

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281a.2
(153 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
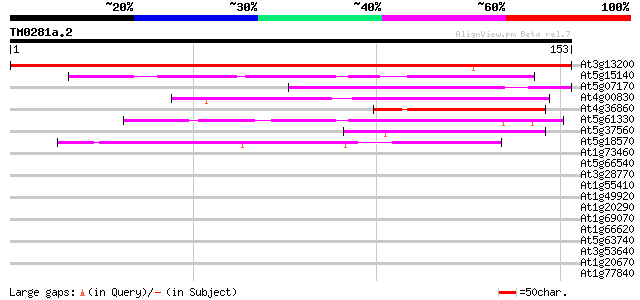
Score E
Sequences producing significant alignments: (bits) Value
At3g13200 unknown protein 250 2e-67
At5g15140 putative aldose 1-epimerase - like protein 49 1e-06
At5g07170 putative protein 43 6e-05
At4g00830 unknown protein 41 3e-04
At4g36860 Unknown protein 40 5e-04
At5g61330 unknown protein 40 6e-04
At5g37560 putative protein 40 6e-04
At5g18570 GTP-binding protein obg -like 39 8e-04
At1g73460 39 0.001
At5g66540 unknown protein 39 0.001
At3g28770 hypothetical protein 39 0.001
At1g55410 hypothetical protein 39 0.001
At1g49920 hypothetical protein 39 0.001
At1g20290 hypothetical protein 39 0.001
At1g69070 hypothetical protein 38 0.002
At1g66620 hypothetical protein 38 0.002
At5g63740 unknown protein 38 0.002
At3g53640 protein kinase -like protein 38 0.002
At1g20670 hypothetical protein 38 0.002
At1g77840 putative eukaryotic translation initiation factor 5 (E... 37 0.003
>At3g13200 unknown protein
Length = 230
Score = 250 bits (638), Expect = 2e-67
Identities = 121/155 (78%), Positives = 140/155 (90%), Gaps = 2/155 (1%)
Query: 1 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR 60
MTTAARPTWAPAKGGNEQGG RIFGPSQKYSSRD+A+HTTLKPRR+GQ TQ+EL++ NLR
Sbjct: 1 MTTAARPTWAPAKGGNEQGGARIFGPSQKYSSRDVAAHTTLKPRREGQHTQEELQKINLR 60
Query: 61 DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD 120
DELEERERRHFSSK+KSY+DDRD + S L LE +KRD E+ I+ RSVDADDSDV++KSD
Sbjct: 61 DELEERERRHFSSKDKSYNDDRDRRRGSQLLLEDSKRDPEERIIPRSVDADDSDVDIKSD 120
Query: 121 EESDD--DDDDEDDTEALLAELEQIKKERAEEKLR 153
++SDD DDDDEDDTEAL+AEL+QIKKER EE+LR
Sbjct: 121 DDSDDESDDDDEDDTEALMAELDQIKKERVEERLR 155
>At5g15140 putative aldose 1-epimerase - like protein
Length = 490
Score = 48.5 bits (114), Expect = 1e-06
Identities = 34/127 (26%), Positives = 57/127 (44%), Gaps = 18/127 (14%)
Query: 17 EQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNK 76
E+GG ++ G D K ++ D Q + K+ +D+ + E++H K
Sbjct: 52 EKGGEKVDGAGSDDEDND------KKEKKKEHDVQKKDKQHENKDK--DDEKKHVDKKKS 103
Query: 77 SYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEAL 136
D D + H + K+D + D DDSD + D++ DDDDDD+D+ +
Sbjct: 104 GGHDKDDDDEKKH---KDKKKDGHN-------DDDDSDDDTDDDDDDDDDDDDDDEVDGD 153
Query: 137 LAELEQI 143
E E+I
Sbjct: 154 DNEKEKI 160
>At5g07170 putative protein
Length = 542
Score = 43.1 bits (100), Expect = 6e-05
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Query: 77 SYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEAL 136
S SD+ D +S+H + D D D DD D + D++ DDDDDD++ ++
Sbjct: 90 SSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDS- 148
Query: 137 LAELEQIKKERAEEKLR 153
++++E ++ LR
Sbjct: 149 -----EVEEEEGDDDLR 160
Score = 26.6 bits (57), Expect = 5.7
Identities = 14/64 (21%), Positives = 27/64 (41%), Gaps = 2/64 (3%)
Query: 73 SKNKSYSDDRDHGKSSHLFLEGTKRD--VEDHIVARSVDADDSDVEVKSDEESDDDDDDE 130
S + Y D D + ++ D ++D D++ + D+ D DDD++
Sbjct: 59 SSDDHYCGDDDENMKIAAAIPSSQDDPLIDDSSSDEEDDSESTHCYAADDDADDTDDDED 118
Query: 131 DDTE 134
DD +
Sbjct: 119 DDDD 122
>At4g00830 unknown protein
Length = 495
Score = 40.8 bits (94), Expect = 3e-04
Identities = 26/105 (24%), Positives = 48/105 (44%), Gaps = 7/105 (6%)
Query: 45 RDGQDTQD--ELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDH 102
RD D D E + DE+EE + + + + DD D G + +R+VED+
Sbjct: 5 RDNDDRVDFEEGSYSEMEDEVEEEQVEEYEEEEEEDDDDDDVGNQN-----AEEREVEDY 59
Query: 103 IVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKER 147
+ D +D E+ D+++ D + DD E + ++ +E+
Sbjct: 60 GDTKGGDMEDVQEEIAEDDDNHIDIETADDDEKPPSPIDDEDREK 104
>At4g36860 Unknown protein
Length = 542
Score = 40.0 bits (92), Expect = 5e-04
Identities = 17/47 (36%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query: 100 EDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKE 146
++H++ + D +E KS+ E DDDDD+++D E + A+LE ++E
Sbjct: 81 QEHVIPQD-DKGKKIIEYKSETEEDDDDDEDEDEEYMRAQLEAAEEE 126
>At5g61330 unknown protein
Length = 436
Score = 39.7 bits (91), Expect = 6e-04
Identities = 33/122 (27%), Positives = 57/122 (46%), Gaps = 11/122 (9%)
Query: 32 SRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLF 91
S DI+ LK D +D D+L DE++ E + +S DD +
Sbjct: 17 SEDISDQENLKAESDNED--DQLPDGIEDDEVDSME----DDEGESEEDDEGDTEEDD-- 68
Query: 92 LEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDT-EALLAELE-QIKKERAE 149
EG + ++ D + D E +D+ES+ D+ DD +A + ELE ++K+ R++
Sbjct: 69 -EGDSEEDDEGENKEDEDGESEDFEDGNDKESESGDEGNDDNKDAQMEELEKEVKELRSQ 127
Query: 150 EK 151
E+
Sbjct: 128 EQ 129
Score = 26.6 bits (57), Expect = 5.7
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query: 86 KSSHLFLEGTKRDVED--HIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
+S L+ D+ D ++ A S + DD + D+E D +DDE ++E
Sbjct: 7 RSKRARLDSESEDISDQENLKAESDNEDDQLPDGIEDDEVDSMEDDEGESE 57
>At5g37560 putative protein
Length = 408
Score = 39.7 bits (91), Expect = 6e-04
Identities = 20/58 (34%), Positives = 31/58 (52%), Gaps = 3/58 (5%)
Query: 92 LEGTKRDVED---HIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKE 146
L + D ED H+ A + DD D + DE+ ++DDDD+DD + LE ++E
Sbjct: 349 LMDNREDEEDYNLHVDAEVNNDDDDDEDYVFDEDYEEDDDDDDDDDGTTYSLEDFRRE 406
>At5g18570 GTP-binding protein obg -like
Length = 681
Score = 39.3 bits (90), Expect = 8e-04
Identities = 36/123 (29%), Positives = 55/123 (44%), Gaps = 12/123 (9%)
Query: 14 GGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDE-LEERERRHFS 72
GG TR+ P + Y + S + LK + + ++ + R + E L E E
Sbjct: 64 GGEATTYTRL-PPREDYYDVSLLSSSYLKVSEEVKLSESNVARVEEKIETLREIEEEEKE 122
Query: 73 SKNKSYSDDRDHGKSSHL-FLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDED 131
+ KSY DD G L EG+ +RS+D DD D E + E D+ D ++D
Sbjct: 123 KEVKSYDDDDIWGNYRRLDVFEGS---------SRSIDEDDEDWEDEVFEYGDETDAEKD 173
Query: 132 DTE 134
D+E
Sbjct: 174 DSE 176
>At1g73460
Length = 1157
Score = 38.9 bits (89), Expect = 0.001
Identities = 36/137 (26%), Positives = 61/137 (44%), Gaps = 14/137 (10%)
Query: 24 FGPSQK-------YSSRDIAS--HTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSK 74
FG SQK SS+ + S H T+ R+ + L D++ RR S
Sbjct: 679 FGSSQKDGQSMHAESSKSLWSGNHETVTRDRN----TERLSASTAMDDMVATWRRKSSDS 734
Query: 75 NKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
+ S+S +D+ +S L + + ++ AD D S E DD+ DD E
Sbjct: 735 SSSHSSVKDNNATSIKSLNSSPSSLSNYACEERKHADKEDDRNDSSEIEDDNATALDDEE 794
Query: 135 ALLAELEQIKKERAEEK 151
A+ + EQ+++ +A+E+
Sbjct: 795 AVAVQ-EQVRQIKAQEE 810
>At5g66540 unknown protein
Length = 524
Score = 38.5 bits (88), Expect = 0.001
Identities = 26/91 (28%), Positives = 41/91 (44%), Gaps = 3/91 (3%)
Query: 46 DGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVE--DHI 103
+G+D ++E + +E EE E N+ D K FLE + + DH
Sbjct: 137 EGEDEEEEEEDEEEEEEEEEEEEEEKDGDNEGIEDKFFKIKELEEFLEEGEAEEYGIDHK 196
Query: 104 VARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
+ V A + DE+ +DDDD+E+D E
Sbjct: 197 NKKGV-AQRKKQNLSDDEDEEDDDDEEEDVE 226
Score = 36.2 bits (82), Expect = 0.007
Identities = 22/66 (33%), Positives = 34/66 (51%)
Query: 86 KSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKK 145
K L L+ + D D + D+DD D E K E +D + +DE++ E E E+ ++
Sbjct: 97 KLGKLALKVSHEDDIDEMDMDGFDSDDVDDEDKEIESNDSEGEDEEEEEEDEEEEEEEEE 156
Query: 146 ERAEEK 151
E EEK
Sbjct: 157 EEEEEK 162
Score = 29.6 bits (65), Expect = 0.67
Identities = 23/91 (25%), Positives = 42/91 (45%), Gaps = 10/91 (10%)
Query: 61 DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD 120
D+++E + F S + DD D +E + ED + ++ + E + +
Sbjct: 109 DDIDEMDMDGFDSDDV---DDEDKE------IESNDSEGEDEEEEEEDEEEEEEEEEEEE 159
Query: 121 EESDDDDDDEDDTEALLAELEQIKKE-RAEE 150
EE D D++ +D + ELE+ +E AEE
Sbjct: 160 EEKDGDNEGIEDKFFKIKELEEFLEEGEAEE 190
>At3g28770 hypothetical protein
Length = 2081
Score = 38.5 bits (88), Expect = 0.001
Identities = 25/116 (21%), Positives = 58/116 (49%), Gaps = 8/116 (6%)
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
K +++ ++++D ++ + E++E + SK K D ++H + + E K++ +
Sbjct: 1047 KSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKE--DKKEHEDNKSMKKEEDKKEKKK 1104
Query: 102 H--IVARSVDADDSDVEVKSDEESDDDDDDEDDTE----ALLAELEQIKKERAEEK 151
H +R + D D+E D+ S+ +D+++ + L + E KKE+ E +
Sbjct: 1105 HEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENE 1160
Score = 31.6 bits (70), Expect = 0.18
Identities = 25/111 (22%), Positives = 49/111 (43%), Gaps = 2/111 (1%)
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
KP+ D ++T + + E E +E + + D D K+ L ++ D
Sbjct: 1344 KPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHS 1403
Query: 102 HIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIK-KERAEEK 151
A S D +++ +++D ++ ++E+D + + E K KE EEK
Sbjct: 1404 DSQADS-DESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEK 1453
Score = 31.6 bits (70), Expect = 0.18
Identities = 25/111 (22%), Positives = 49/111 (43%), Gaps = 2/111 (1%)
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
KP+ D ++T + + E E +E + + D D K+ L ++ D
Sbjct: 1233 KPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHS 1292
Query: 102 HIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIK-KERAEEK 151
A S D +++ +++D ++ ++E+D + + E K KE EEK
Sbjct: 1293 DSQADS-DESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEK 1342
Score = 30.4 bits (67), Expect = 0.39
Identities = 33/148 (22%), Positives = 61/148 (40%), Gaps = 13/148 (8%)
Query: 14 GGNEQGGTRIFGPSQKYSSRDIASHTTLKPR-----RDGQDTQDELKRRNLRDELEERER 68
GG + + +K S++ T + R + Q + E ++ ++ E ++
Sbjct: 737 GGESKDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDA 796
Query: 69 RHFSSK-NKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDS------DVEVKSDE 121
+ +K NK S + ++ E K D E+ +SV+A + D V + E
Sbjct: 797 KSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNKE 856
Query: 122 ESDDDDDDEDDTEALLAELEQIKKERAE 149
+S D DD E + E +KK+R E
Sbjct: 857 DSKDLKDDR-SVEVKANKEESMKKKREE 883
Score = 27.7 bits (60), Expect = 2.5
Identities = 21/114 (18%), Positives = 46/114 (39%)
Query: 38 HTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKR 97
HT K + + +K NL ++ +++E + S +++ + +G
Sbjct: 581 HTKEKREETQGNNGESVKNENLENKEDKKELKDDESVGAKTNNETSLEEKREQTQKGHDN 640
Query: 98 DVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
+ IV DS+ E + +D++ + E +E+E K + + EK
Sbjct: 641 SINSKIVDNKGGNADSNKEKEVHVGDSTNDNNMESKEDTKSEVEVKKNDGSSEK 694
>At1g55410 hypothetical protein
Length = 672
Score = 38.5 bits (88), Expect = 0.001
Identities = 15/27 (55%), Positives = 18/27 (66%)
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTEA 135
DADD D D++ DDDDDD+DD A
Sbjct: 159 DADDDDANDDDDDDDDDDDDDDDDANA 185
Score = 37.4 bits (85), Expect = 0.003
Identities = 15/31 (48%), Positives = 20/31 (64%)
Query: 105 ARSVDADDSDVEVKSDEESDDDDDDEDDTEA 135
A VD D D + D++ DDDDDD+DD +A
Sbjct: 153 AADVDNDADDDDANDDDDDDDDDDDDDDDDA 183
Score = 34.7 bits (78), Expect = 0.021
Identities = 17/44 (38%), Positives = 27/44 (60%), Gaps = 2/44 (4%)
Query: 93 EGTKRDVEDHIVARSVDADDSDVEVKSDEE--SDDDDDDEDDTE 134
E D D + A + D D +DV+ +D++ +DDDDDD+DD +
Sbjct: 134 EDDDADNNDVVAAAADDDDAADVDNDADDDDANDDDDDDDDDDD 177
Score = 32.3 bits (72), Expect = 0.10
Identities = 16/57 (28%), Positives = 28/57 (49%)
Query: 79 SDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEA 135
+DD D ++ + D + V D DD++ + D++ DDDDDD+ + A
Sbjct: 131 ADDEDDDADNNDVVAAAADDDDAADVDNDADDDDANDDDDDDDDDDDDDDDDANAYA 187
>At1g49920 hypothetical protein
Length = 785
Score = 38.5 bits (88), Expect = 0.001
Identities = 21/45 (46%), Positives = 26/45 (57%), Gaps = 5/45 (11%)
Query: 93 EGTKRDVEDHIVARSVDADDSDVE----VKSDEESDDD-DDDEDD 132
+G ++D ED + D DD + E V DEE DDD DDDEDD
Sbjct: 739 KGKEKDTEDDHLEEDEDGDDDEEEEDDDVDDDEEDDDDVDDDEDD 783
Score = 30.0 bits (66), Expect = 0.51
Identities = 10/30 (33%), Positives = 19/30 (63%)
Query: 103 IVARSVDADDSDVEVKSDEESDDDDDDEDD 132
+ + + D D ++ DE+ DDD+++EDD
Sbjct: 736 VSGKGKEKDTEDDHLEEDEDGDDDEEEEDD 765
>At1g20290 hypothetical protein
Length = 497
Score = 38.5 bits (88), Expect = 0.001
Identities = 18/117 (15%), Positives = 55/117 (46%)
Query: 35 IASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEG 94
+ T ++ + ++ ++E + +E EE E + + ++ + + E
Sbjct: 355 VGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 414
Query: 95 TKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
+ + E+ + ++ + E + +EE ++++++E++ E E E+ ++E EE+
Sbjct: 415 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 471
Score = 37.7 bits (86), Expect = 0.002
Identities = 20/128 (15%), Positives = 56/128 (43%)
Query: 24 FGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRD 83
F PS + + + ++ ++E + +E EE E + + ++ +
Sbjct: 348 FTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 407
Query: 84 HGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQI 143
+ E + + E+ + ++ + E + +EE ++++++E++ E E E+
Sbjct: 408 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 467
Query: 144 KKERAEEK 151
++E EE+
Sbjct: 468 EEEEEEEE 475
Score = 37.7 bits (86), Expect = 0.002
Identities = 18/104 (17%), Positives = 50/104 (47%)
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ ++E + +E EE E + + ++ + + E + + E+
Sbjct: 379 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 438
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
+ ++ + E + +EE ++++++E++ E E E+ ++E EEK
Sbjct: 439 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEK 482
Score = 36.2 bits (82), Expect = 0.007
Identities = 17/104 (16%), Positives = 50/104 (47%)
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ ++E + +E EE E + + ++ + + E + + E+
Sbjct: 378 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 437
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
+ ++ + E + +EE ++++++E++ E E E+ ++E EE+
Sbjct: 438 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 481
Score = 36.2 bits (82), Expect = 0.007
Identities = 17/104 (16%), Positives = 50/104 (47%)
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ ++E + +E EE E + + ++ + + E + + E+
Sbjct: 374 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 433
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
+ ++ + E + +EE ++++++E++ E E E+ ++E EE+
Sbjct: 434 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 477
Score = 36.2 bits (82), Expect = 0.007
Identities = 17/104 (16%), Positives = 50/104 (47%)
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ ++E + +E EE E + + ++ + + E + + E+
Sbjct: 376 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 435
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
+ ++ + E + +EE ++++++E++ E E E+ ++E EE+
Sbjct: 436 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 479
Score = 33.5 bits (75), Expect = 0.046
Identities = 15/100 (15%), Positives = 48/100 (48%)
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ ++E + +E EE E + + ++ + + E + + E+
Sbjct: 395 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 454
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKER 147
+ ++ + E + +EE ++++++E++ E +++ KK+R
Sbjct: 455 EEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKKKR 494
Score = 32.3 bits (72), Expect = 0.10
Identities = 14/104 (13%), Positives = 49/104 (46%)
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ ++E + +E EE E + + ++ + + E + + E+
Sbjct: 392 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 451
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
+ ++ + E + +EE ++++++E++ E + +KK++ + +
Sbjct: 452 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKKKRR 495
>At1g69070 hypothetical protein
Length = 901
Score = 38.1 bits (87), Expect = 0.002
Identities = 29/107 (27%), Positives = 48/107 (44%), Gaps = 10/107 (9%)
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
KP+R D D L+R + D E E S+ + D G G ++ +
Sbjct: 338 KPKRGWID--DVLEREDNVDNSESDEDEDSESEEEEDDDGESDG--------GDEKQRKG 387
Query: 102 HIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERA 148
H + +DD DEE DDD++D+D+ +A L +++K + A
Sbjct: 388 HHLEDWEQSDDELGAELEDEEEDDDEEDDDEEDAELRVHKKLKNDYA 434
Score = 33.1 bits (74), Expect = 0.060
Identities = 20/87 (22%), Positives = 43/87 (48%), Gaps = 3/87 (3%)
Query: 51 QDELKRRNLRDELEERERR---HFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ KR +EL + + S+K + D G S + + KR D ++ R
Sbjct: 292 EERKKRMQETEELSDGDEEIGGEESTKRLTVISGDDLGDSFSVEEDKPKRGWIDDVLERE 351
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTE 134
+ D+S+ + D ES++++DD+ +++
Sbjct: 352 DNVDNSESDEDEDSESEEEEDDDGESD 378
Score = 26.2 bits (56), Expect = 7.4
Identities = 15/42 (35%), Positives = 21/42 (49%)
Query: 86 KSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDD 127
KS + +G + ED + S DD D + SDE+ DDD
Sbjct: 121 KSMYNLSDGEEDVYEDGALGGSSVKDDFDSGLLSDEDLQDDD 162
Score = 26.2 bits (56), Expect = 7.4
Identities = 26/105 (24%), Positives = 47/105 (44%), Gaps = 8/105 (7%)
Query: 55 KRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADD-- 112
+R L EER++R ++ S D+ G+ S L D D + SV+ D
Sbjct: 283 EREKLEALEEERKKRMQETEELSDGDEEIGGEESTKRLTVISGD--DLGDSFSVEEDKPK 340
Query: 113 ----SDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
DV + D + + D+++D+E+ E + + + +EK R
Sbjct: 341 RGWIDDVLEREDNVDNSESDEDEDSESEEEEDDDGESDGGDEKQR 385
>At1g66620 hypothetical protein
Length = 313
Score = 38.1 bits (87), Expect = 0.002
Identities = 16/34 (47%), Positives = 25/34 (73%)
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTEALLAELEQ 142
D +++D + +S+EE DDDDDD+DD E A+ E+
Sbjct: 280 DEEEADEDEESEEEEDDDDDDDDDDEEEDADEEE 313
Score = 29.6 bits (65), Expect = 0.67
Identities = 10/32 (31%), Positives = 19/32 (59%)
Query: 103 IVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
I R + D+ + + + E ++DDDD+DD +
Sbjct: 272 ICIRKLKKDEEEADEDEESEEEEDDDDDDDDD 303
>At5g63740 unknown protein
Length = 226
Score = 37.7 bits (86), Expect = 0.002
Identities = 27/89 (30%), Positives = 37/89 (41%), Gaps = 18/89 (20%)
Query: 46 DGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVA 105
DG DE DE E+ + +++ DD D + D +D
Sbjct: 64 DGDGDGDE-------DEDEDADADEDEDEDEDEDDDDDDDDDD----DDDADDADD---- 108
Query: 106 RSVDADDSDVEVKSDEESDDDDDDEDDTE 134
D DD D + DE+ DDDDDDE+D E
Sbjct: 109 ---DEDDDDEDDDEDEDDDDDDDDENDEE 134
Score = 35.4 bits (80), Expect = 0.012
Identities = 19/60 (31%), Positives = 29/60 (47%), Gaps = 3/60 (5%)
Query: 76 KSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEA 135
K Y D + G+ +G ED D D+ + E + D++ DDDDDD+D +A
Sbjct: 50 KVYDDHNNDGEGDG---DGDGDGDEDEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDA 106
>At3g53640 protein kinase -like protein
Length = 642
Score = 37.7 bits (86), Expect = 0.002
Identities = 32/124 (25%), Positives = 52/124 (41%), Gaps = 16/124 (12%)
Query: 31 SSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRD-HGKSSH 89
SSRD K R ++T E +R +D+ +R RR S + Y D +G+ H
Sbjct: 93 SSRD-------KARSSSRETGRENERERRKDQDRDRGRREDQSDQEIYKSGGDGYGEVRH 145
Query: 90 LFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAE 149
D ED + + + + + + D+DDD +D + E E + +R E
Sbjct: 146 --------DAEDDLDSLKSHKPNGETQGNVERSEVDNDDDGEDVVWGVEEQEAAEMKRIE 197
Query: 150 EKLR 153
E R
Sbjct: 198 ESKR 201
>At1g20670 hypothetical protein
Length = 652
Score = 37.7 bits (86), Expect = 0.002
Identities = 19/91 (20%), Positives = 44/91 (47%)
Query: 39 TTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRD 98
T K ++ G+ + +L++R ++ + ++ +++ KN DD H +++
Sbjct: 7 TMTKKKKKGRPSLLDLQKRAIKQQQQQLQQQQQQHKNNHQDDDDHHHNNNNRSGSKNPNS 66
Query: 99 VEDHIVARSVDADDSDVEVKSDEESDDDDDD 129
+ R+ +++D D DE D+DDD+
Sbjct: 67 LNHRSKRRNPNSNDGDSPWIKDEGEDNDDDE 97
>At1g77840 putative eukaryotic translation initiation factor 5
(EIF-5) sp|P48724; similar to ESTs emb|F19992,
gb|N96933, emb|Z33699, emb|F19991
Length = 437
Score = 37.4 bits (85), Expect = 0.003
Identities = 30/111 (27%), Positives = 51/111 (45%), Gaps = 13/111 (11%)
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
K +D + + K R EL + E+R +K K+ S+ +D S + + D+
Sbjct: 151 KVSKDKKAMRKAEKERLKEGELADEEQRKLKAKKKALSNGKDSKTSKN---HSSDEDI-- 205
Query: 102 HIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAE---LEQIKKERAE 149
S D++ +EV DE+ DD + + DT AE +EQ+ + AE
Sbjct: 206 -----SPKHDENALEVDEDEDDDDGVEWQTDTSREAAEKRMMEQLSAKTAE 251
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.306 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,958,582
Number of Sequences: 26719
Number of extensions: 198068
Number of successful extensions: 5748
Number of sequences better than 10.0: 693
Number of HSP's better than 10.0 without gapping: 434
Number of HSP's successfully gapped in prelim test: 265
Number of HSP's that attempted gapping in prelim test: 1948
Number of HSP's gapped (non-prelim): 1913
length of query: 153
length of database: 11,318,596
effective HSP length: 90
effective length of query: 63
effective length of database: 8,913,886
effective search space: 561574818
effective search space used: 561574818
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0281a.2


