

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269b.9
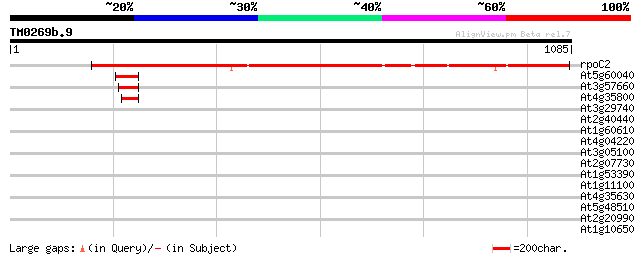
(1085 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2 1187 0.0
At5g60040 DNA-directed RNA polymerase - like protein 48 3e-05
At3g57660 DNA-directed RNA polymerase I 190K chain - like protein 46 1e-04
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 45 2e-04
At3g29740 hypothetical protein 36 0.14
At2g40440 hypothetical protein 32 2.6
At1g60610 unknown protein 32 2.6
At4g04220 putative disease resistance protein, predicted GPI-anc... 31 3.3
At3g05100 unknown protein 31 3.3
At2g07730 putative non-LTR retroelement reverse transcriptase 31 3.3
At1g53390 putative ABC transporter gb|AAD31586.1 31 3.3
At1g11100 transcription factor RUSH-1alpha isolog 31 4.4
At4g35630 phosphoserine aminotransferase 30 5.7
At5g48510 putative protein 30 7.4
At2g20990 unknown protein 30 9.7
At1g10650 similar to putative inhibitor of apoptosis gi|3924605;... 30 9.7
>rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2
Length = 1376
Score = 1187 bits (3071), Expect = 0.0
Identities = 621/958 (64%), Positives = 734/958 (75%), Gaps = 50/958 (5%)
Query: 159 IYIGSRCIVVRNQDIGIGLINRFINFQTQPIFIRTPFTCRNTSWICRLCYGRSPIHGDLV 218
IYIGSRC+ RNQD+GIGL+NR I F TQ I IRTPFTCR+TSWICRLCYGRSP HGDLV
Sbjct: 255 IYIGSRCVAFRNQDLGIGLVNRLITFGTQSISIRTPFTCRSTSWICRLCYGRSPTHGDLV 314
Query: 219 ELGEAVGIIAGQSIGEPGTQLTLITFHTGGVFTGGTAEYVRSPSNGKIKFNENSAYPTRT 278
ELGEAVGIIAGQSIGEPGTQLTL TFHTGGVFTGGTAE+VR+P NGKIKFNE+ +PTRT
Sbjct: 315 ELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAEHVRAPYNGKIKFNEDLVHPTRT 374
Query: 279 RHGHPAFLCYIDLYVTIESNDIMHNVIIPPKSFLLVQNDQYVKSEQIIAEIRAGTYTLNL 338
RHGHPAFLCYIDL V IES DI+H+V IPPKSFLLVQNDQYV+SEQ+IAEIR GTYT +
Sbjct: 375 RHGHPAFLCYIDLSVIIESEDIIHSVTIPPKSFLLVQNDQYVESEQVIAEIREGTYTFHF 434
Query: 339 KEKIRKHIFSDSEGEMHWSTNIYHVSEFAYSNVHILPKTSHLWILSGNSHKSDTVSLSLL 398
KE++RK+I+SDSEGEMHWST++ H EF YSNVH+LPKTSHLWILSG S S + S+
Sbjct: 435 KERVRKYIYSDSEGEMHWSTDVSHAPEFTYSNVHLLPKTSHLWILSGGSCGSSLIRFSIH 494
Query: 399 KDQDQMSTHSLPTAKRNTSNFLVSNNQV-------------------------RLCPDHC 433
KDQDQM+ L +++ S+ V+N+QV L H
Sbjct: 495 KDQDQMNIPFLSAERKSISSLSVNNDQVSQKFFSSDFADPKKLGIYDYSELNGNLGTSHY 554
Query: 434 HFMHPTISPDTSNLLAKKRRNRFIIPFLFRSIRERNNELMPD--ISVEIPIDGIIHKNSI 491
+ ++ I + S+LLAK+RRNRF+IP F+SI+E+ E +P ISVEIPI+GI +NSI
Sbjct: 555 NLIYSAIFHENSDLLAKRRRNRFLIP--FQSIQEQEKEFIPQSGISVEIPINGIFRRNSI 612
Query: 492 LAYFDDPQYRTQSSGIAKYKTIGIHSIFQKEDLIEYRGIREFKPKYQIKVDRFFFIPQEV 551
A+FDDP+YR +SSGI KY T+ SI QKED+IEYRG+++ K KY++KVDRFFFIP+EV
Sbjct: 613 FAFFDDPRYRRKSSGILKYGTLKADSIIQKEDMIEYRGVQKIKTKYEMKVDRFFFIPEEV 672
Query: 552 HILSESSSIMVRNNSIIGVNTPITLNKKSRVGGLVRVEKNKKKIELKIFSGDIHFPGEID 611
HIL ESS+IMV+N SIIGV+T +TLN +S+VGGL+RVEK KK+IELKIFSGDIHFP + D
Sbjct: 673 HILPESSAIMVQNYSIIGVDTRLTLNIRSQVGGLIRVEKKKKRIELKIFSGDIHFPDKTD 732
Query: 612 KISQHSAILIPPEMVKKKILRNQKKKTNWRYIQWITTTKKKYFVLVRPVILYDIADSINL 671
KIS+HS ILIPP KK + KK NW Y+Q IT TKKK+FVLVRPV Y+IADSINL
Sbjct: 733 KISRHSGILIPPGRGKKN-SKESKKFKNWIYVQRITPTKKKFFVLVRPVATYEIADSINL 791
Query: 672 VKLFPQDLFKEWDNLELKVLNFILYGNGKSIRGILDTSIQLVRTCLVLNWNEDEKSSSIE 731
LFPQDLF+E DN++L+V N+ILYGNGK RGI DTSIQLVRTCLVLNW +K+SS+E
Sbjct: 792 ATLFPQDLFREKDNIQLRVFNYILYGNGKPTRGISDTSIQLVRTCLVLNW---DKNSSLE 848
Query: 732 EALASFVEVSTNGLIRYFLRIDLVKSHISYIRKRNDPSSSGLISYNESDRININPFFSIY 791
E A FVEVST GLI+ F+RI LVKSHISYIRKRN+ SGLIS + ++NPF+SI
Sbjct: 849 EVRAFFVEVSTKGLIQDFIRIGLVKSHISYIRKRNNSPDSGLISAD-----HMNPFYSIS 903
Query: 792 KEN--IQQSLSQKHGTIRMLLNRNKENRSFIILSSSNCFQMGPFNNVKYHNGIKEEINQF 849
++ +QQSL Q HGTIRM LNRNKE++S +ILSSSNCF+MGPFN+VK+HN I + I
Sbjct: 904 PKSGILQQSLRQNHGTIRMFLNRNKESQSLLILSSSNCFRMGPFNHVKHHNVINQSI--- 960
Query: 850 KRNHKIPIKISLGPLGVAPQIANFFSFYHLITHNKISSIKKNLQLNKFKETFQVIKYYLM 909
K+N I IK S GPLG A I+NF+SF L+T+N+IS I K QL+ K FQ I YL+
Sbjct: 961 KKNTLITIKNSSGPLGTATPISNFYSFLPLLTYNQISLI-KYFQLDNLKYIFQKINSYLI 1019
Query: 910 DENERIYKPDLYNNIILNPFHLNWDFIHP----NYCEKTFPIISLGQFICENVCIVQTKN 965
DEN I D Y+N++LNPF LNW F+H NYCE+T IISLGQF CENVCI K
Sbjct: 1020 DENGIILNLDPYSNVVLNPFKLNWYFLHQNYHHNYCEETSTIISLGQFFCENVCI--AKK 1077
Query: 966 GPNLKSGQVITVQMDFVGIRLANPYLATPGATIHGHYGEMLYEGDILVTFIYEKSRSGDI 1025
P+LKSGQV+ VQ D IR A PYLATPGA +HGHY E+LYEGD LVTFIYEKSRSGDI
Sbjct: 1078 EPHLKSGQVLIVQRDSAVIRSAKPYLATPGAKVHGHYSEILYEGDTLVTFIYEKSRSGDI 1137
Query: 1026 TQGLPKVEQVLEVRSIDSISMNLEKRIDAWNERITGILGIPWSRKISMSRKLLQDNIS 1083
TQGLPKVEQVLEVRSIDSIS+NLEKRI WN+ IT ILGIPW I ++Q IS
Sbjct: 1138 TQGLPKVEQVLEVRSIDSISLNLEKRIKGWNKCITRILGIPWGFLIGAELTIVQSRIS 1195
>At5g60040 DNA-directed RNA polymerase - like protein
Length = 1328
Score = 47.8 bits (112), Expect = 3e-05
Identities = 23/45 (51%), Positives = 29/45 (64%)
Query: 205 RLCYGRSPIHGDLVELGEAVGIIAGQSIGEPGTQLTLITFHTGGV 249
++ Y S + +E G A+G I QSIGEPGTQ+TL TFH GV
Sbjct: 965 QVLYKASGVTDKQLEAGTAIGTIGAQSIGEPGTQMTLKTFHFAGV 1009
>At3g57660 DNA-directed RNA polymerase I 190K chain - like protein
Length = 1670
Score = 46.2 bits (108), Expect = 1e-04
Identities = 20/39 (51%), Positives = 26/39 (66%)
Query: 210 RSPIHGDLVELGEAVGIIAGQSIGEPGTQLTLITFHTGG 248
+S L + GE VG++A QS+GEP TQ+TL TFH G
Sbjct: 1144 KSKFFASLAQPGEPVGVLAAQSVGEPSTQMTLNTFHLAG 1182
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 45.1 bits (105), Expect = 2e-04
Identities = 21/33 (63%), Positives = 24/33 (72%)
Query: 217 LVELGEAVGIIAGQSIGEPGTQLTLITFHTGGV 249
LV GE +G +A QSIGEP TQ+TL TFH GV
Sbjct: 1078 LVAPGEMIGCVAAQSIGEPATQMTLNTFHYAGV 1110
>At3g29740 hypothetical protein
Length = 87
Score = 35.8 bits (81), Expect = 0.14
Identities = 25/81 (30%), Positives = 43/81 (52%), Gaps = 3/81 (3%)
Query: 577 NKKSRVGGLVRVEKNKKKIELKIFSGDIHFPGEIDKISQHSAILIPPEMVKKKILRNQKK 636
NK+ VGGL R+ K ++++++++ +GD G IS H +L V K IL ++
Sbjct: 6 NKELFVGGLARILKEQRQVDVRLKAGDSDQKGV--SISAHKLVLSARSEVFKMILETEEI 63
Query: 637 KTNWRYIQWITTTKKKYFVLV 657
K + IT ++ K+ LV
Sbjct: 64 KAT-TTLDTITLSELKHTELV 83
>At2g40440 hypothetical protein
Length = 194
Score = 31.6 bits (70), Expect = 2.6
Identities = 20/77 (25%), Positives = 41/77 (52%), Gaps = 3/77 (3%)
Query: 577 NKKSRVGGLVRVEKNKKKIELKIFSGDIHFPGEIDKISQHSAILIPPEMVKKKILRNQKK 636
NK+ VGG ++ K ++++++++ +GD G S H +L V KK+L + +
Sbjct: 6 NKELFVGGFAKILKEQRQVDVRLKAGDSGDEGA--STSAHKLVLSARSEVFKKMLESDEI 63
Query: 637 KTNWRYIQWITTTKKKY 653
K + ++ IT + K+
Sbjct: 64 KAS-AQLETITLCEMKH 79
>At1g60610 unknown protein
Length = 340
Score = 31.6 bits (70), Expect = 2.6
Identities = 24/107 (22%), Positives = 55/107 (50%), Gaps = 7/107 (6%)
Query: 712 LVRTCLVLNWNEDEKSSSIEEALASFVEVSTNGLIRYFLRIDLVKSH---ISYIRKRNDP 768
LV T L L++++DE++SS+ A S + + +R+DL + + +I+ R D
Sbjct: 104 LVSTGLRLSYDDDERNSSVTSANGS-ITTPVYQSLGDNIRLDLNRQNDELDQFIKFRADQ 162
Query: 769 SSSGLISYNESDRININPFFSIYKENIQQSLSQKHGTIRMLLNRNKE 815
+ G+ + + ++ F + ++++ + L +K I + +N+E
Sbjct: 163 MAKGV---RDIKQRHVTSFVTALEKDVSKKLQEKDHEIESMNKKNRE 206
>At4g04220 putative disease resistance protein, predicted
GPI-anchored protein
Length = 766
Score = 31.2 bits (69), Expect = 3.3
Identities = 35/145 (24%), Positives = 66/145 (45%), Gaps = 17/145 (11%)
Query: 664 DIADSINLVKLFPQDLFK---EWDNLELKVL--NFILYG----NGKSIRGILDTSIQLVR 714
DI + L+++ +D+F W N + + NF LY + + G + TS+ ++
Sbjct: 562 DIPNIERLIEIESEDIFSLVVNWKNSKQVLFDRNFYLYTLLDLSKNKLHGEIPTSLGNLK 621
Query: 715 TCLVLNWNEDEKSSSIEEALASFVEVSTNGLIRYFLRIDLVK-----SHISYIRKRNDPS 769
+ VLN + +E S I ++ +V + L L ++ K S ++ + RN+
Sbjct: 622 SLKVLNLSNNEFSGLIPQSFGDLEKVESLDLSHNNLTGEIPKTLSKLSELNTLDLRNNKL 681
Query: 770 SSGLISYNESDRININPFFSIYKEN 794
+ + DR+N NP +IY N
Sbjct: 682 KGRIPESPQLDRLN-NP--NIYANN 703
>At3g05100 unknown protein
Length = 336
Score = 31.2 bits (69), Expect = 3.3
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query: 455 RFIIPFLFRSIRERNNELMPDISVEIPIDGIIHKNSILAYFDDPQYRTQSSGIAKYKTIG 514
R++ P F + + LM + E+P G++HK ++ DD ++ S A Y I
Sbjct: 194 RYLSPQHFHCLEKDELSLMAALRYELPSQGLLHKRPLIVRGDDMEFSKFGSDTA-YDLIY 252
Query: 515 IHSIF 519
++F
Sbjct: 253 ASAVF 257
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 31.2 bits (69), Expect = 3.3
Identities = 26/107 (24%), Positives = 46/107 (42%), Gaps = 12/107 (11%)
Query: 299 DIMHNVIIPPKSFLLVQNDQYVKSEQIIAEIRAGTYTLNLKEKIRKHIFSDSEGEMHWST 358
D+MH+ LL Q + +K ++ E + +EK R I S +G+ W T
Sbjct: 72 DVMHS------DELLAQEEILLKDLDLVLEQEETLWFQKSREKKRNRIESPKDGDNRWMT 125
Query: 359 NIYHVSEFA---YSNVHILPKTSHLW---ILSGNSHKSDTVSLSLLK 399
+ + A Y+ ++ L S +W + G + +D +LLK
Sbjct: 126 DKVELETMALNYYTRLYSLEDVSAVWNMLPIGGFATLTDGEKAALLK 172
>At1g53390 putative ABC transporter gb|AAD31586.1
Length = 1096
Score = 31.2 bits (69), Expect = 3.3
Identities = 18/62 (29%), Positives = 26/62 (41%), Gaps = 5/62 (8%)
Query: 144 NFDTNINW--SCISRRYIYIGSRCIVVRNQDIGIGLINRFINFQTQPIFIRTPFTCRNTS 201
NF +N+N+ SCI + IG R N F N P +++ C TS
Sbjct: 88 NFSSNLNFLSSCIKKTQGSIGKRICTAAEMKF---YFNGFFNKTNNPGYLKPNVNCNLTS 144
Query: 202 WI 203
W+
Sbjct: 145 WV 146
>At1g11100 transcription factor RUSH-1alpha isolog
Length = 1227
Score = 30.8 bits (68), Expect = 4.4
Identities = 26/99 (26%), Positives = 48/99 (48%), Gaps = 19/99 (19%)
Query: 550 EVHILSESSSIMVRNNSIIGVNTPITLNKKS-------RVGGLVRVEKNKKKIELKIFSG 602
E+H + ES V + + V P+ ++ S GGL + E NK G
Sbjct: 465 EIHKI-ESYGEFVNPHQYLPVQRPVFSSEHSTGSQTLNNCGGL-KFESNK---------G 513
Query: 603 DIHFPGEIDKISQHSAILIPPE-MVKKKILRNQKKKTNW 640
+++F ++ +SQHS+ PP+ ++ +LR+Q+ +W
Sbjct: 514 NMNFHADLQDLSQHSSEASPPDGVLAVSLLRHQRIALSW 552
>At4g35630 phosphoserine aminotransferase
Length = 430
Score = 30.4 bits (67), Expect = 5.7
Identities = 21/80 (26%), Positives = 36/80 (44%), Gaps = 9/80 (11%)
Query: 522 EDLIEYRGIREFKPKYQIKVDRFFFIPQEVHILSESSSIM---VRNNSIIGVNTPITLNK 578
EDL+E G++E + K Q K D + + + ES+ V + +N P TL K
Sbjct: 321 EDLLEQGGLKEVEKKNQRKADLLY------NAIEESNGFFRCPVEKSVRSLMNVPFTLEK 374
Query: 579 KSRVGGLVRVEKNKKKIELK 598
++ +K ++LK
Sbjct: 375 SELEAEFIKEAAKEKMVQLK 394
>At5g48510 putative protein
Length = 224
Score = 30.0 bits (66), Expect = 7.4
Identities = 24/79 (30%), Positives = 39/79 (48%), Gaps = 7/79 (8%)
Query: 577 NKKSRVGGLVRV--EKNKKKIELKIFSGDIHFPGEIDKISQHSAILIPPEMVKKKILRNQ 634
NK+ GGL +V EK + + LK + D E IS H IL V K +
Sbjct: 6 NKEHFSGGLAKVLAEKWQVDVMLKAKNSD-----EGSAISAHKLILASRSEVFKNMFELD 60
Query: 635 KKKTNWRYIQWITTTKKKY 653
+ KT+ ++++ IT ++ K+
Sbjct: 61 EFKTSTKHVETITLSEMKH 79
>At2g20990 unknown protein
Length = 541
Score = 29.6 bits (65), Expect = 9.7
Identities = 17/38 (44%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query: 884 KISSIK-KNLQLNKFKETFQVIKYYLMDENERIYKPDL 920
KI S++ + L L TFQ +K YL DE E I +P L
Sbjct: 109 KIDSVEFETLTLGSLPPTFQGMKVYLTDEKELIMEPCL 146
>At1g10650 similar to putative inhibitor of apoptosis gi|3924605;
similar to ESTs gb|AI994578.1, gb|T04171, gb|AA728525
Length = 339
Score = 29.6 bits (65), Expect = 9.7
Identities = 25/105 (23%), Positives = 49/105 (45%), Gaps = 1/105 (0%)
Query: 712 LVRTCLVLNWNEDEKSSSIEEALASFVEVS-TNGLIRYFLRIDLVKSHISYIRKRNDPSS 770
LV T L L++++DE +SS+ A S + S + LRIDL + + + ++
Sbjct: 103 LVSTGLRLSYDDDEHNSSVTSASGSILAASPIFQSLDDSLRIDLHRQKDEFDQFIKIQAA 162
Query: 771 SGLISYNESDRININPFFSIYKENIQQSLSQKHGTIRMLLNRNKE 815
+ + +I F + ++ + + L +K I + +NKE
Sbjct: 163 QMAKGVRDMKQRHIASFLTTLEKGVSKKLQEKDHEINDMNKKNKE 207
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.142 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,863,304
Number of Sequences: 26719
Number of extensions: 1109939
Number of successful extensions: 3016
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 2998
Number of HSP's gapped (non-prelim): 21
length of query: 1085
length of database: 11,318,596
effective HSP length: 110
effective length of query: 975
effective length of database: 8,379,506
effective search space: 8170018350
effective search space used: 8170018350
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0269b.9


