

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0276.6
(483 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
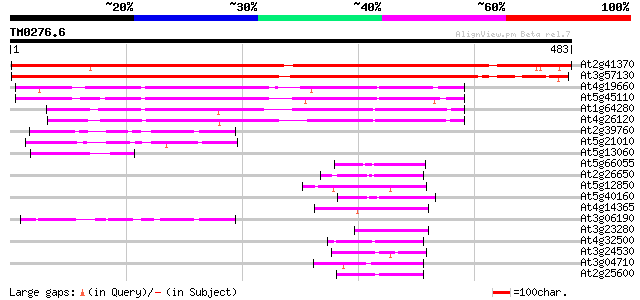
Score E
Sequences producing significant alignments: (bits) Value
At2g41370 unknown protein (At2g41370) 761 0.0
At3g57130 putative protein 713 0.0
At4g19660 putative protein 164 1e-40
At5g45110 regulatory protein NPR1-like; transcription factor inh... 154 1e-37
At1g64280 ranscription factor inhibitor I kappa B-like protein 134 1e-31
At4g26120 NPR1 like protein 120 2e-27
At2g39760 unknown protein 50 3e-06
At5g21010 Unknown protein 48 1e-05
At5g13060 unknown protein 48 1e-05
At5g66055 unknown protein 47 2e-05
At2g26650 K+ transporter, AKT1 46 4e-05
At5g12850 zinc finger transcription factor -like protein 46 5e-05
At5g40160 ankyrin repeat protein EMB506 45 7e-05
At4g14365 C3HC4-type zinc finger ankyrin repeat protein - like 45 7e-05
At3g06190 unknown protein 45 1e-04
At3g23280 unknown protein 44 2e-04
At4g32500 potassium channel - protein 44 3e-04
At3g24530 unknown protein 43 3e-04
At3g04710 ankyrin-like protein 43 4e-04
At2g25600 potassium transporter/channel like protein 42 6e-04
>At2g41370 unknown protein (At2g41370)
Length = 491
Score = 761 bits (1964), Expect = 0.0
Identities = 394/502 (78%), Positives = 426/502 (84%), Gaps = 33/502 (6%)
Query: 2 SLEDSLRSLSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPP 61
+LE+SLRSLSLD+LNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCG D P
Sbjct: 3 NLEESLRSLSLDFLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGTDSPQ 62
Query: 62 PSGNLDS------PGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPR 115
P +D P P S P G+IPVNSVGYEVFLL+LQFLYSGQVSIVPQKHEPR
Sbjct: 63 PVTGIDPTQHGSVPASPTRGSTAPAGIIPVNSVGYEVFLLLLQFLYSGQVSIVPQKHEPR 122
Query: 116 PNCGERACWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLL 175
PNCGER CWHTHC++AVDLALDTLAA+RYFGVEQLALLTQKQLASMVEKASIEDVMKVL+
Sbjct: 123 PNCGERGCWHTHCSAAVDLALDTLAASRYFGVEQLALLTQKQLASMVEKASIEDVMKVLI 182
Query: 176 ASRKQDMHQLWTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHH 235
ASRKQDMHQLWTTCSHLVAKSGLPPE+LAKHLPID+V KIEELRLKS++ARRSL+PH+HH
Sbjct: 183 ASRKQDMHQLWTTCSHLVAKSGLPPEILAKHLPIDVVTKIEELRLKSSIARRSLMPHNHH 242
Query: 236 HHHHHGHHDMGAAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENC 295
H D+ A DLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDE+LAL YAVE+C
Sbjct: 243 H-------DLSVAQDLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDESLALHYAVESC 295
Query: 296 SREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTP 355
SREVVKALLELGAADVNYP+GP+GKTPLHIAAEMVSPDMVAVLLDHHADPNVRTV G+TP
Sbjct: 296 SREVVKALLELGAADVNYPAGPAGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVGGITP 355
Query: 356 LDILRTLTSDFLFKGAVPGLTHIEPNKLRLCLELVQSAALVMSREEGNANANSSNNNNAP 415
LDILRTLTSDFLFKGAVPGLTHIEPNKLRLCLELVQSAA+V+SREEGN + N +N+NN
Sbjct: 356 LDILRTLTSDFLFKGAVPGLTHIEPNKLRLCLELVQSAAMVISREEGNNSNNQNNDNN-- 413
Query: 416 CSAATPIYPPMNEDHNS-SSSNANLNLDSRLVYLNLGA--AQM--SGDDDSSHGSHREAM 470
T IYP MNE+HNS SS +N NLDSRLVYLNLGA QM D H S RE M
Sbjct: 414 ----TGIYPHMNEEHNSGSSGGSNNNLDSRLVYLNLGAGTGQMGPGRDQGDDHNSQREGM 469
Query: 471 NQ---------AMYHHHHSHDY 483
++ MYHHHH H +
Sbjct: 470 SRHHHHHQDPSTMYHHHHQHHF 491
>At3g57130 putative protein
Length = 467
Score = 713 bits (1841), Expect = 0.0
Identities = 372/488 (76%), Positives = 406/488 (82%), Gaps = 33/488 (6%)
Query: 2 SLEDSLRSLSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPP 61
+ E+SL+S+SLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFC DP
Sbjct: 4 TFEESLKSMSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCESDPSQ 63
Query: 62 PSGNLDSPGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGER 121
P + G + GGVIPVNSVGYEVFLL+LQFLYSGQVSIVP KHEPR NCG+R
Sbjct: 64 PGAEPANQTGSGARAAAVGGVIPVNSVGYEVFLLLLQFLYSGQVSIVPHKHEPRSNCGDR 123
Query: 122 ACWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQD 181
CWHTHCT+AVDL+LD LAAARYFGVEQLALLTQK L SMVEKASIEDVMKVL+ASRKQD
Sbjct: 124 GCWHTHCTAAVDLSLDILAAARYFGVEQLALLTQKHLTSMVEKASIEDVMKVLIASRKQD 183
Query: 182 MHQLWTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHG 241
MHQLWTTCS+L+AKSGLP E+LAKHLPI++VAKIEELRLKS++ RSL+PH
Sbjct: 184 MHQLWTTCSYLIAKSGLPQEILAKHLPIELVAKIEELRLKSSMPLRSLMPH--------- 234
Query: 242 HHDMGAAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVVK 301
HHD+ + DLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDE+LAL YAVENCSREVVK
Sbjct: 235 HHDLTSTLDLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDESLALIYAVENCSREVVK 294
Query: 302 ALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILRT 361
ALLELGAADVNYP+GP+GKT LHIAAEMVSPDMVAVLLDHHADPNV+TVDG+TPLDILRT
Sbjct: 295 ALLELGAADVNYPAGPTGKTALHIAAEMVSPDMVAVLLDHHADPNVQTVDGITPLDILRT 354
Query: 362 LTSDFLFKGAVPGLTHIEPNKLRLCLELVQSAALVMSREEGNANANSSNNNNAPCSAATP 421
LTSDFLFKGA+PGLTHIEPNKLRLCLELVQSAALV+SREEGN N+SN+NN T
Sbjct: 355 LTSDFLFKGAIPGLTHIEPNKLRLCLELVQSAALVISREEGN---NNSNDNN------TM 405
Query: 422 IYPPMNEDHNSSSSNANLNLDSRLVYLNLGAAQMSGDDDSSHGSHREAMN--------QA 473
IYP M ++H S SS LDSRLVYLNLGA DD+S + RE MN
Sbjct: 406 IYPRMKDEHTSGSS-----LDSRLVYLNLGATNRDIGDDNS--NQREGMNLHHHHHDPST 458
Query: 474 MYHHHHSH 481
MYHHHH H
Sbjct: 459 MYHHHHHH 466
>At4g19660 putative protein
Length = 601
Score = 164 bits (414), Expect = 1e-40
Identities = 122/392 (31%), Positives = 194/392 (49%), Gaps = 46/392 (11%)
Query: 6 SLRSLSLDYLNLLINGQAF---SDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPP 62
+L LS + LL N +++ E V HRC+LAARS FF F
Sbjct: 34 NLEELSSNLEQLLTNPDCDYTDAEIIIEEEANPVSVHRCVLAARSKFFLDLF-------- 85
Query: 63 SGNLDSPGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERA 122
D + + ++P +VG E FL L ++Y+G++ P + C +
Sbjct: 86 --KKDKDSSEKKPKYQMKDLLPYGNVGREAFLHFLSYIYTGRLKPFPIEVS---TCVDSV 140
Query: 123 CWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDM 182
C H C A+D A++ + A+ F + L Q++L + VEK+ +E+V+ +LL + D+
Sbjct: 141 CAHDSCKPAIDFAVELMYASFVFQIPDLVSSFQRKLRNYVEKSLVENVLPILLVAFHCDL 200
Query: 183 HQLWTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGH 242
QL C VA+S L + K LP++++ KI++LR+KS IP
Sbjct: 201 TQLLDQCIERVARSDLDRFCIEKELPLEVLEKIKQLRVKSV-----NIP----------- 244
Query: 243 HDMGAAADLEDQKIRR---MRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREV 299
++ED+ I R + +ALDS DVELVKL++ + LD+A L YAV +V
Sbjct: 245 -------EVEDKSIERTGKVLKALDSDDVELVKLLLTESDITLDQANGLHYAVAYSDPKV 297
Query: 300 VKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDIL 359
V +L+L ADVN+ + G T LHIAA P ++ L+ A+ + T DG + ++I
Sbjct: 298 VTQVLDLDMADVNFRNS-RGYTVLHIAAMRREPTIIIPLIQKGANASDFTFDGRSAVNIC 356
Query: 360 RTLTSDFLFKGAVPGLTHIEPNKLRLCLELVQ 391
R LT K + EP+K RLC+++++
Sbjct: 357 RRLTRP---KDYHTKTSRKEPSKYRLCIDILE 385
>At5g45110 regulatory protein NPR1-like; transcription factor
inhibitor I kappa B-like
Length = 586
Score = 154 bits (388), Expect = 1e-37
Identities = 119/392 (30%), Positives = 190/392 (48%), Gaps = 42/392 (10%)
Query: 6 SLRSLSLDYLNLLINGQA-FSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSG 64
SL LS + LL N +SD V+G V HRCILAARS FF+ F
Sbjct: 40 SLTKLSSNLEQLLSNSDCDYSDAEIIVDGVPVGVHRCILAARSKFFQDLF------KKEK 93
Query: 65 NLDSPGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACW 124
+ P+ ++P +V +E FL L ++Y+G++ P + C + C
Sbjct: 94 KISKTEKPKYQLRE---MLPYGAVAHEAFLYFLSYIYTGRLKPFPLEVS---TCVDPVCS 147
Query: 125 HTHCTSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQ 184
H C A+D + + A+ V +L Q++L + VEK +E+V+ +L+ + + Q
Sbjct: 148 HDCCRPAIDFVVQLMYASSVLQVPELVSSFQRRLCNFVEKTLVENVLPILMVAFNCKLTQ 207
Query: 185 LWTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHHD 244
L C VA+S L + K +P ++ KI++LRL S D
Sbjct: 208 LLDQCIERVARSDLYRFCIEKEVPPEVAEKIKQLRLISP-------------------QD 248
Query: 245 MGAAADLED---QKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVVK 301
+ + + ++I ++ +ALDS DVELVKL++ + LD+A L Y+V +VV
Sbjct: 249 EETSPKISEKLLERIGKILKALDSDDVELVKLLLTESDITLDQANGLHYSVVYSDPKVVA 308
Query: 302 ALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILRT 361
+L L DVNY + G T LH AA P ++ L+D A+ + T DG + ++ILR
Sbjct: 309 EILALDMGDVNYRNS-RGYTVLHFAAMRREPSIIISLIDKGANASEFTSDGRSAVNILRR 367
Query: 362 LTS--DFLFKGAVPGLTHIEPNKLRLCLELVQ 391
LT+ D+ K A E +K RLC+++++
Sbjct: 368 LTNPKDYHTKTA----KGRESSKARLCIDILE 395
>At1g64280 ranscription factor inhibitor I kappa B-like protein
Length = 593
Score = 134 bits (337), Expect = 1e-31
Identities = 103/362 (28%), Positives = 164/362 (44%), Gaps = 37/362 (10%)
Query: 32 EGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSPRPGGVIPVNSVGYE 91
+GR V HRC+L+ARS FF+ S N + + VG++
Sbjct: 73 DGREVSFHRCVLSARSSFFKSALAAAKKEKDSNNTAAV------KLELKEIAKDYEVGFD 126
Query: 92 VFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDTLAAARYFGVEQLA 151
+ +L ++YS +V P+ C + C H C AVD L+ L A F + +L
Sbjct: 127 SVVTVLAYVYSSRVRPPPKGVS---ECADENCCHVACRPAVDFMLEVLYLAFIFKIPELI 183
Query: 152 LLTQKQLASMVEKASIEDVMKVLLASR--KQDMHQLWTTCSHLVAKSGLPPEVLAKHLPI 209
L Q+ L +V+K IED + +L + + +L C ++ KS + L K LP
Sbjct: 184 TLYQRHLLDVVDKVVIEDTLVILKLANICGKACMKLLDRCKEIIVKSNVDMVSLEKSLPE 243
Query: 210 DIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRMRRALDSSDVE 269
++V +I + R ++G + + + +ALDS D+E
Sbjct: 244 ELVKEIIDRR-----------------------KELGLEVPKVKKHVSNVHKALDSDDIE 280
Query: 270 LVKLMVMGEGLNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEM 329
LVKL++ + NLD+A AL +AV C+ + LL+L ADVN+ P G T LH+AA
Sbjct: 281 LVKLLLKEDHTNLDDACALHFAVAYCNVKTATDLLKLDLADVNH-RNPRGYTVLHVAAMR 339
Query: 330 VSPDMVAVLLDHHADPNVRTVDGVTPLDILRTLTSDFLFKGAVPGLTHIEPNKLRLCLEL 389
P ++ LL+ A + T++G T L I + T H K RLC+E+
Sbjct: 340 KEPQLILSLLEKGASASEATLEGRTALMIAKQATMAVECNNIPEQCKH--SLKGRLCVEI 397
Query: 390 VQ 391
++
Sbjct: 398 LE 399
>At4g26120 NPR1 like protein
Length = 600
Score = 120 bits (300), Expect = 2e-27
Identities = 102/361 (28%), Positives = 163/361 (44%), Gaps = 40/361 (11%)
Query: 33 GRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSPRPGGVIPVNSVGYEV 92
GR V HRCIL+AR F+ S + + + VG++
Sbjct: 76 GREVSFHRCILSARIPVFKSALATVKEQKSSTTVKL---------QLKEIARDYEVGFDS 126
Query: 93 FLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDTLAAARYFGVEQLAL 152
+ +L ++YSG+V P+ C + C H C S VD ++ L + F +++L
Sbjct: 127 VVAVLAYVYSGRVRSPPKGASA---CVDDDCCHVACRSKVDFMVEVLYLSFVFQIQELVT 183
Query: 153 LTQKQLASMVEKASIEDVMKVLLASRK--QDMHQLWTTCSHLVAKSGLPPEVLAKHLPID 210
L ++Q +V+K +ED++ + +L C ++ KS + L K LP
Sbjct: 184 LYERQFLEIVDKVVVEDILVIFKLDTLCGTTYKKLLDRCIEIIVKSDIELVSLEKSLPQH 243
Query: 211 IVAKIEELRLKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRMRRALDSSDVEL 270
I +I ++R L L H ++ + +ALDS DVEL
Sbjct: 244 IFKQIIDIREALCLEPPKLERH-----------------------VKNIYKALDSDDVEL 280
Query: 271 VKLMVMGEGLNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMV 330
VK++++ NLDEA AL +A+ +C+ + LLEL ADVN P G T LH+AA
Sbjct: 281 VKMLLLEGHTNLDEAYALHFAIAHCAVKTAYDLLELELADVNL-RNPRGYTVLHVAAMRK 339
Query: 331 SPDMVAVLLDHHADPNVRTVDGVTPLDILRTLTSDFLFKGAVPGLTHIEPNKLRLCLELV 390
P ++ LL A+ T+DG T L I++ LT +K + T K LC+E++
Sbjct: 340 EPKLIISLLMKGANILDTTLDGRTALVIVKRLTKADDYKTSTEDGT--PSLKGGLCIEVL 397
Query: 391 Q 391
+
Sbjct: 398 E 398
>At2g39760 unknown protein
Length = 408
Score = 50.1 bits (118), Expect = 3e-06
Identities = 46/177 (25%), Positives = 74/177 (40%), Gaps = 31/177 (17%)
Query: 18 LINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSP 77
L++ + D+ F V AH+ ILAARS FR F G P + N+D
Sbjct: 187 LLDSEVGCDIAFQVGDETYKAHKLILAARSPVFRAQFFG---PIGNNNVDR--------- 234
Query: 78 RPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALD 137
I ++ + +F ML F+Y+ ++P HE T +S ++
Sbjct: 235 -----IVIDDIEPSIFKAMLSFIYT---DVLPNVHE--------ITGSTSASSFTNMIQH 278
Query: 138 TLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVA 194
LAAA + + +L +L + L EK +++V L + + QL C VA
Sbjct: 279 LLAAADLYDLARLKILCEVLLC---EKLDVDNVATTLALAEQHQFLQLKAFCLEFVA 332
>At5g21010 Unknown protein
Length = 410
Score = 48.1 bits (113), Expect = 1e-05
Identities = 48/189 (25%), Positives = 85/189 (44%), Gaps = 38/189 (20%)
Query: 14 YLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFR-KFFCGPDPPPPSGNLDSPGGP 72
+ +L++ SD+TF++ G AH+ +LAARS FF+ KFF + + N +
Sbjct: 187 HFGVLLDSMEGSDITFNIAGEKFLAHKLVLAARSPFFKSKFFSEFE----ANNTE----- 237
Query: 73 RVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAV 132
+ +N + +VF +LQF+Y +P+ EP A HT +
Sbjct: 238 ----------VTINDLEPKVFKALLQFMYKDS---LPEDVEP-------ATAHTFERLKL 277
Query: 133 D-----LALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWT 187
L + LAAA + + +L LL + + V S++ V K+L + + + +L
Sbjct: 278 SEIYETLIVKVLAAADKYDLIRLRLLCESHICKGV---SVKSVAKILALADRYNAKELKG 334
Query: 188 TCSHLVAKS 196
C A++
Sbjct: 335 VCLKFTAEN 343
>At5g13060 unknown protein
Length = 709
Score = 48.1 bits (113), Expect = 1e-05
Identities = 28/89 (31%), Positives = 46/89 (51%), Gaps = 17/89 (19%)
Query: 19 INGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSPR 78
+N SDVTF ++G+ +AH+ L A S FR F G + N++ P
Sbjct: 534 VNNPTMSDVTFLIDGKQFYAHKIGLVASSDIFRAMFDGLYKERNAQNVEIP--------- 584
Query: 79 PGGVIPVNSVGYEVFLLMLQFLYSGQVSI 107
++ +EVF LM++F+YSG+++I
Sbjct: 585 --------NIRWEVFELMMKFIYSGRINI 605
>At5g66055 unknown protein
Length = 435
Score = 47.4 bits (111), Expect = 2e-05
Identities = 31/79 (39%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Query: 280 LNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLL 339
L+ + A + YAV+ S +K LL L AD+N G TPLH+A + D+V +LL
Sbjct: 322 LDDEGATLMHYAVQTASAPTIKLLL-LYNADIN-AQDRDGWTPLHVAVQARRSDIVKLLL 379
Query: 340 DHHADPNVRTVDGVTPLDI 358
AD V+ DG+TPL +
Sbjct: 380 IKGADIEVKNKDGLTPLGL 398
>At2g26650 K+ transporter, AKT1
Length = 857
Score = 46.2 bits (108), Expect = 4e-05
Identities = 32/89 (35%), Positives = 50/89 (55%), Gaps = 5/89 (5%)
Query: 268 VELVKLMVMGEGLNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAA 327
+E+ ++ G+ +D L L +A ++ LL+ G D N S +G+TPLHIAA
Sbjct: 505 LEIENMLARGK---MDLPLNLCFAAIREDDLLLHQLLKRGL-DPN-ESDNNGRTPLHIAA 559
Query: 328 EMVSPDMVAVLLDHHADPNVRTVDGVTPL 356
+ + V +LL++HADPN R +G PL
Sbjct: 560 SKGTLNCVLLLLEYHADPNCRDAEGSVPL 588
Score = 31.2 bits (69), Expect = 1.3
Identities = 27/92 (29%), Positives = 42/92 (45%), Gaps = 3/92 (3%)
Query: 269 ELVKLMVMGEGLNLDEALALPYAVENCSREVVKALLE--LGAADVNYPSGPSGKTPLHIA 326
E V +++ G +D +A + +K L E L DV P +G + LH A
Sbjct: 597 EKVVKVLLEHGSTIDAGDVGHFACTAAEQGNLKLLKEIVLHGGDVTRPRA-TGTSALHTA 655
Query: 327 AEMVSPDMVAVLLDHHADPNVRTVDGVTPLDI 358
+ +MV LL+ AD N + + G TP D+
Sbjct: 656 VCEENIEMVKYLLEQGADVNKQDMHGWTPRDL 687
>At5g12850 zinc finger transcription factor -like protein
Length = 706
Score = 45.8 bits (107), Expect = 5e-05
Identities = 38/117 (32%), Positives = 54/117 (45%), Gaps = 12/117 (10%)
Query: 253 DQKIRRMRRALDSSDVELVKLMVMG-------EGLNLDEALALPYAVENCSREVVKALLE 305
D + RR L SDV + M + + L++ L A S +VVK +L
Sbjct: 48 DNDVEGFRRQL--SDVSCINQMGLWYRRQRFVRRMVLEQRTPLMVASLYGSLDVVKFILS 105
Query: 306 LGAADVNYPSGPSGKTPLHIA---AEMVSPDMVAVLLDHHADPNVRTVDGVTPLDIL 359
A++N GP T LH A A + S D+V +LL ADPN+ G P+D+L
Sbjct: 106 FPEAELNLSCGPDKSTALHCAASGASVNSLDVVKLLLSVGADPNIPDAHGNRPVDVL 162
>At5g40160 ankyrin repeat protein EMB506
Length = 315
Score = 45.4 bits (106), Expect = 7e-05
Identities = 31/84 (36%), Positives = 42/84 (49%), Gaps = 2/84 (2%)
Query: 283 DEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHH 342
D A + YAV+ + + VK L + DVN G TPLHIA + + D+ +LL +
Sbjct: 217 DGAAPIHYAVQVGALQTVKLLFKYNV-DVNVADN-EGWTPLHIAVQSRNRDITKILLTNG 274
Query: 343 ADPNVRTVDGVTPLDILRTLTSDF 366
AD RT DG LD+ DF
Sbjct: 275 ADKTRRTKDGKLALDLALCFGRDF 298
>At4g14365 C3HC4-type zinc finger ankyrin repeat protein - like
Length = 376
Score = 45.4 bits (106), Expect = 7e-05
Identities = 37/103 (35%), Positives = 53/103 (50%), Gaps = 5/103 (4%)
Query: 263 LDSSDVELVK-LMVMGEGLNLDEALALPYAVENCSRE----VVKALLELGAADVNYPSGP 317
+ +++VE +K L G GL + L + C+ + V K LLELG+ Y SG
Sbjct: 17 VSNNNVEGIKSLHHEGAGLEGVDKLGRTPLILACTNDDLYDVAKTLLELGSNVNAYRSGC 76
Query: 318 SGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLDILR 360
+G TPLH AA+ V +LL H A+P V D T L++ R
Sbjct: 77 NGGTPLHHAAKRGLVHTVKLLLSHGANPLVLDDDVKTALEVAR 119
>At3g06190 unknown protein
Length = 406
Score = 44.7 bits (104), Expect = 1e-04
Identities = 47/185 (25%), Positives = 77/185 (41%), Gaps = 32/185 (17%)
Query: 10 LSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSP 69
L + LL +G+ +DVTF V+G AH+ +LAARS FR GP
Sbjct: 188 LGQQFGKLLESGKG-ADVTFEVDGETFPAHKLVLAARSAVFRAQLFGP------------ 234
Query: 70 GGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCT 129
+ S +I + V +F ++L F+Y ++ + + G W
Sbjct: 235 ----LRSENTNCII-IEDVQAPIFKMLLHFIYWDEMP------DMQDLIGTDLKW----- 278
Query: 130 SAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTC 189
++ +A LAAA + +E+L + + +L E SI V L + + QL C
Sbjct: 279 ASTLVAQHLLAAADRYALERLRTICESKLC---EGISINTVATTLALAEQHHCFQLKAAC 335
Query: 190 SHLVA 194
+A
Sbjct: 336 LKFIA 340
>At3g23280 unknown protein
Length = 462
Score = 44.3 bits (103), Expect = 2e-04
Identities = 26/63 (41%), Positives = 35/63 (55%)
Query: 298 EVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPLD 357
+V K L+ELG+ Y G TPLH AA+ + V +LL H A+P V D TPL+
Sbjct: 55 DVAKTLIELGSNVNAYRPGRHAGTPLHHAAKRGLENTVKLLLSHGANPLVLNDDCQTPLE 114
Query: 358 ILR 360
+ R
Sbjct: 115 VAR 117
>At4g32500 potassium channel - protein
Length = 880
Score = 43.5 bits (101), Expect = 3e-04
Identities = 28/83 (33%), Positives = 46/83 (54%), Gaps = 3/83 (3%)
Query: 274 MVMGEGLNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPD 333
+++ +G +D L+L +A ++ LL+ G+ + +G+T LHIAA S
Sbjct: 533 LMLAQG-KMDLPLSLCFAAARGDDLLLHQLLKRGSNPNE--TDKNGRTALHIAASKGSQY 589
Query: 334 MVAVLLDHHADPNVRTVDGVTPL 356
V +LL+H ADPN+R +G PL
Sbjct: 590 CVVLLLEHGADPNIRDSEGSVPL 612
>At3g24530 unknown protein
Length = 481
Score = 43.1 bits (100), Expect = 3e-04
Identities = 34/88 (38%), Positives = 45/88 (50%), Gaps = 10/88 (11%)
Query: 278 EGLNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIA------AEMVS 331
E +N L A +N E K LLE GA S +G TPLH+A A+ +S
Sbjct: 81 EAMNTYGETPLHMAAKNGCNEAAKLLLESGAFIEAKAS--NGMTPLHLAVWYSITAKEIS 138
Query: 332 PDMVAVLLDHHADPNVRTVDGVTPLDIL 359
V LLDH+AD + + +G+TPLD L
Sbjct: 139 T--VKTLLDHNADCSAKDNEGMTPLDHL 164
>At3g04710 ankyrin-like protein
Length = 456
Score = 42.7 bits (99), Expect = 4e-04
Identities = 36/99 (36%), Positives = 47/99 (47%), Gaps = 7/99 (7%)
Query: 262 ALDSSDVELVK-LMVMGEGLNLDEAL---ALPYAVENCSREVVKALLELGAADVNYPSGP 317
A +E VK L+ G N+ L AL +A E++K LL G V S
Sbjct: 96 AARQGQIETVKYLLEQGADPNIASELGATALHHAAGTGEIELLKELLSRG---VPVDSES 152
Query: 318 SGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPL 356
TPL AA + V VLL+H+A+PN T D +TPL
Sbjct: 153 ESGTPLIWAAGHDQKNAVEVLLEHNANPNAETEDNITPL 191
Score = 38.9 bits (89), Expect = 0.006
Identities = 27/76 (35%), Positives = 42/76 (54%), Gaps = 3/76 (3%)
Query: 283 DEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHH 342
D L AV S ++ L++ GA N +G G TPLHIAA++ + +++ LL
Sbjct: 186 DNITPLLSAVAAGSLSCLELLVKAGAK-ANVFAG--GATPLHIAADIGNLELINCLLKAG 242
Query: 343 ADPNVRTVDGVTPLDI 358
ADPN + +G PL++
Sbjct: 243 ADPNQKDEEGNRPLEV 258
Score = 33.5 bits (75), Expect = 0.27
Identities = 30/115 (26%), Positives = 54/115 (46%), Gaps = 13/115 (11%)
Query: 254 QKIRRMRRALDSSDVELVKLMV--MGEGLNLDEAL----------ALPYAVENCSREVVK 301
+K++++ A + ++E +K + + EG +L + + AL +A E+ +
Sbjct: 13 EKVQQILNAACTGNLEFLKNVAKQLDEGKDLTKTVESIKDANKRGALHFAAREGQTEICR 72
Query: 302 ALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPL 356
LLE + + +G TPL AA + V LL+ ADPN+ + G T L
Sbjct: 73 YLLEELKLNAD-AKDETGDTPLVHAARQGQIETVKYLLEQGADPNIASELGATAL 126
>At2g25600 potassium transporter/channel like protein
Length = 888
Score = 42.4 bits (98), Expect = 6e-04
Identities = 27/75 (36%), Positives = 39/75 (52%), Gaps = 2/75 (2%)
Query: 282 LDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDH 341
+D L+L +A ++ LL G++ G+T LHIAA S V +LL+H
Sbjct: 542 MDLPLSLCFAAARGDDLLLHQLLRRGSSPNEMDK--DGRTALHIAASKGSHYCVVLLLEH 599
Query: 342 HADPNVRTVDGVTPL 356
ADPN+R +G PL
Sbjct: 600 GADPNIRDSEGNVPL 614
Score = 30.8 bits (68), Expect = 1.7
Identities = 30/89 (33%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Query: 269 ELVKLMVM-GEGLNLDEALALP-YAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIA 326
E+ KL+ G L+LD AVE + +K +++ G DV P G +G T LH A
Sbjct: 624 EIAKLLAENGAKLSLDSVSYFSGLAVEKNCLDALKDIIKYGG-DVTLPDG-NGTTALHRA 681
Query: 327 AEMVSPDMVAVLLDHHADPNVRTVDGVTP 355
++V LLD AD + G TP
Sbjct: 682 VSEGHLEIVKFLLDQGADLDWPDSYGWTP 710
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,873,745
Number of Sequences: 26719
Number of extensions: 474156
Number of successful extensions: 3927
Number of sequences better than 10.0: 151
Number of HSP's better than 10.0 without gapping: 107
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 2822
Number of HSP's gapped (non-prelim): 596
length of query: 483
length of database: 11,318,596
effective HSP length: 103
effective length of query: 380
effective length of database: 8,566,539
effective search space: 3255284820
effective search space used: 3255284820
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0276.6


