYRQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTLPEGTRIHPVFHVSLLKPWV 866
Query: 1315 KPTQQPQPLPPMLNEELELEVVPEDVIDHR---EDAQGNLEVLIKWEQLPTCDNTWELAA 1371
+ P L EL++ P V++ R +D + ++L++WE L D TWE
Sbjct: 867 GDGEPDMGQLPPLRNNGELKLQPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEEYD 926
Query: 1372 VIQDKFPLF--PLEDKVKL 1388
+ FP F LEDKV+L
Sbjct: 927 QLAASFPEFVLNLEDKVRL 945
Score = 235 bits (600), Expect = 1e-61
Identities = 177/639 (27%), Positives = 282/639 (43%), Gaps = 115/639 (17%)
Query: 2 KLEIPIFSGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWERCNPSPT 61
KL+ P F G D W K ++YF E ++ ++ T L+G A W+Q
Sbjct: 105 KLDFPRFHGGDLTKWLSKAKQYFEYHEGPVEQAVRFTTFHLDGVANGWWQ---------- 154
Query: 62 WEAFKLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALKEIDHDFVRGIFL 121
A +RR ++ Q G++ EY EFE + + + G F+
Sbjct: 155 ------ATLRRSA------------TIGQTGSLSEYQHEFEWLQNKVYGWTQEVLVGAFM 196
Query: 122 NGLKEEIKAEVRLIQKSLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVVTVEPQNNTDQ 181
NGL I +R+ Q L E N V KG R ++ ++ +
Sbjct: 197 NGLYYSISNGIRMFQPRSLREVINLRVGWKGKSKDRRRKRATHHLHAVL----------- 245
Query: 182 KQDSSSVGSVAMPSRNNAGIDSSRN---RGGEFKHLTGAQMREKRDKNLCFRCDEPYSRE 238
S +V A N DS R R G + +++ E D+
Sbjct: 246 ---SLTVWWCAKRQSNRRQRDSVRKNYERKGVWAYVSAVMKGENDDE------------- 289
Query: 239 HKCRNKQLRMIIMEDDDDADCGEFEDALSGEFNSLQLSLCSMAGLTSTKSWKMVGQIGNQ 298
+ E+ +A+ +++L ++ G+ +T + ++ I
Sbjct: 290 ----------AVEEESTEAE------------GKPEITLYALEGVDTTSTIRVRATIHRN 327
Query: 299 SIVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKCSQLGIMIQALE 358
++ LI+ G++HNFI K V +NL T+ +TV V +G + C+ + + +++ +
Sbjct: 328 RLIALINSGSTHNFIGEKAVRGMNLKATTTKPFTVRVVNGMPLVCRSRYEAIPVVMGGVV 387
Query: 359 IVQNFYLFGLNGMDVVLGIEWLASLGDIKANFGKMELTLK-RGKEVYTLKGDPSLTKSEL 417
Y L G+D+ +G++WL++LG N+ + L G EV + P+ +
Sbjct: 388 FPVTLYALPLMGLDLAMGVQWLSTLGPTLCNWKEQTLQFHWAGDEVRLMGIKPTGLRGVE 447
Query: 418 SYGALMQILLEEGEGLLVHCDTGEKVNKKECEIPEIITGVLLQFDTVFQDPQGLPPRRRH 477
+ + + N + EI ++I +F +F+ P LP R
Sbjct: 448 HKTITKKARMGHTIFAITMAHNSSDPNTPDGEIKKLIE----EFAGLFETPTQLPSERPI 503
Query: 478 DHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISPFSSPIILVKKKDG 537
H I LK+G N+RPY+ YQK+E+ + AG+IRPS SPF SP++L
Sbjct: 504 VHRIALKKGTDPVNVRPYRYAFYQKDEMSR-------AGIIRPSSSPFLSPVLL------ 550
Query: 538 GWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKT 597
+FPIP ++++LDEL GA F+KLDL +GY Q+RM DI K
Sbjct: 551 -----------------ERFPIPTVDDMLDELNGAVYFTKLDLTAGYQQVRMHSPDIPKI 593
Query: 598 AFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRK 636
AF+TH GHYE+LVMPFGL NAPSTFQ+LMNE+ P L +
Sbjct: 594 AFQTHNGHYEYLVMPFGLCNAPSTFQALMNEIFWPLLSR 632
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 286 bits (733), Expect = 5e-77
Identities = 199/606 (32%), Positives = 301/606 (48%), Gaps = 65/606 (10%)
Query: 802 RGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQ 861
RG+ +A+ S+ L + ++ E+ ++ A++ WR YL G V TD KSLK++ +Q
Sbjct: 831 RGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKYIFNQ 890
Query: 862 RLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSILTSA---------- 911
+ Q +W + D EI Y PG N VADALSRK + +A A
Sbjct: 891 PELNLRQMRWMELVADYDLEIAYHPGKANVVADALSRKRVGAAPGQSVEALVSEIGALRL 950
Query: 912 ----------EKEDWNSELQQDSKLVKVIQDLLISPTSHPGFEFK---KGLLYYKNRLVL 958
E D ++L ++L + + LI+ + G E++ G ++ R+ +
Sbjct: 951 CVVAREPLGLEAVD-RADLLTRARLAQEKDEGLIAASKAEGSEYQFAANGTIFVYGRVCV 1009
Query: 959 SKASNRIPMIISECHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKY 1018
K I+SE H + H G + Y+ + + W GMK D+ N+V +C++CQ K
Sbjct: 1010 PKDEELRREILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWVAECDVCQLVKA 1069
Query: 1019 STLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYS 1078
P D + V++DRLTK AHF A+
Sbjct: 1070 EHQVP-------------------------------DAIWVIMDRLTKSAHFLAIRKTDG 1098
Query: 1079 AKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQT 1138
A +A ++ E+VKLHG P +IVSDRD F FW+ GT++++ST+YHPQTDGQ+
Sbjct: 1099 AAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQS 1158
Query: 1139 EVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKA 1198
E T + LE LR V W D L EF +N +Y +S GM+PF+ALYGR L+
Sbjct: 1159 ERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPLRW 1218
Query: 1199 GAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQP 1258
+ R Q+ + LK N+ +AQ + + ADK RR +EF GD VYLK+
Sbjct: 1219 TQVEERSIYGADYVQETTERIRVLKLNMKEAQARQRSYADKRRRELEFEVGDRVYLKMAM 1278
Query: 1259 YK--LKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELP-AHSKIHPVFHVSLLKQAL- 1314
+ +SI KL+PR+ GP++I++RVGPVAY+LELP H VFHV +L++ L
Sbjct: 1279 LRGPNRSILE---TKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVLMLRKCLH 1335
Query: 1315 KPTQQPQPLPPMLNEELELEVVPEDVIDHR-EDAQGNLEVLIK--WEQLPTCDNTWELAA 1371
K + +P L + LE P V++ R ++ + LIK W+ + TWE A
Sbjct: 1336 KDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLWDCDGVTEETWEPEA 1395
Query: 1372 VIQDKF 1377
++ +F
Sbjct: 1396 RMKARF 1401
Score = 262 bits (670), Expect = 9e-70
Identities = 149/373 (39%), Positives = 217/373 (57%), Gaps = 8/373 (2%)
Query: 333 VEVGDGYKIRCKGKCSQLGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLG-DIKANFG 391
V+V G + G+ + I I + + + + DV+LG++WL + + G
Sbjct: 365 VKVAGGQFLAVLGRAKGVDIQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRG 424
Query: 392 KMELTLKRGKEVYTLKGDPSLTKSELSYGALMQILLEEG-EGLLVHCDTGEKVNKKECEI 450
++ G+ VY +G + S + + ++E+G E LV E + + +
Sbjct: 425 RVSFERPEGRLVY--QGVRPTSGSLVISAVQAKKMIEKGCEAYLVTISMPESLG--QVAV 480
Query: 451 PEIITGVLLQFDTVFQDPQGLPPRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVA 510
+I V+ +F+ VFQ QGLPP R I L+ G + PY+ + E++K +
Sbjct: 481 SDI--RVIQEFEDVFQSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLE 538
Query: 511 EMLEAGVIRPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELG 570
++L G IRPS SP+ +P++ VKKKDG +RLC+DYR LN VT+ NK+P+P I+ELLD+L
Sbjct: 539 DLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLR 598
Query: 571 GAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVL 630
GA FSK+DL SGYHQI + +AD+ KTAFRT GH+EF+VMPF LTNAP+ F LMN V
Sbjct: 599 GATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVF 658
Query: 631 KPYLRKFALVFFDDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGH 690
+ +L +F ++F DDILVYS H HL++V+ L + L A KCSF Q + +LGH
Sbjct: 659 QEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGH 718
Query: 691 IVSGEGGGC*PIK 703
IVS EG P K
Sbjct: 719 IVSAEGVSVDPEK 731
Score = 39.3 bits (90), Expect = 0.016
Identities = 52/214 (24%), Positives = 84/214 (38%), Gaps = 29/214 (13%)
Query: 9 SGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQ--WWERCNPSPTWEAFK 66
S ++A W +++R F + R+ + LEG A W++ R +W F
Sbjct: 143 SPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQADMSWADFV 202
Query: 67 LAVVRRFQPSMIQNPFEL-LLSLKQ-VGTVDEYVEEFEK---YAGALKEIDHDFVRGIFL 121
++ P + E L L Q +V EY EF + YAG E D +R FL
Sbjct: 203 AEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNRLLAYAGRGMEDDQAQMRR-FL 261
Query: 122 NGLKEEIKAEVRLIQ---KSLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVVTVEP--- 175
GL+ +++ + R+ Q K+ L+E ++ + +VV V P
Sbjct: 262 RGLRPDLRVQCRVSQYATKAALVETAAEV---------------EEDLQRQVVGVSPAVQ 306
Query: 176 QNNTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGG 209
NT Q+ S G A + S +GG
Sbjct: 307 TKNTQQQVTPSKGGKPAQGQKRKWDHPSRAGQGG 340
>At4g07830 putative reverse transcriptase
Length = 611
Score = 274 bits (701), Expect = 2e-73
Identities = 203/605 (33%), Positives = 293/605 (47%), Gaps = 71/605 (11%)
Query: 802 RGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQ 861
RG+ +A+ S+ L + ++ E+ A V TD KSLK++ Q
Sbjct: 3 RGKVIAYGSRQLKKHEGNYPTHDLEMAA------------------VFTDHKSLKYIFTQ 44
Query: 862 RLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSA---------LSI----L 908
+ +W + D EI Y PG N V DALSRK + +A + I L
Sbjct: 45 LELNLRLRRWMKLVADYDLEIAYHPGKANVVTDALSRKRVGAAPGQSVETLVIEIGALRL 104
Query: 909 TSAEKEDWNSELQQDSKLVKVIQ------DLLISPTSHPGFEFK---KGLLYYKNRLVLS 959
+ +E E + L+ +Q + LI+ + GFE++ G + R+ +
Sbjct: 105 CAVAREPLGLEAVDQTDLLSRVQLAQEKDEGLIAASKAEGFEYQFAANGTILVHGRVCVP 164
Query: 960 KASNRIPMIISECHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYS 1019
K I+SE H + H G + Y+ + + W GMK D+ N+VE+C++CQ K
Sbjct: 165 KDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEECDVCQLVKIE 224
Query: 1020 TLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSA 1079
G LLQ LPIP W I+MDFV GLP R KD + V+VDRLTK AHF A+ A
Sbjct: 225 HQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGA 284
Query: 1080 KDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTE 1139
+A ++ E+VKLHG P +IVSDRD F S FW+ GT++++ST+YHPQTDGQ+E
Sbjct: 285 AVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTDGQSE 344
Query: 1140 VTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAG 1199
T + LE LR V W D L EF +N +Y +S GM+PF+ LYGR P L
Sbjct: 345 RTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMAPFEVLYGR-PCRTLCWT 403
Query: 1200 AIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPY 1259
+ R Q+ + LK N+ +AQ++ + ADK R+ +EF GD V
Sbjct: 404 QVGERSIYGADYVQEITERIRVLKLNMKEAQNRQRSYADKRRKELEFEVGDSVSQDGHVA 463
Query: 1260 KLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLEL-PAHSKIHPVFHVSLLKQAL-KPT 1317
+ + RVGPVA++LEL H VFHVS+L++ L K
Sbjct: 464 RSE---------------------QRVGPVAFRLELSDVMRAFHKVFHVSMLRKCLHKDD 502
Query: 1318 QQPQPLPPMLNEELELEVVPEDVIDHR-EDAQGNLEVLIKWEQLPTCD----NTWELAAV 1372
+ +P L + LE P V++ R ++ + LIK L CD TWE A
Sbjct: 503 EVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIK--VLRNCDGVTEETWEPEAR 560
Query: 1373 IQDKF 1377
++ +F
Sbjct: 561 LKARF 565
>At4g07850 putative polyprotein
Length = 1138
Score = 268 bits (684), Expect = 2e-71
Identities = 175/491 (35%), Positives = 234/491 (47%), Gaps = 100/491 (20%)
Query: 806 LAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQRLFS 865
+A++S+ L + Y++EL ALV+A+Q W+HYL + F++ TD +SLK L Q+ +
Sbjct: 647 IAYFSEKLGGATLNYPTYDKELYALVRALQTWQHYLWPKEFVIHTDHESLKHLKGQQKLN 706
Query: 866 *DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSILTSAEKEDWNSELQQDSK 925
+W + + I+YK G +N VADALSR+
Sbjct: 707 KRHARWVEFIETFPYVIKYKKGKDNVVADALSRR-------------------------- 740
Query: 926 LVKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTGGHSGHFR 985
H GF L+Y NRL + +S R + I E H GH G +
Sbjct: 741 --------------HEGF------LFYDNRLCIPNSSLR-ELFIREAHGGGLMGHFGVSK 779
Query: 986 TYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFV 1045
T K + +W MK D++ E C C+++K + P GL PLPIP W DISMDFV
Sbjct: 780 TLKVMQDHFHWPHMKRDVERMCERCTTCKQAKAKS-QPHGLCTPLPIPLHPWNDISMDFV 838
Query: 1046 GGLPKVR-GKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDR 1104
GLP+ R GKD++ VVVDR +K AHF A +A LF +E+V+LHG P TIVSDR
Sbjct: 839 VGLPRTRTGKDSIFVVVDRFSKMAHFIPCHKTDDAMHIANLFFREVVRLHGMPKTIVSDR 898
Query: 1105 DPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWL 1164
D FLS FWK L+ GT+L ST+ HPQTDGQTEV NR L T LR + K W D L
Sbjct: 899 DTKFLSYFWKTLWSKLGTKLLFSTTCHPQTDGQTEVVNRTLSTLLRALIKKNLKTWEDCL 958
Query: 1165 PWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQ 1224
P EF +N + +S+ SPF+ +YG +P L +P
Sbjct: 959 PHVEFAYNHSVHSATKFSPFQIVYGFNPITPLDLMPLP---------------------- 996
Query: 1225 NLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILD 1284
H R F P K KS KL PR G +K+L
Sbjct: 997 -----------LISIHLRKERF-----------PNKRKS-------KLMPRMDGSFKVLK 1027
Query: 1285 RVGPVAYKLEL 1295
R+ AY L+L
Sbjct: 1028 RINDNAYSLDL 1038
Score = 68.9 bits (167), Expect = 2e-11
Identities = 33/75 (44%), Positives = 47/75 (62%)
Query: 466 QDPQGLPPRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISPF 525
++P+GLPP R +H I GA +PN Y+ + E+EK V E++E G I S+SP
Sbjct: 455 ENPEGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELEKQVNELMERGHICESMSPC 514
Query: 526 SSPIILVKKKDGGWR 540
+ P++LV KKDG WR
Sbjct: 515 AVPVLLVPKKDGSWR 529
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 258 bits (658), Expect = 2e-68
Identities = 154/379 (40%), Positives = 214/379 (55%), Gaps = 5/379 (1%)
Query: 968 IISECHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLL 1027
I+ E H + H G + Y+ + + +W GM+ D+ +V C CQ K P GLL
Sbjct: 555 ILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWVAKCPTCQLVKAEHQVPSGLL 614
Query: 1028 QPLPIPTQVWMDISMDFVGGLPK-VRGK-DTVLVVVDRLTKFAHFYALGHPYSAKDVAVL 1085
Q LPI W I+MDFV LP ++ K + V VVVDRLTK AHF A+ A+ +A
Sbjct: 615 QNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEK 674
Query: 1086 FIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCL 1145
+I E+++LHG P +IVSDRD F S FW K GT++ LST+YHPQTDGQ+E T + L
Sbjct: 675 YIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTL 734
Query: 1146 ETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRV 1205
E LR V W +L EF +N ++ +S GMSP++ALYGR L + R
Sbjct: 735 EDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERR 794
Query: 1206 EEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIA 1265
+ + + LK L +AQ + K A+K R+ +EF GD VYLK YK
Sbjct: 795 LFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRF 854
Query: 1266 TRPCAKLAPRFYGPYKILDRVGPVAYKLELPAHSKI-HPVFHVSLLKQALKPTQQP-QPL 1323
T KL+PR+ GPYK+++RVG VAYKL+LP + H VFHVS L++ L ++ + +
Sbjct: 855 TSR-KKLSPRYVGPYKVIERVGAVAYKLDLPPKLNVFHNVFHVSQLRKYLSDQEESVEDI 913
Query: 1324 PPMLNEELELEVVPEDVID 1342
PP L E + +E P ++D
Sbjct: 914 PPGLKENMTVEAWPVRIMD 932
Score = 254 bits (650), Expect = 2e-67
Identities = 137/355 (38%), Positives = 213/355 (59%), Gaps = 6/355 (1%)
Query: 350 LGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASL-GDIKANFGKMELTLKRGKEVYTLKG 408
+ +++ + + + + L DV+LG++WL G I + + + +R + + +G
Sbjct: 19 ISVVVNGVNMPADLIIVPLKKHDVILGMDWLGKYKGHIDCH--RERIQFERDEGMLKFQG 76
Query: 409 DPSLTKSELSYGALMQILLEEGEGLLVHCDTGEKVNKKECEIPEIITGVLLQFDTVFQDP 468
+ + S + + +L +G + T ++V E+ +I+ ++ +F VF
Sbjct: 77 IRTTSGSLVISAIQAERMLGKGCEAYLATITTKEVGAS-AELKDIL--IVNEFSDVFAAV 133
Query: 469 QGLPPRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISPFSSP 528
G+PP R I L+ G + PY+ + E++K + E+L G IRPS SP+ +P
Sbjct: 134 SGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAP 193
Query: 529 IILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIR 588
++ VKKKDG +RLC+DYR LNKVT+ NK+P+P I+EL+D+LGGA+ FSK+DL SGYHQI
Sbjct: 194 VLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIP 253
Query: 589 MKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVY 648
++ D+ KTAFRT GH+EF+VMPFGLTNAP+ F +MN V + +L +F ++F DDILV+
Sbjct: 254 IEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVH 313
Query: 649 SPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEGGGC*PIK 703
S H HL+ VL L +H L A K SF Q S+ +LGH++S +G P K
Sbjct: 314 SKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEK 368
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 255 bits (652), Expect = 1e-67
Identities = 145/361 (40%), Positives = 213/361 (58%), Gaps = 8/361 (2%)
Query: 345 GKCSQLGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLG-DIKANFGKMELTLKRGKEV 403
G+ + I I + + + + DV+LG++WL + + G++ G+ V
Sbjct: 351 GRAKGVDIQIAGESMPADLIISPVELYDVILGMDWLDYYRVHLDWHRGRVFFERPEGRLV 410
Query: 404 YTLKGDPSLTKSELSYGALMQILLEEG-EGLLVHCDTGEKVNKKECEIPEIITGVLLQFD 462
Y +G ++ S + + ++E+G E LV E V + + +I V+ +F
Sbjct: 411 Y--QGVRPISGSLVISAVQAEKMIEKGCEAYLVTISMPESVG--QVAVSDI--RVVQEFQ 464
Query: 463 TVFQDPQGLPPRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSI 522
VFQ QGLPP + I L+ G + PY+ + E++K + ++L G IRPS
Sbjct: 465 DVFQSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPST 524
Query: 523 SPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKS 582
SP+ +P++ VKKKDG +RLC+DYR LN+VT+ N++P+P I+ELLD+L GA FSK+DL S
Sbjct: 525 SPWGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTS 584
Query: 583 GYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFF 642
GYHQI + +AD+ KTAFRT GH+EF+VMPFGLTNAP+ F LMN V + +L +F ++F
Sbjct: 585 GYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFI 644
Query: 643 DDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEGGGC*PI 702
DDILVYS HL++V+ L + L A KCSF Q + +LGHIVS EG P
Sbjct: 645 DDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPE 704
Query: 703 K 703
K
Sbjct: 705 K 705
Score = 144 bits (362), Expect = 5e-34
Identities = 97/303 (32%), Positives = 144/303 (47%), Gaps = 28/303 (9%)
Query: 803 GQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQR 862
G+ +A+ S+ L + ++ E+ A++ A++ WR YL G V TD KSLK++ Q
Sbjct: 806 GKVIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQP 865
Query: 863 LFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSILTSAEKEDWNSELQQ 922
+ Q +W + D EI Y PG N V DALSRK + +AL Q
Sbjct: 866 ELNLRQRRWMELVADYDLEIAYHPGKANVVVDALSRKRVGAALG--------------QS 911
Query: 923 DSKLVKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTGGHSG 982
LV I L + + + + +A + +++ D
Sbjct: 912 VEVLVSEIGALRLCAVAREPLGLE----------AVDRADLLTRVRLAQKKDEGLRAT-- 959
Query: 983 HFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISM 1042
+ Y+ + + W GMK D+ N+V +C++CQ K GG+LQ LPIP W I+M
Sbjct: 960 --KMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITM 1017
Query: 1043 DFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVS 1102
D V GL R KD + V+VDRLTK AHF A+ A +A F+ E+VKLHG P +
Sbjct: 1018 DLVVGLRVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMKE 1077
Query: 1103 DRD 1105
+D
Sbjct: 1078 AQD 1080
Score = 89.7 bits (221), Expect = 1e-17
Identities = 60/160 (37%), Positives = 92/160 (57%), Gaps = 10/160 (6%)
Query: 1225 NLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYK--LKSIATRPCAKLAPRFYGPYKI 1282
N+ +AQ + + ADK RR +EF GD VYLK+ + +SI+ KL+PR+ GP+KI
Sbjct: 1074 NMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISE---TKLSPRYMGPFKI 1130
Query: 1283 LDRVGPVAYKLELP-AHSKIHPVFHVSLLKQAL-KPTQQPQPLPPMLNEELELEVVPEDV 1340
++RV PVAY+LELP H VFHVS+L++ L K + +P L + LE P V
Sbjct: 1131 VERVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAKIPEDLQPNMTLEARPVRV 1190
Query: 1341 IDHR--EDAQGNLEVL-IKWEQLPTCDNTWELAAVIQDKF 1377
++ R E Q + ++ + W+ + TWE A ++ +F
Sbjct: 1191 LERRIKELRQKKIPLIKVLWDCDGVTEETWEPEARMKARF 1230
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 234 bits (596), Expect = 3e-61
Identities = 180/581 (30%), Positives = 262/581 (44%), Gaps = 81/581 (13%)
Query: 802 RGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQ 861
RG+ + + S+ L + ++ E+ A++ A++ W YL G V TD KSLK + Q
Sbjct: 721 RGKVIVYASRQLRKHEGNYPTHDLEMAAIIFALKIWGSYLYGGKVQVFTDHKSLKDIFTQ 780
Query: 862 RLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSILTSAEKEDWNSELQ 921
+ Q +W + D EI Y PG N VADALSRK + +A E SE+
Sbjct: 781 PELNLRQRRWMELVADYDLEIAYHPGKTNVVADALSRKRVGAA----PGQSVEALVSEIG 836
Query: 922 QDSKLVKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTGGHS 981
V + L + G + R+ L K I+SE H + H
Sbjct: 837 ALRLCVVAREPLKLEAVDRAA----NGTILVHERVCLPKDEELRREILSEAHASMFSIHP 892
Query: 982 GHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDIS 1041
G + Y+ + W GMK D+ N+V +C++CQ K PGGLLQ LPI W
Sbjct: 893 GATKMYRDLKRHYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPISE--WK--K 948
Query: 1042 MDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIV 1101
D LPK ++ E+VKLHG P +I+
Sbjct: 949 TDGAAVLPKK----------------------------------YVSEIVKLHGVPVSIL 974
Query: 1102 SDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWL 1161
S RD F S FW+ GT++++ST+YHPQTDGQ+E T + LE L+ V W
Sbjct: 975 SHRDSKFTSAFWRAFQVEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLQMCVLDWGGHWA 1034
Query: 1162 DWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGE 1221
D L +F +N +Y +S GM+PF+ALYGR LL + + Q+ +
Sbjct: 1035 DHLSLVKFAYNNSYQASIGMAPFEALYGRPCRTLLCWTQVGEKSIYGADYVQETTERIRV 1094
Query: 1222 LKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYK 1281
LK N+ +AQ + + ADK RR +EF G +
Sbjct: 1095 LKLNMKEAQDRQRSYADKRRRELEFEVGT------------------------------E 1124
Query: 1282 ILDRVGPVAYKLELP-AHSKIHPVFHVSLLKQAL-KPTQQPQPLPPMLNEELELEVVPED 1339
I++RVGPVAY+LELP H VFHVS+L++ L K + +P L + LE P
Sbjct: 1125 IVERVGPVAYRLELPDVMRAFHNVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVR 1184
Query: 1340 VIDHR-EDAQGNLEVLIK--WEQLPTCDNTWELAAVIQDKF 1377
V++ R ++ + +IK W+ + TWE A I+ +F
Sbjct: 1185 VLERRIKEVRRKKIPMIKVLWDCDGVTEETWEPEARIKARF 1225
Score = 188 bits (478), Expect = 2e-47
Identities = 120/350 (34%), Positives = 188/350 (53%), Gaps = 27/350 (7%)
Query: 302 VLIDCGASHNFISHKLVEQLNLTVAETQTY-TVEVGDGYKIRCKGKCSQLGIMIQALEIV 360
VL D GASH FI+ + + N+ + + V++ G + G+ + I I+ +
Sbjct: 351 VLFDSGASHCFITPESASRDNIRGEPGEQFGAVKIAGGQFLTVLGRAKGVNIQIEEESMP 410
Query: 361 QNFYLFGLNGMDVVLGIEWLASLG-DIKANFGKMELTLKRGKEVYTLKGDPSLTKSELSY 419
+ + + D +LG++WL + + G++ G+ VY +G ++S +
Sbjct: 411 ADLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFERPEGRLVY--QGVRPTSRSLVIS 468
Query: 420 GALMQILLEEG-EGLLVHCDTGEKVNKKECEIPEIITGVLLQFDTVFQDPQGLPPRRRHD 478
++ ++E+G E LV E V + + +I V+ +F+ VFQ QGLPP R
Sbjct: 469 EVQVEKMIEKGCEAYLVTISMSESVG--QVAVSDI--WVVQEFEDVFQSLQGLPPSRSDP 524
Query: 479 HAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISPFSSPIILVKKKDGG 538
I L+ G + PY+ + E++K + ++L G IRPS S + +P
Sbjct: 525 FTIELELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGAP---------- 574
Query: 539 WRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTA 598
LN+VT+ NK+P+P I+ELLD+L GA FSK+DL GYHQ + +AD+ KTA
Sbjct: 575 --------GLNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEADVRKTA 626
Query: 599 FRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVY 648
FRT GH+EF+VMPFGLTNAP+ LMN V + +L +F ++F DDILVY
Sbjct: 627 FRTRYGHFEFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVY 676
>At2g06890 putative retroelement integrase
Length = 1215
Score = 211 bits (537), Expect = 2e-54
Identities = 194/687 (28%), Positives = 307/687 (44%), Gaps = 118/687 (17%)
Query: 66 KLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALK-----EIDHDFVRGIF 120
K + +RF PS EL L L+ + + VEE+ K L D + F
Sbjct: 2 KAIMRKRFVPSHYHR--ELHLKLRNLTQGNRSVEEYYKEMETLMLRADISEDREATLSRF 59
Query: 121 LNGLKEEIKAEVR---------LIQKSLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVV 171
L L +I+ + ++ K++L EQ+ + R SS+ ++Y
Sbjct: 60 LGDLNRDIQDRLETQYYVQIEEMLHKAILFEQQVK----------RKSSS--RSSYGSGT 107
Query: 172 TVEPQNNTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFKHLTGAQMREKRDKNLCFRC 231
+P +++ S + P + + ++ G+ + ++ +++R+ R C++C
Sbjct: 108 IAKPTYQREERTSSYHNKPIVSPRAESKPYAAVQDHKGKAE-ISTSRVRDVR----CYKC 162
Query: 232 DEPYSREHKCRNKQLRMIIMEDDDDADCGEFEDALS-----------GEFNSLQLSLCSM 280
++C NK++ MI++++ + E D+ S GE + +L
Sbjct: 163 QGKGHYANECPNKRV-MILLDNGEIEPEEEIPDSPSSLKENEELPAQGELLVARRTLSVQ 221
Query: 281 AGLTSTKSWKMVGQ----IGNQSIVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVG 336
+ K + + + ++ID G+ N S +V++L L +
Sbjct: 222 TKTDEQEQRKNLFHTRCHVHGKVCSLIIDGGSCTNVASETMVKKLGLKW---------LN 272
Query: 337 DGYKIRCKGKCSQLGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLGDIKANF------ 390
D K+R K + + I+I E + + ++LG W + + F
Sbjct: 273 DSGKMRVKNQVV-VPIVIGKYEDEILCDVLPMEAGHILLGRPWQSDRKVMHDGFTNRHSF 331
Query: 391 ----GKMELTLKRGKEVY------------TLKGDPSLTKS---ELSYGALMQILLEEGE 431
GK L EVY +K KS + +Y + +LL +
Sbjct: 332 EFKGGKTILVSMTPHEVYQDQIHLKQKKEQVVKQPNFFAKSGEVKSAYSSKQPMLLFVFK 391
Query: 432 GLLVHCDTGEKVNKKECEIPEIITGVLLQFDTVFQD--PQGLPPRRRHDHAIHLKEGAQV 489
L V +P +T +L + VF + P+GLPP R +H I GA +
Sbjct: 392 EALTSLTNFAPV------LPSEMTSLLQDYKDVFPEDNPKGLPPIRGIEHQIDFVPGASL 445
Query: 490 PNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISPFSSPIILVKKKDGGWRLCVDYRALN 549
PN P Y+ N VE L ++KDG WR+C D RA+N
Sbjct: 446 PNR-----PAYRTNPVETKE---------------------LQRQKDGSWRMCFDCRAIN 479
Query: 550 KVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHEGHYEFL 609
VT+ PIP ++++LDEL G+ +FSK+DLKSGYHQIRM + D KTAF+T G YE+L
Sbjct: 480 NVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWL 539
Query: 610 VMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPDLLTHTTHLQQVLTLLADH 669
VMPFGLT+APSTF LMN VL+ ++ F +V+FDDILVYS L H HL VL +L
Sbjct: 540 VMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKE 599
Query: 670 ALKANRKKCSFGQTSLEYLGHIVSGEG 696
L AN KKC+F +L +LG +VS +G
Sbjct: 600 ELYANLKKCTFCTDNLVFLGFVVSADG 626
Score = 146 bits (368), Expect = 1e-34
Identities = 102/293 (34%), Positives = 152/293 (51%), Gaps = 15/293 (5%)
Query: 806 LAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQRLFS 865
+AF+S+ L + Y++EL ALV+A+Q+W+HYL + F++ TD +SLK L Q+ +
Sbjct: 737 IAFFSEKLGGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLN 796
Query: 866 *DQFKWAVKLVGLDFEIQYKPGSENTVADALS-RKMMYSALSI-LTSAEKEDWNSELQQD 923
+W + + I+YK G +N VADALS R + S L++ L E+ E D
Sbjct: 797 KRHARWVEFIETFAYVIKYKKGKDNVVADALSQRYTLLSTLNVKLMGFEQIKEVYETDHD 856
Query: 924 SKLVKVIQDLLISPTSHPGFEFKKG-LLYYKNRLVLSKASNRIPMIISECHDTPTGGHSG 982
+ V + S G F++ L+Y+NRL + S R + + E H GH G
Sbjct: 857 FQEVYKACEKFAS-----GRYFRQDKFLFYENRLCVPNCSLR-DLFVREAHGGGLMGHFG 910
Query: 983 HFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISM 1042
+T + + W MK D+K C C+++K S + P GL PLPIP W DISM
Sbjct: 911 IAKTLEVMTEHFRWPHMKCDVKRICGRCNTCKQAK-SKIQPNGLYTPLPIPKHPWNDISM 969
Query: 1043 DFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHG 1095
DFV GLP+ GKD++ VV + F Y +P S D+ L + E V + G
Sbjct: 970 DFVMGLPRT-GKDSIFVV---YSPFQIVYGF-NPISPFDLIPLPLSERVSIDG 1017
Score = 45.1 bits (105), Expect = 3e-04
Identities = 37/120 (30%), Positives = 62/120 (50%), Gaps = 12/120 (10%)
Query: 1182 SPFKALYGRDP--PLLLKAGAIPSRV----EEVNQLFQQRDGVLGELKQNLLKAQHQMKV 1235
SPF+ +YG +P P L + RV ++ +L QQ + ++N+ +
Sbjct: 988 SPFQIVYGFNPISPFDLIPLPLSERVSIDGKKKAELVQQ---IHENARRNIEEKTKLYAK 1044
Query: 1236 QADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLEL 1295
QA+K RR F GD V++ L+ + + + +KL PR GP+KI+ R+ AY+L+L
Sbjct: 1045 QANKGRREQIFEVGDMVWIHLRKERFPA---QRKSKLMPRIDGPFKIIKRINDNAYQLDL 1101
>At4g04230 putative transposon protein
Length = 315
Score = 194 bits (493), Expect = 3e-49
Identities = 91/168 (54%), Positives = 123/168 (73%)
Query: 513 LEAGVIRPSISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGA 572
+ G IR S+SP + P++LV KKDG WR+C+D RA+N +TI + PIP + ++LDEL GA
Sbjct: 125 IHKGYIRESLSPCAVPVLLVPKKDGTWRMCLDCRAINNITIKYRHPIPRLYDMLDELSGA 184
Query: 573 KVFSKLDLKSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKP 632
+FSK+DL+SGYHQ+RM++ D KTAF+T +G YE LVMPFGLTNAPSTF LMN+VL+
Sbjct: 185 IIFSKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQVLRS 244
Query: 633 YLRKFALVFFDDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSF 680
++ KF +V+FDDIL+Y+ H L+ +L L L AN KKC+F
Sbjct: 245 FIGKFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTF 292
>At3g29490 hypothetical protein
Length = 438
Score = 170 bits (431), Expect = 5e-42
Identities = 128/386 (33%), Positives = 185/386 (47%), Gaps = 62/386 (16%)
Query: 999 MKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGKDTVL 1058
MK D+ N+V +C++CQ K PGG+LQ LPIP
Sbjct: 1 MKRDVANWVAECDVCQLVKAEHQVPGGMLQSLPIP------------------------- 35
Query: 1059 VVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFK 1118
++ F A+ A +A ++ E+VKLHG P +
Sbjct: 36 -------EWNTFLAIRKTDGAAVLAKKYVSEIVKLHGVPAEM------------------ 70
Query: 1119 MAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSS 1178
GT++++ST YHPQTDGQ E T + LE LR V W D L EF +N +Y +
Sbjct: 71 --GTKVQMSTPYHPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQAG 128
Query: 1179 AGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQAD 1238
GM+PF+ALYGR L + R Q+ + LK N+ +AQ + AD
Sbjct: 129 IGMAPFEALYGRPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYAD 188
Query: 1239 KHRRLVEFNEGDWVYLKLQPYK--LKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELP 1296
K RR +EF GD VYLK+ + +SI+ KL+ R+ GP++I++RVGPVAY LELP
Sbjct: 189 KRRRELEFEVGDRVYLKMAMLRGPNRSISE---TKLSLRYMGPFRIVERVGPVAYMLELP 245
Query: 1297 -AHSKIHPVFHVSLLKQAL-KPTQQPQPLPPMLNEELELEVVPEDVIDHR-EDAQGNLEV 1353
H VFHVS+L++ L K + +P L + LE V++ R ++ Q
Sbjct: 246 DVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKIS 305
Query: 1354 LIK--WEQLPTCDNTWELAAVIQDKF 1377
LIK W+ + TW+ A ++ +F
Sbjct: 306 LIKVLWDCDGVTEETWQPEARMKARF 331
>At4g08100 putative polyprotein
Length = 1054
Score = 167 bits (424), Expect = 3e-41
Identities = 82/153 (53%), Positives = 110/153 (71%)
Query: 544 DYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHE 603
D RA+N +T+ + PIP +++ LD+L G+ +FSK+DLKSGYHQ RMK+ D KTA +T +
Sbjct: 527 DCRAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQ 586
Query: 604 GHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPDLLTHTTHLQQVL 663
YE+LVMPFGLTNAP+TF LMN VL+ ++ F +V+FDDILVYS +L H HL+ VL
Sbjct: 587 RLYEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVL 646
Query: 664 TLLADHALKANRKKCSFGQTSLEYLGHIVSGEG 696
LL L AN KKC+F +L +LG +VS +G
Sbjct: 647 DLLRKEKLYANLKKCTFCTDNLVFLGFVVSADG 679
Score = 66.6 bits (161), Expect = 1e-10
Identities = 54/167 (32%), Positives = 84/167 (49%), Gaps = 11/167 (6%)
Query: 1176 NSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKV 1235
+S+ SPF+ +YG P L IP + E + L ++ L + NL Q K
Sbjct: 851 HSATKFSPFEIVYGFKPTSPLDL--IPLPLSERSSLDGKKKDDLVQQVLNLEARTKQYKK 908
Query: 1236 QADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLEL 1295
A+K R+ V FNEGD V++ L+ + + + +KL PR GP+K+L R+ AYKL+L
Sbjct: 909 YANKGRKEVIFNEGDQVWVHLRKKRFPEVRS---SKLMPRIDGPFKVLKRINNNAYKLDL 965
Query: 1296 PAHSKIHP-VFHVSLLKQALKPTQQPQPLPPMLNEELELEVVPEDVI 1341
S + F V + T +PL P +EE ++VPE+ +
Sbjct: 966 QDTSDLRTNPFQVG--EDDENMTNATKPLEPQEDEE---QLVPEEAL 1007
Score = 58.5 bits (140), Expect = 3e-08
Identities = 27/80 (33%), Positives = 47/80 (58%)
Query: 806 LAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFLTDQRLFS 865
+A++S+ L + Y++EL ALV+A+Q W+HYL + F++ TD +SLK L Q+ +
Sbjct: 760 IAYFSEKLGGATLNYPTYDKELYALVRALQTWQHYLWPKKFVIHTDHESLKHLKGQQKLN 819
Query: 866 *DQFKWAVKLVGLDFEIQYK 885
+W + + I+YK
Sbjct: 820 KRHARWVEFIETFPYVIKYK 839
Score = 50.8 bits (120), Expect = 5e-06
Identities = 74/313 (23%), Positives = 115/313 (36%), Gaps = 56/313 (17%)
Query: 2 KLEIPIFSGDDA----VGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWW---- 53
KL+IP F G D + W QK+E F +E + +++ ALSW+
Sbjct: 93 KLKIPAFHGTDNPDTYLEWEQKIELVFLCQECLQSNKVKIAATKFYNYALSWWDQLVTSR 152
Query: 54 --ERCNPSPTWEAFKLAVVRRFQPSMIQNPFELLLSLKQVG--TVDEYVEEFEK-YAGAL 108
R P TW K + +RF PS L G TV+EY E E A
Sbjct: 153 RRTRDYPIKTWNQLKFVMRKRFVPSYYHRELHQRLRNLVQGSKTVEEYFLEMETLMLRAD 212
Query: 109 KEIDHDFVRGIFLNGLKEEIKAEV---------RLIQKSLLIEQKNQIVTKKGVGFTRPS 159
+ D + V F+ GL EI+ + ++ K+++ EQ QI K
Sbjct: 213 LQEDGEAVMSRFMGGLNREIQDRLETQHYVELEEMLHKAVMFEQ--QIKRKNA------R 264
Query: 160 SAFRNNNYSKVVTVEPQNNTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFKHLTGAQM 219
S+ NYS + QK++ + + P +D G+ +
Sbjct: 265 SSHTKTNYSS------GKPSYQKEEKFGYQNDSKPFVKPKPVDLDPKGKGKEVITRARDI 318
Query: 220 REKRDKNLCFRCDEPYSREHKCRNKQLRMIIMED-----------DDDADCGEFEDALSG 268
R CF+ KC NK R+++ D ++D+ + E G
Sbjct: 319 R-------CFKSQGLRHYASKCSNK--RIMVHRDNGEVESKDESAEEDSMEEDVESPARG 369
Query: 269 EFNSLQLSLCSMA 281
EF LS+ + A
Sbjct: 370 EFARRSLSVLTKA 382
>At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 280
Score = 161 bits (408), Expect = 2e-39
Identities = 111/268 (41%), Positives = 141/268 (52%), Gaps = 21/268 (7%)
Query: 1042 MDFVGGLPKV-RGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTI 1100
MDFV GLP+ RG D+V VVVDR +K HF A A ++A LF KE+V+LHG P +I
Sbjct: 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADASNIAKLFFKEVVRLHGVPKSI 60
Query: 1101 VSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQW 1160
SDRD FLS+FW L++M GT L S++ H Q DGQTEVTNR L +R G PKQW
Sbjct: 61 TSDRDTKFLSHFWSTLWRMFGTALNRSSTPHTQIDGQTEVTNRTLGNMVRSICGDNPKQW 120
Query: 1161 LDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLG 1220
LP EF YNS + L P L A A + EE+ V
Sbjct: 121 DLALPQIEF----AYNSVVHVVDLVKL-----PKALGASA-ETMAEEILV-------VKE 163
Query: 1221 ELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPY 1280
+K L + KV ADK RR F EGD V + L+ + A K+ PR YGP+
Sbjct: 164 VVKAKLEATGKKNKVAADKRRRFKVFKEGDDVMVLLRKGR---FAVGTYNKVKPRKYGPF 220
Query: 1281 KILDRVGPVAYKLELPAHSKIHPVFHVS 1308
K+L ++ AY + LP I F+V+
Sbjct: 221 KVLRKINDNAYVVALPKSMNISNTFNVA 248
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,033,422
Number of Sequences: 26719
Number of extensions: 1476105
Number of successful extensions: 3689
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 3382
Number of HSP's gapped (non-prelim): 201
length of query: 1430
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1318
effective length of database: 8,326,068
effective search space: 10973757624
effective search space used: 10973757624
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0281a.3


