

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
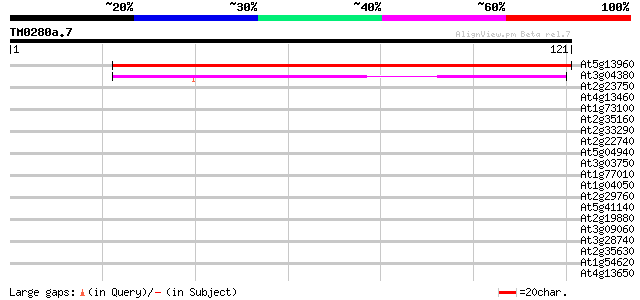
Query= TM0280a.7
(121 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g13960 unknown protein (At5g13960) 73 3e-14
At3g04380 Su(VAR)3-9-related protein 4 38 7e-04
At2g23750 putative SET-domain transcriptional regulator 35 0.005
At4g13460 putative protein 33 0.023
At1g73100 putative protein 32 0.038
At2g35160 similar to mammalian MHC III region protein G9a 32 0.050
At2g33290 similar to mammalian MHC III region protein G9a 32 0.050
At2g22740 similar to mammalian MHC III region protein G9a 32 0.066
At5g04940 SET-domain protein-like 29 0.33
At3g03750 unknown protein 29 0.33
At1g77010 29 0.43
At1g04050 hypothetical protein 28 0.56
At2g29760 hypothetical protein 27 1.2
At5g41140 putative protein 27 1.6
At2g19880 unknown protein 27 1.6
At3g09060 hypothetical protein 27 2.1
At3g28740 cytochrome P450 like protein 26 2.8
At2g35630 similar to ch-TOG protein from Homo sapiens 26 2.8
At1g54620 26 2.8
At4g13650 unknown protein 26 3.6
>At5g13960 unknown protein (At5g13960)
Length = 624
Score = 72.8 bits (177), Expect = 3e-14
Identities = 38/99 (38%), Positives = 61/99 (61%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR SQK L++ L+V+ KGWAV + ++I G+PV EY+ +R+ ++ S +E FE
Sbjct: 437 NRTSQKRLRFNLEVFRSAKKGWAVRSWEYIPAGSPVCEYIGVVRRTADVDTISDNEYIFE 496
Query: 83 IDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETME 121
IDC + + + GR R L +V++P + V +S +DE E
Sbjct: 497 IDCQQTMQGLGGRQRRLRDVAVPMNNGVSQSSEDENAPE 535
>At3g04380 Su(VAR)3-9-related protein 4
Length = 492
Score = 38.1 bits (87), Expect = 7e-04
Identities = 28/99 (28%), Positives = 45/99 (45%), Gaps = 16/99 (16%)
Query: 23 NRISQKDLQYCLQVYL-KISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGF 81
NR+ Q+ ++ LQVY + KGW + T + GT + EY+ E+ N EL +V
Sbjct: 293 NRVVQRGIRCQLQVYFTQEGKGWGLRTLQDLPKGTFICEYIGEILTNTELYDRNVR---- 348
Query: 82 EIDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETM 120
R + V+L A + E+ + DEE +
Sbjct: 349 -----------SSSERHTYPVTLDADWGSEKDLKDEEAL 376
>At2g23750 putative SET-domain transcriptional regulator
Length = 203
Score = 35.4 bits (80), Expect = 0.005
Identities = 17/43 (39%), Positives = 26/43 (59%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR+ Q ++ L+V+ SKGW + + IL GT V EY+ E+
Sbjct: 36 NRVLQNGIRAKLEVFRTESKGWGLRACEHILRGTFVCEYIGEV 78
>At4g13460 putative protein
Length = 650
Score = 33.1 bits (74), Expect = 0.023
Identities = 14/39 (35%), Positives = 23/39 (58%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEY 61
NR++QK L+ L+V+ + GW V + D + G + EY
Sbjct: 484 NRVTQKGLRNRLEVFRSLETGWGVRSLDVLHAGAFICEY 522
>At1g73100 putative protein
Length = 669
Score = 32.3 bits (72), Expect = 0.038
Identities = 15/49 (30%), Positives = 28/49 (56%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVEL 71
NR+ Q L+ L+V+ ++GW + + D + G+ + EY E++ N L
Sbjct: 485 NRVIQTGLKSRLEVFKTRNRGWGLRSWDSLRAGSFICEYAGEVKDNGNL 533
>At2g35160 similar to mammalian MHC III region protein G9a
Length = 794
Score = 32.0 bits (71), Expect = 0.050
Identities = 14/42 (33%), Positives = 25/42 (59%)
Query: 24 RISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
R+SQ ++ L+++ S+GW V + + I G+ + EY EL
Sbjct: 639 RVSQHGIKIKLEIFKTESRGWGVRSLESIPIGSFICEYAGEL 680
>At2g33290 similar to mammalian MHC III region protein G9a
Length = 651
Score = 32.0 bits (71), Expect = 0.050
Identities = 15/39 (38%), Positives = 22/39 (55%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEY 61
+R++QK L+ L+V+ GW V T D I G + EY
Sbjct: 486 SRVTQKGLRNRLEVFRSKETGWGVRTLDLIEAGAFICEY 524
>At2g22740 similar to mammalian MHC III region protein G9a
Length = 790
Score = 31.6 bits (70), Expect = 0.066
Identities = 15/47 (31%), Positives = 27/47 (56%)
Query: 24 RISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVE 70
R++Q ++ L+++ S+GW V I G+ + EYV EL ++ E
Sbjct: 608 RVTQHGIKLPLEIFKTKSRGWGVRCLKSIPIGSFICEYVGELLEDSE 654
>At5g04940 SET-domain protein-like
Length = 670
Score = 29.3 bits (64), Expect = 0.33
Identities = 16/68 (23%), Positives = 35/68 (50%), Gaps = 1/68 (1%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
N+++Q ++ L+V+ ++GW + + D I G+ + YV E K+ V +++ +
Sbjct: 486 NKVTQMGVKVRLEVFKTANRGWGLRSWDAIRAGSFICIYVGE-AKDKSKVQQTMANDDYT 544
Query: 83 IDCLKIIN 90
D + N
Sbjct: 545 FDTTNVYN 552
>At3g03750 unknown protein
Length = 354
Score = 29.3 bits (64), Expect = 0.33
Identities = 14/43 (32%), Positives = 21/43 (48%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR++QK + L++ KGW + I G + EY EL
Sbjct: 182 NRVTQKGVSVSLKIVRDEKKGWCLYADQLIKQGQFICEYAGEL 224
>At1g77010
Length = 695
Score = 28.9 bits (63), Expect = 0.43
Identities = 14/50 (28%), Positives = 25/50 (50%)
Query: 72 VIDSVSECGFEIDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETME 121
++D S+CG ++ K+ +EVE + +L N + F R D + E
Sbjct: 359 LLDMYSKCGSPMEACKLFSEVESYDTILLNSMIKVYFSCGRIDDAKRVFE 408
>At1g04050 hypothetical protein
Length = 468
Score = 28.5 bits (62), Expect = 0.56
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query: 23 NRISQKDLQYCLQVYLKIS-KGWAVNTRDFILFGTPVLEYVKELRKNVEL 71
NR+ Q+ + LQV+ + KGW + T + + G + EY+ E+ EL
Sbjct: 320 NRVVQRGMHNKLQVFFTPNGKGWGLRTLEKLPKGAFICEYIGEILTIPEL 369
>At2g29760 hypothetical protein
Length = 738
Score = 27.3 bits (59), Expect = 1.2
Identities = 14/50 (28%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query: 54 FGTPVLEYVKELRKNVEL-----VIDSVSECGFEIDCLKIINEVEGRNRL 98
FG V Y++E R NV L ++D ++CG D ++ + +E ++ +
Sbjct: 250 FGRQVCSYIEENRVNVNLTLANAMLDMYTKCGSIEDAKRLFDAMEEKDNV 299
>At5g41140 putative protein
Length = 983
Score = 26.9 bits (58), Expect = 1.6
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 2/63 (3%)
Query: 59 LEYVKELRKNVELVIDSVSECGFE-IDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDE 117
L+Y K+L N+ L + E E I ++ + +EG+ R V LP ER+ ++
Sbjct: 385 LDYEKDLNSNLRLQLQKTQESNTELILAVQDLEAMEGQ-RTKKTVDLPGPRTCERNTEES 443
Query: 118 ETM 120
M
Sbjct: 444 RRM 446
>At2g19880 unknown protein
Length = 519
Score = 26.9 bits (58), Expect = 1.6
Identities = 14/54 (25%), Positives = 28/54 (50%), Gaps = 1/54 (1%)
Query: 32 YCLQVY-LKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFEID 84
YC+ Y + S G+A R F L+G ++ + + R++ V+ + + G+ D
Sbjct: 222 YCIYEYHMPCSMGFATGGRTFFLWGGCMMMHADDFRQDRYGVVSGLRDGGYSDD 275
>At3g09060 hypothetical protein
Length = 687
Score = 26.6 bits (57), Expect = 2.1
Identities = 18/57 (31%), Positives = 27/57 (46%), Gaps = 3/57 (5%)
Query: 67 KNVELVIDSVSECGFEIDCLKIINEVEGRNR---LLHNVSLPASFCVERSVDDEETM 120
K ++I +S+CG DCLKI ++ R L SL C +VD E++
Sbjct: 256 KTHNIMISGLSKCGRVDDCLKIWERMKQNEREKDLYTYSSLIHGLCDAGNVDKAESV 312
>At3g28740 cytochrome P450 like protein
Length = 509
Score = 26.2 bits (56), Expect = 2.8
Identities = 18/65 (27%), Positives = 30/65 (45%), Gaps = 4/65 (6%)
Query: 40 ISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFEIDCLKIINEVEGRNRLL 99
+ G A N D+ P+L YV K+V+ + V E + K + +V+G +
Sbjct: 225 VVSGGAGNAADYF----PILRYVTNYEKHVKKLAGRVDEFLQSLVNEKRVEKVKGNTMID 280
Query: 100 HNVSL 104
H +SL
Sbjct: 281 HLLSL 285
>At2g35630 similar to ch-TOG protein from Homo sapiens
Length = 2021
Score = 26.2 bits (56), Expect = 2.8
Identities = 10/39 (25%), Positives = 24/39 (60%)
Query: 58 VLEYVKELRKNVELVIDSVSECGFEIDCLKIINEVEGRN 96
+LE +KE +++V + + ++ CL +++ +EG+N
Sbjct: 372 LLEKLKEKKQSVTDPLTQTLQTMYKAGCLNLVDVIEGKN 410
>At1g54620
Length = 182
Score = 26.2 bits (56), Expect = 2.8
Identities = 28/101 (27%), Positives = 46/101 (44%), Gaps = 22/101 (21%)
Query: 21 FDNRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVK---------ELRKNVEL 71
+ R+S+K LQ CL++Y S+G+ T L+Y+K L+K E
Sbjct: 93 YKGRLSKKLLQKCLKLY---SEGYDAL--------TSALKYIKVRDLFKVSVHLQKAKE- 140
Query: 72 VIDSVSECGFEIDCLKIINEVEGRNRLLHNVSLPASFCVER 112
S+ E GF D +I + + L +++P SF + R
Sbjct: 141 -APSICEMGFNGDNKQISPMKKENDVLFDVINIPYSFNMNR 180
>At4g13650 unknown protein
Length = 1024
Score = 25.8 bits (55), Expect = 3.6
Identities = 15/53 (28%), Positives = 25/53 (46%), Gaps = 11/53 (20%)
Query: 60 EYVKELRKNVELVIDSVSECGFEIDCLKIINEVEGRNR----LLHNVSLPASF 108
EY ++L K SE G+ DC ++NE++ + +H+ L SF
Sbjct: 919 EYFQDLTKRA-------SEIGYVQDCFSLLNELQHEQKDPIIFIHSEKLAISF 964
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.148 0.478
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,560,942
Number of Sequences: 26719
Number of extensions: 94667
Number of successful extensions: 379
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 361
Number of HSP's gapped (non-prelim): 32
length of query: 121
length of database: 11,318,596
effective HSP length: 97
effective length of query: 24
effective length of database: 8,726,853
effective search space: 209444472
effective search space used: 209444472
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0280a.7


