

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
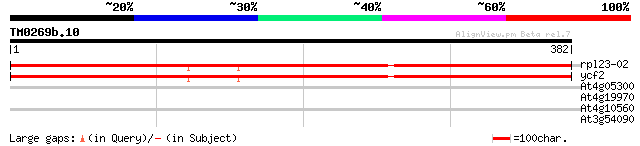
Query= TM0269b.10
(382 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
rpl23-02 -chloroplast genome- 639 0.0
ycf2 -chloroplast genome- 639 0.0
At4g05300 putative protein 30 2.9
At4g19970 unknown protein 29 3.8
At4g10560 putative protein 29 3.8
At3g54090 fructokinase - like protein 28 6.4
>rpl23-02 -chloroplast genome-
Length = 2294
Score = 639 bits (1648), Expect = 0.0
Identities = 323/387 (83%), Positives = 336/387 (86%), Gaps = 8/387 (2%)
Query: 1 KPFLLDDHDTSQKSRFLINGGTISPFLFNKIPKWMIDSFDTIKNRRKFFDNTDS-YFSMI 59
KPFLLDDH+TSQKS+FLINGGTISPFLFNKIPKWMIDSF T KNRRK FDNTDS YFS++
Sbjct: 945 KPFLLDDHNTSQKSKFLINGGTISPFLFNKIPKWMIDSFHTRKNRRKSFDNTDSAYFSIV 1004
Query: 60 SHDEDNWLNPVKPFHRSSLISSFYKANRLRFLNNRYHFCFYCNKRLPFYVEKACINNYDF 119
SHD+DNWLNPVKPF RSSLISSF KANRLRFLNN +HFCFYCNKR PFYVEKA +NN DF
Sbjct: 1005 SHDQDNWLNPVKPFQRSSLISSFSKANRLRFLNNPHHFCFYCNKRFPFYVEKARLNNSDF 1064
Query: 120 T--YGQFLNILFIRNKRFSLCGGKKKHAFLERDTISP--IESRVFNILILNDFPQSGDEG 175
T YGQFL ILFI NK FS CGGKKKHAFLERDTISP IES+V NI I NDFPQSGDE
Sbjct: 1065 TFTYGQFLTILFIHNKTFSSCGGKKKHAFLERDTISPSSIESQVSNIFISNDFPQSGDER 1124
Query: 176 YNLYKSFHFPIRSDPFVHRAIYSIADISVTPLTEGQIVNFERTYCQPLSDMNLPDSEGKN 235
YNLYKSFHFPIRSDP V RAIYSIADIS TPL EGQ VNFERTYCQ LSDMNL DSE K+
Sbjct: 1125 YNLYKSFHFPIRSDPLVRRAIYSIADISGTPLIEGQRVNFERTYCQTLSDMNLSDSEEKS 1184
Query: 236 LHQYLKFNSNMGLIHIPCSEKYLPSENRKKGIPCLKKCLEKGQMYRTFQRDSVFSTLSKW 295
LHQYL FNSNMGLIH PCSEKYL RKK CLKKC++KGQM RTFQRDS FSTLSKW
Sbjct: 1185 LHQYLNFNSNMGLIHTPCSEKYL---QRKKRSLCLKKCVDKGQMDRTFQRDSAFSTLSKW 1241
Query: 296 NLFQTYIPWFLTSTGYKYLNFIFLDTFSDLLPILSSSQKFVSIFHDIMHRSDISWRILQK 355
NLFQTY+PWF TSTGYKYLN IFLDTFSDLL ILSSSQKFVSIFHDIMH DISWRILQK
Sbjct: 1242 NLFQTYMPWFFTSTGYKYLNLIFLDTFSDLLRILSSSQKFVSIFHDIMHGLDISWRILQK 1301
Query: 356 KWCLSQWNLISEISSKCFHNLLLSEEI 382
K CL Q NLISEISSK HNLLLSEE+
Sbjct: 1302 KLCLPQRNLISEISSKSLHNLLLSEEM 1328
>ycf2 -chloroplast genome-
Length = 2294
Score = 639 bits (1648), Expect = 0.0
Identities = 323/387 (83%), Positives = 336/387 (86%), Gaps = 8/387 (2%)
Query: 1 KPFLLDDHDTSQKSRFLINGGTISPFLFNKIPKWMIDSFDTIKNRRKFFDNTDS-YFSMI 59
KPFLLDDH+TSQKS+FLINGGTISPFLFNKIPKWMIDSF T KNRRK FDNTDS YFS++
Sbjct: 945 KPFLLDDHNTSQKSKFLINGGTISPFLFNKIPKWMIDSFHTRKNRRKSFDNTDSAYFSIV 1004
Query: 60 SHDEDNWLNPVKPFHRSSLISSFYKANRLRFLNNRYHFCFYCNKRLPFYVEKACINNYDF 119
SHD+DNWLNPVKPF RSSLISSF KANRLRFLNN +HFCFYCNKR PFYVEKA +NN DF
Sbjct: 1005 SHDQDNWLNPVKPFQRSSLISSFSKANRLRFLNNPHHFCFYCNKRFPFYVEKARLNNSDF 1064
Query: 120 T--YGQFLNILFIRNKRFSLCGGKKKHAFLERDTISP--IESRVFNILILNDFPQSGDEG 175
T YGQFL ILFI NK FS CGGKKKHAFLERDTISP IES+V NI I NDFPQSGDE
Sbjct: 1065 TFTYGQFLTILFIHNKTFSSCGGKKKHAFLERDTISPSSIESQVSNIFISNDFPQSGDER 1124
Query: 176 YNLYKSFHFPIRSDPFVHRAIYSIADISVTPLTEGQIVNFERTYCQPLSDMNLPDSEGKN 235
YNLYKSFHFPIRSDP V RAIYSIADIS TPL EGQ VNFERTYCQ LSDMNL DSE K+
Sbjct: 1125 YNLYKSFHFPIRSDPLVRRAIYSIADISGTPLIEGQRVNFERTYCQTLSDMNLSDSEEKS 1184
Query: 236 LHQYLKFNSNMGLIHIPCSEKYLPSENRKKGIPCLKKCLEKGQMYRTFQRDSVFSTLSKW 295
LHQYL FNSNMGLIH PCSEKYL RKK CLKKC++KGQM RTFQRDS FSTLSKW
Sbjct: 1185 LHQYLNFNSNMGLIHTPCSEKYL---QRKKRSLCLKKCVDKGQMDRTFQRDSAFSTLSKW 1241
Query: 296 NLFQTYIPWFLTSTGYKYLNFIFLDTFSDLLPILSSSQKFVSIFHDIMHRSDISWRILQK 355
NLFQTY+PWF TSTGYKYLN IFLDTFSDLL ILSSSQKFVSIFHDIMH DISWRILQK
Sbjct: 1242 NLFQTYMPWFFTSTGYKYLNLIFLDTFSDLLRILSSSQKFVSIFHDIMHGLDISWRILQK 1301
Query: 356 KWCLSQWNLISEISSKCFHNLLLSEEI 382
K CL Q NLISEISSK HNLLLSEE+
Sbjct: 1302 KLCLPQRNLISEISSKSLHNLLLSEEM 1328
>At4g05300 putative protein
Length = 387
Score = 29.6 bits (65), Expect = 2.9
Identities = 22/84 (26%), Positives = 40/84 (47%), Gaps = 10/84 (11%)
Query: 305 FLTSTGYKYLNFIFL-DTFSDLLPILSSSQKFVSI--FH------DIMHRSDISWRILQK 355
FLT TG +L+F+ + + F ++ P L + + +H + R + SWR L+
Sbjct: 124 FLTCTG-NHLDFLPISEKFWEMTPTLEAGHAVAEMVMYHISQLHTENARRWERSWRCLRV 182
Query: 356 KWCLSQWNLISEISSKCFHNLLLS 379
+ S NL+ C HN+ ++
Sbjct: 183 RTLYSTLNLVYRTLGNCVHNISIN 206
>At4g19970 unknown protein
Length = 715
Score = 29.3 bits (64), Expect = 3.8
Identities = 20/61 (32%), Positives = 27/61 (43%), Gaps = 5/61 (8%)
Query: 34 WMIDSFDTIKNRRKFFDNTDSYFSMISHDEDNWLNP----VKPFHRSSLISSFYKANRLR 89
W+ D F + F D +F HD DNW+N VK HRS F+ +RL
Sbjct: 563 WLRDPFPRLYPDGDFQMACDRFFGD-PHDSDNWVNGGFTYVKSNHRSIEFYKFWYNSRLD 621
Query: 90 F 90
+
Sbjct: 622 Y 622
>At4g10560 putative protein
Length = 703
Score = 29.3 bits (64), Expect = 3.8
Identities = 13/34 (38%), Positives = 18/34 (52%), Gaps = 4/34 (11%)
Query: 80 SSFYKANRLRFLNNRYHF----CFYCNKRLPFYV 109
SSFY + L+N +H C YC KR P+ +
Sbjct: 649 SSFYLEKTVDVLSNNHHMTRPICCYCEKRCPYKI 682
>At3g54090 fructokinase - like protein
Length = 471
Score = 28.5 bits (62), Expect = 6.4
Identities = 11/35 (31%), Positives = 18/35 (51%)
Query: 29 NKIPKWMIDSFDTIKNRRKFFDNTDSYFSMISHDE 63
N P++ + FD KNRR ++ T + HD+
Sbjct: 327 NYTPQYFAEDFDQTKNRRDYYHYTPEEIKSLWHDK 361
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.141 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,520,876
Number of Sequences: 26719
Number of extensions: 432462
Number of successful extensions: 1073
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1055
Number of HSP's gapped (non-prelim): 11
length of query: 382
length of database: 11,318,596
effective HSP length: 101
effective length of query: 281
effective length of database: 8,619,977
effective search space: 2422213537
effective search space used: 2422213537
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0269b.10


