

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
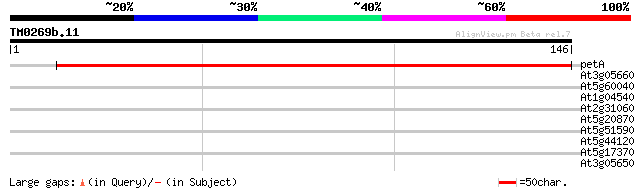
Query= TM0269b.11
(146 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
petA -chloroplast genome- cytochrome f 241 1e-64
At3g05660 putative disease resistance protein 29 0.78
At5g60040 DNA-directed RNA polymerase - like protein 28 1.7
At1g04540 hypothetical protein 28 1.7
At2g31060 putative GTP-binding protein 27 2.9
At5g20870 beta-1,3-glucanase - like protein 26 6.6
At5g51590 unknown protein 26 8.6
At5g44120 legumin-like protein 26 8.6
At5g17370 unknown protein 26 8.6
At3g05650 putative disease resistance protein 26 8.6
>petA -chloroplast genome- cytochrome f
Length = 320
Score = 241 bits (614), Expect = 1e-64
Identities = 113/134 (84%), Positives = 127/134 (94%)
Query: 13 RILANGKRGALNVGAVLILPEGFELAPTDRISPEIKEKMGNLSFQSYRPTKKNILVVGPV 72
++LANGK+GALNVGAVLILPEGFELAP DRISPE+KEK+GNLSFQ+YRP KKNILV+GPV
Sbjct: 94 QVLANGKKGALNVGAVLILPEGFELAPPDRISPEMKEKIGNLSFQNYRPNKKNILVIGPV 153
Query: 73 PGQKYSEITFPILSPDPATNRDVNFLKYPIYVGGNRGRGQIYPDGSKSNNNVYNATKSGI 132
PGQKYSEITFPIL+PDPATN+DV+FLKYPIYVGGNRGRGQIYPDGSKSNN VYNAT GI
Sbjct: 154 PGQKYSEITFPILAPDPATNKDVHFLKYPIYVGGNRGRGQIYPDGSKSNNTVYNATAGGI 213
Query: 133 INKIIRKDKGGIHL 146
I+KI+RK+KGG +
Sbjct: 214 ISKILRKEKGGYEI 227
>At3g05660 putative disease resistance protein
Length = 883
Score = 29.3 bits (64), Expect = 0.78
Identities = 15/34 (44%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query: 41 DRISPEIKEKMGNLSFQSYRPTKKNILVVGPVPG 74
+++S EI +++GNLS+ +Y N L VG VPG
Sbjct: 745 NKLSGEIPQELGNLSYLAYMNFSHNQL-VGQVPG 777
>At5g60040 DNA-directed RNA polymerase - like protein
Length = 1328
Score = 28.1 bits (61), Expect = 1.7
Identities = 27/93 (29%), Positives = 40/93 (42%), Gaps = 15/93 (16%)
Query: 39 PTDRISPEIKEKMG----NLSFQSYRPTKKNILVVGPVPGQKYSEITFPILS------PD 88
P I +K K G NLS + T + V+ P P K +E+ PIL P+
Sbjct: 338 PLSGILQRLKGKGGRFRANLSGKRVEFTGRT--VISPDPNLKITEVGIPILMAQILTFPE 395
Query: 89 PATNRDVNFLKYPIYVGGNR---GRGQIYPDGS 118
+ ++ L+ + G N+ R YPDGS
Sbjct: 396 CVSRHNIEKLRQCVRNGPNKYPGARNVRYPDGS 428
>At1g04540 hypothetical protein
Length = 601
Score = 28.1 bits (61), Expect = 1.7
Identities = 21/69 (30%), Positives = 28/69 (40%), Gaps = 11/69 (15%)
Query: 27 AVLILPEGFELAPTDRISPEIKEKMGNLSFQSYRPTKKNILVVGPVPGQKYSEITFPILS 86
A I+ E PT S E K Y P KN+ V P Q+Y+ I +++
Sbjct: 243 ASAIVSSESETLPTTTDSDEKKSS-------EYTPPSKNLRV----PRQRYNSIESDLIN 291
Query: 87 PDPATNRDV 95
P P N V
Sbjct: 292 PSPMENHHV 300
>At2g31060 putative GTP-binding protein
Length = 664
Score = 27.3 bits (59), Expect = 2.9
Identities = 11/44 (25%), Positives = 25/44 (56%)
Query: 33 EGFELAPTDRISPEIKEKMGNLSFQSYRPTKKNILVVGPVPGQK 76
+G +L P + ++ EI ++ L ++ + ++ +GPVPG +
Sbjct: 450 KGQKLEPIEEVTIEINDEHVGLVMEALSHRRAEVIDMGPVPGNE 493
>At5g20870 beta-1,3-glucanase - like protein
Length = 501
Score = 26.2 bits (56), Expect = 6.6
Identities = 31/118 (26%), Positives = 53/118 (44%), Gaps = 6/118 (5%)
Query: 24 NVGAVLILPE-GFELAPTDRISPEIKEKMGNL-SFQSYRPTKKNILV-VGPVPGQKYSEI 80
NV A L+ G ++ T ++ ++ E L S +R K +++ + S I
Sbjct: 145 NVQAALVKAGLGRQVKVTVPLNADVYESSDGLPSSGDFRSDIKTLMISIVRFLADSVSPI 204
Query: 81 TFPILSPDPATNRDVNFLKYPIYV--GGNRGRGQIYPDGSKSNNNVYNATKSGIINKI 136
TF I P + N D NF + + GG G + DGS S NV++A +++ +
Sbjct: 205 TFNIY-PFLSLNADPNFPREYAFFPNGGGGGGAKPVVDGSISYTNVFDANFDTLVSAL 261
>At5g51590 unknown protein
Length = 419
Score = 25.8 bits (55), Expect = 8.6
Identities = 23/88 (26%), Positives = 33/88 (37%), Gaps = 11/88 (12%)
Query: 39 PTDRISPEIKEKMGNLSFQSYRPTKKNILVVGPVPGQKYSEITFPILSPDPATNRDVNFL 98
P + S E+K+K G + Y P + + P+P +T S R
Sbjct: 69 PVENSSSELKKKRGRP--RKYNPDGSLAVTLSPMPISSSVPLTSEFGSRKRGRGRGR--- 123
Query: 99 KYPIYVGGNRGRGQIYPDGSKSNNNVYN 126
G RGRG+ GS+ NN N
Sbjct: 124 ------GRGRGRGRGQGQGSREPNNNNN 145
>At5g44120 legumin-like protein
Length = 472
Score = 25.8 bits (55), Expect = 8.6
Identities = 11/42 (26%), Positives = 22/42 (52%), Gaps = 2/42 (4%)
Query: 101 PIYVGGNRGRGQIYPDG--SKSNNNVYNATKSGIINKIIRKD 140
P Y+ GN +GQ++ G + N++N +I + ++ D
Sbjct: 190 PFYLAGNNPQGQVWLQGREQQPQKNIFNGFGPEVIAQALKID 231
>At5g17370 unknown protein
Length = 467
Score = 25.8 bits (55), Expect = 8.6
Identities = 10/32 (31%), Positives = 17/32 (52%)
Query: 68 VVGPVPGQKYSEITFPILSPDPATNRDVNFLK 99
+ GP+PG K S + P+P ++ N+ K
Sbjct: 39 IKGPIPGAKSSSSSRTKQKPEPKPEQETNYQK 70
>At3g05650 putative disease resistance protein
Length = 868
Score = 25.8 bits (55), Expect = 8.6
Identities = 14/34 (41%), Positives = 22/34 (64%), Gaps = 1/34 (2%)
Query: 41 DRISPEIKEKMGNLSFQSYRPTKKNILVVGPVPG 74
+++S EI +++GNLS+ +Y N L G VPG
Sbjct: 734 NKLSGEIPQELGNLSYLAYMNFSHNQL-GGLVPG 766
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.141 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,569,484
Number of Sequences: 26719
Number of extensions: 163612
Number of successful extensions: 335
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 332
Number of HSP's gapped (non-prelim): 10
length of query: 146
length of database: 11,318,596
effective HSP length: 90
effective length of query: 56
effective length of database: 8,913,886
effective search space: 499177616
effective search space used: 499177616
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0269b.11


