

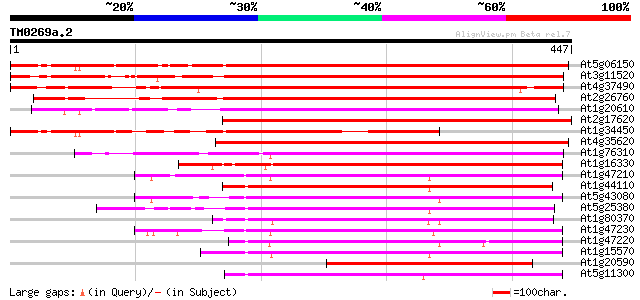
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269a.2
(447 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g06150 mitosis-specific cyclin 1b 476 e-134
At3g11520 cyclin box 459 e-129
At4g37490 cyclin cyc1 440 e-124
At2g26760 putative cyclin 352 2e-97
At1g20610 hypothetical protein 274 6e-74
At2g17620 putative cyclin 2 263 1e-70
At1g34450 cyclin, putative 261 4e-70
At4g35620 cyclin 2b protein 259 2e-69
At1g76310 putative G2/mitotic-specific cyclin 1 (B-like cyclin) 259 2e-69
At1g16330 putative mitotic cyclin 246 1e-65
At1g47210 cyclin like protein 182 3e-46
At1g44110 mitotic cyclin a2-type, putative 180 2e-45
At5g43080 cyclin A-type 175 4e-44
At5g25380 cyclin 3a 172 4e-43
At1g80370 putative cyclin 170 1e-42
At1g47230 cyclin like protein 170 2e-42
At1g47220 cyclin, putative 169 4e-42
At1g15570 putative cyclin (At1g15570) 167 1e-41
At1g20590 hypothetical protein 165 5e-41
At5g11300 cyclin 3b 161 6e-40
>At5g06150 mitosis-specific cyclin 1b
Length = 445
Score = 476 bits (1224), Expect = e-134
Identities = 260/450 (57%), Positives = 324/450 (71%), Gaps = 18/450 (4%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIE---VKP--N 55
MA+R VP+Q RG +V G K Q KNGA ++RRALGDIGNL V G++ +P N
Sbjct: 1 MATRANVPEQVRGAPLVDGLKIQ-NKNGAV--KSRRALGDIGNLVSVPGVQGGKAQPPIN 57
Query: 56 RPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVT 115
RPITRSF AQLLANAQ N + P + P+AA + K + +
Sbjct: 58 RPITRSFRAQLLANAQLERKPINGDNKV-PALGPKRQPLAARNPEAQRAVQKKNLVVKQQ 116
Query: 116 GKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVD 175
KP VEVIE + E KKE ++ K +KK T +SVL+ARSKAACG+ NKPK I+D
Sbjct: 117 TKP---VEVIE-TKKEVTKKEVAMSPK---NKKVTYSSVLSARSKAACGIVNKPK--IID 167
Query: 176 IDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTK 235
ID SD DN LAAVEY++D+Y FYK VE ES+P YM Q E+NEKMRAIL+DWL++VH K
Sbjct: 168 IDESDKDNHLAAVEYVDDMYSFYKEVEKESQPKMYMHIQTEMNEKMRAILIDWLLEVHIK 227
Query: 236 FELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR 295
FEL+LETLYLT+NI+DRFL+VK VP+RELQLVGIS++L+A+KYEEIWPP+VND V ++D
Sbjct: 228 FELNLETLYLTVNIIDRFLSVKAVPKRELQLVGISALLIASKYEEIWPPQVNDLVYVTDN 287
Query: 296 AYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYD 355
AYS QILVMEK ILG LEW LTVPT +VFL RFIKAS+ D + NM HFL+ELGMMHYD
Sbjct: 288 AYSSRQILVMEKAILGNLEWYLTVPTQYVFLVRFIKASMSDPEMENMVHFLAELGMMHYD 347
Query: 356 TLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGK 415
TL +CPSM+AASAVY ARC+LNKSPAW +TL+ HT Y+E ++MDC++LL H G +
Sbjct: 348 TLTFCPSMLAASAVYTARCSLNKSPAWTDTLQFHTGYTESEIMDCSKLLAFLHSRCGESR 407
Query: 416 LKVVFRKYSDPERGAVAVLPPAKNLMPQEA 445
L+ V++KYS E G VA++ PAK+L+ A
Sbjct: 408 LRAVYKKYSKAENGGVAMVSPAKSLLSAAA 437
>At3g11520 cyclin box
Length = 414
Score = 459 bits (1181), Expect = e-129
Identities = 256/446 (57%), Positives = 318/446 (70%), Gaps = 41/446 (9%)
Query: 1 MASRPVVPQQP-RGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEV-KPNRPI 58
MA+ PVV QP RGD + KN AA +NRRALGDIGN+D + G+E K NRPI
Sbjct: 1 MATGPVVHPQPVRGDPI-------DLKNAAA--KNRRALGDIGNVDSLIGVEGGKLNRPI 51
Query: 59 TRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTG-- 116
TR+F AQLL NAQ A AA NKK AP+ GV K+E + V K+ G
Sbjct: 52 TRNFRAQLLENAQVAAAA--NKK----------APIL-DGVT-KKQEVVRAVQKKARGDK 97
Query: 117 -KPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVD 175
+P +EVI ISPD V K KE+ KK T +SVL ARSKAA + +D
Sbjct: 98 REPSKPIEVIVISPDTN-----EVAKAKENKKKVTYSSVLDARSKAA--------SKTLD 144
Query: 176 IDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTK 235
ID D +N+LAAVEY+ED+Y FYK V NES+P YM +QPEI+EKMR+IL+DWL++VH K
Sbjct: 145 IDYVDKENDLAAVEYVEDMYIFYKEVVNESKPQMYMHTQPEIDEKMRSILIDWLVEVHVK 204
Query: 236 FELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR 295
F+LS ETLYLT+NI+DRFL++KTVPRRELQLVG+S++L+A+KYEEIWPP+VND V ++D
Sbjct: 205 FDLSPETLYLTVNIIDRFLSLKTVPRRELQLVGVSALLIASKYEEIWPPQVNDLVYVTDN 264
Query: 296 AYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYD 355
+Y+ QILVMEK ILG LEW LTVPT +VFL RFIKAS D+ + N+ HFL+ELG+MH+D
Sbjct: 265 SYNSRQILVMEKTILGNLEWYLTVPTQYVFLVRFIKASGSDQKLENLVHFLAELGLMHHD 324
Query: 356 TLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGK 415
+LM+CPSM+AASAVY ARC LNK+P W +TLK HT YSE QLMDC++LL H G K
Sbjct: 325 SLMFCPSMLAASAVYTARCCLNKTPTWTDTLKFHTGYSESQLMDCSKLLAFIHSKAGESK 384
Query: 416 LKVVFRKYSDPERGAVAVLPPAKNLM 441
L+ V +KYS RGAVA++ PAK+LM
Sbjct: 385 LRGVLKKYSKLGRGAVALISPAKSLM 410
>At4g37490 cyclin cyc1
Length = 428
Score = 440 bits (1132), Expect = e-124
Identities = 254/455 (55%), Positives = 310/455 (67%), Gaps = 48/455 (10%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITR 60
M SR +VPQQ D VV GK A GRNR+ LGDIGN+ VRG K N P
Sbjct: 2 MTSRSIVPQQSTDDVVVVDGKN------VAKGRNRQVLGDIGNV--VRGNYPKNNEPEKI 53
Query: 61 SFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREA-PKPVTKRVTGKPK 119
+ + + + +N KK V KR A PKP K+V GKPK
Sbjct: 54 NHRPRTRSQNPTLLVEDNLKKP------------------VVKRNAVPKP--KKVAGKPK 93
Query: 120 PVVEVIEISPDEQIKKEKSVQKKKEDSKKK--TLTSVLTARSKAACGLTNKPKEEIVDID 177
VV+VIEIS D + ++K+ +KKK T TSVLTARSKAACGL K KE+IVDID
Sbjct: 94 -VVDVIEISSDSDEELGLVAAREKKATKKKATTYTSVLTARSKAACGLEKKQKEKIVDID 152
Query: 178 ASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFE 237
++DV+N+LAAVEY+EDIY FYK VE+E RP YMASQP+INEKMR ILV+WLIDVH +FE
Sbjct: 153 SADVENDLAAVEYVEDIYSFYKSVESEWRPRDYMASQPDINEKMRLILVEWLIDVHVRFE 212
Query: 238 LSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAY 297
L+ ET YLT+NI+DRFL+VK VPR+ELQLVG+S++LM+AKYEEIWPP+V D V ++D AY
Sbjct: 213 LNPETFYLTVNILDRFLSVKPVPRKELQLVGLSALLMSAKYEEIWPPQVEDLVDIADHAY 272
Query: 298 SHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDT- 356
SH+QILVMEK IL LEW LTVPT +VFL RFIKAS+ DE + NM H+L+ELG+MHYDT
Sbjct: 273 SHKQILVMEKTILSTLEWYLTVPTHYVFLARFIKASIADEKMENMVHYLAELGVMHYDTM 332
Query: 357 LMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLV----------S 406
+M+ PSM+AASA+YAAR +L + P W TLK HT YSE QLMDCA+LL S
Sbjct: 333 IMFSPSMVAASAIYAARSSLRQVPIWTSTLKHHTGYSETQLMDCAKLLAYQQWKQQEEGS 392
Query: 407 FHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLM 441
T G + +KYS ER AVA++PPAK L+
Sbjct: 393 ESSTKG-----ALRKKYSKDERFAVALIPPAKALL 422
>At2g26760 putative cyclin
Length = 387
Score = 352 bits (903), Expect = 2e-97
Identities = 198/418 (47%), Positives = 260/418 (61%), Gaps = 59/418 (14%)
Query: 20 GKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITRSFCAQLLANAQAAVAAENN 79
G+ +PK +NR+ LGDIGNL V G +V + + +
Sbjct: 17 GEIKPKNVAGHGRQNRKVLGDIGNL--VTGRDVATGKDVAK------------------- 55
Query: 80 KKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEISPDEQIKKEKSV 139
K + P+ TK EVI ISPDE K +
Sbjct: 56 -----------------------KAKQPQQQTK---------AEVIVISPDENEKCKPHF 83
Query: 140 QKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYK 199
++ KT T+ L ARSKAA GL K+ ++DIDA D +NELAAVEY+EDI+KFY+
Sbjct: 84 SRRTHIRGTKTFTATLRARSKAASGL----KDAVIDIDAVDANNELAAVEYVEDIFKFYR 139
Query: 200 MVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTV 259
VE E Y+ SQPEINEKMR+IL+DWL+DVH KFEL ETLYLTIN+VDRFL++ V
Sbjct: 140 TVEEEGGIKDYIGSQPEINEKMRSILIDWLVDVHRKFELMPETLYLTINLVDRFLSLTMV 199
Query: 260 PRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTV 319
RRELQL+G+ +ML+A KYEEIW PEVNDFVC+SD AY+ +Q+L MEK ILG++EW +TV
Sbjct: 200 HRRELQLLGLGAMLIACKYEEIWAPEVNDFVCISDNAYNRKQVLAMEKSILGQVEWYITV 259
Query: 320 PTPFVFLTRFIKASVP-DEGVTNMAHFLSELGMMHYD-TLMYCPSMIAASAVYAARCTLN 377
PTP+VFL R++KA+VP D + + +L+ELG+M Y ++ PSM+AASAVYAAR L
Sbjct: 260 PTPYVFLARYVKAAVPCDAEMEKLVFYLAELGLMQYPIVVLNRPSMLAASAVYAARQILK 319
Query: 378 KSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLP 435
K+P W ETLK HT YSE+++M+ A++L+ + KL VF+KYS E VA+LP
Sbjct: 320 KTPFWTETLKHHTGYSEDEIMEHAKMLMKLRDSASESKLIAVFKKYSVSENAEVALLP 377
>At1g20610 hypothetical protein
Length = 429
Score = 274 bits (701), Expect = 6e-74
Identities = 168/439 (38%), Positives = 246/439 (55%), Gaps = 51/439 (11%)
Query: 18 GGGKQQPKKNGAAAGRNRRALGDIG----------------NLDPVRGIEVKP--NRPIT 59
GGG K A G RRAL I ++ GI KP +RP+T
Sbjct: 19 GGGVVGKIKTTATTGPTRRALSTINKNITEAPSYPYAVNKRSVSERDGICNKPPVHRPVT 78
Query: 60 RSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPK 119
R F AQL A+ + + E KK + P + V + +E +
Sbjct: 79 RKFAAQL-ADHKPHIRDEETKKPDSVSSEEPETIII--DVDESDKEGGDSNEPMFVQHTE 135
Query: 120 PVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDAS 179
++E IE + EK ++ + D K +E ++DIDA
Sbjct: 136 AMLEEIE-------QMEKEIEMEDAD----------------------KEEEPVIDIDAC 166
Query: 180 DVDNELAAVEYIEDIYKFYKMVENESR-PHCYMASQPEINEKMRAILVDWLIDVHTKFEL 238
D +N LAAVEYI D++ FYK E S P YM +Q ++NE+MR IL+DWLI+VH KFEL
Sbjct: 167 DKNNPLAAVEYIHDMHTFYKNFEKLSCVPPNYMDNQQDLNERMRGILIDWLIEVHYKFEL 226
Query: 239 SLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYS 298
ETLYLTIN++DRFLAV + R++LQLVG++++L+A KYEE+ P V+D + +SD+AYS
Sbjct: 227 MEETLYLTINVIDRFLAVHQIVRKKLQLVGVTALLLACKYEEVSVPVVDDLILISDKAYS 286
Query: 299 HEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLM 358
++L MEK++ L++ ++PTP+VF+ RF+KA+ D+ + ++ F+ EL ++ Y+ L
Sbjct: 287 RREVLDMEKLMANTLQFNFSLPTPYVFMKRFLKAAQSDKKLEILSFFMIELCLVEYEMLE 346
Query: 359 YCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKV 418
Y PS +AASA+Y A+CTL W++T + HT Y+E+QL+ CAR +V+FH G GKL
Sbjct: 347 YLPSKLAASAIYTAQCTLKGFEEWSKTCEFHTGYNEKQLLACARKMVAFHHKAGTGKLTG 406
Query: 419 VFRKYSDPERGAVAVLPPA 437
V RKY+ + A PA
Sbjct: 407 VHRKYNTSKFCHAARTEPA 425
>At2g17620 putative cyclin 2
Length = 429
Score = 263 bits (673), Expect = 1e-70
Identities = 133/279 (47%), Positives = 191/279 (67%), Gaps = 1/279 (0%)
Query: 170 KEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESR-PHCYMASQPEINEKMRAILVDW 228
+E IVDID D N LAAVEY++D+Y FY+ +E S P YM Q ++NEKMRAIL+DW
Sbjct: 151 EEPIVDIDVLDSKNSLAAVEYVQDLYAFYRTMERFSCVPVDYMMQQIDLNEKMRAILIDW 210
Query: 229 LIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVND 288
LI+VH KF+L ETL+LT+N++DRFL+ + V R++LQLVG+ ++L+A KYEE+ P V D
Sbjct: 211 LIEVHDKFDLINETLFLTVNLIDRFLSKQNVMRKKLQLVGLVALLLACKYEEVSVPVVED 270
Query: 289 FVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSE 348
V +SD+AY+ +L MEK +L L++ +++PT + FL RF+KA+ D+ +A FL E
Sbjct: 271 LVLISDKAYTRNDVLEMEKTMLSTLQFNISLPTQYPFLKRFLKAAQADKKCEVLASFLIE 330
Query: 349 LGMMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFH 408
L ++ Y+ L + PS++AA++VY A+CTL+ S WN T + H YSE+QLM+C+R LVS H
Sbjct: 331 LALVEYEMLRFPPSLLAATSVYTAQCTLDGSRKWNSTCEFHCHYSEDQLMECSRKLVSLH 390
Query: 409 CTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQEASS 447
G L V+RKYS + G +A A L+ + S
Sbjct: 391 QRAATGNLTGVYRKYSTSKFGYIAKCEAAHFLVSESHHS 429
>At1g34450 cyclin, putative
Length = 504
Score = 261 bits (668), Expect = 4e-70
Identities = 167/347 (48%), Positives = 209/347 (60%), Gaps = 60/347 (17%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIE---VKP--N 55
MA+R +P+Q RG +V G K Q KNGA +NRRALGDIGNL V G++ +P N
Sbjct: 113 MATRANIPEQVRGAPLVDGLKIQ-NKNGAV--KNRRALGDIGNLVSVPGVQGGKAQPPIN 169
Query: 56 RPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVT 115
RPIT SF AQLLANAQ N + P + P+AA P +R
Sbjct: 170 RPITLSFRAQLLANAQLERKPINGDNKV-PALGPKSQPLAARN----------PEAQRAV 218
Query: 116 GKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVD 175
K VV+ +Q K + ++ K+ A+SKAACG+ NKPK I+D
Sbjct: 219 QKKNLVVK-------QQTKPVEVIETKRN------------AQSKAACGIVNKPK--ILD 257
Query: 176 IDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTK 235
ID SD DN +AAVEY++D+Y FYK VE ES+P YM Q E+NEKMRAIL+DWL++VH K
Sbjct: 258 IDESDKDNHVAAVEYVDDMYSFYKEVEKESQPKMYMHIQTEMNEKMRAILIDWLLEVHIK 317
Query: 236 FELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR 295
FEL+LETLYLT+NI+DRFL VK VP+RELQ VND V ++D
Sbjct: 318 FELNLETLYLTVNIIDRFLYVKAVPKRELQ--------------------VNDLVYVTDN 357
Query: 296 AYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNM 342
AYS QILVM+K ILG LEW LT+PT +VFL FIKAS+ D V ++
Sbjct: 358 AYSSRQILVMKKAILGNLEWYLTIPTQYVFLFCFIKASISDPEVLHV 404
>At4g35620 cyclin 2b protein
Length = 429
Score = 259 bits (663), Expect = 2e-69
Identities = 128/282 (45%), Positives = 192/282 (67%), Gaps = 1/282 (0%)
Query: 165 LTNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESR-PHCYMASQPEINEKMRA 223
+ + +E ++DID D +N LAAVEY++D+Y FY+ E S P YMA Q +I++KMRA
Sbjct: 147 MEEEQEEPVLDIDEYDANNSLAAVEYVQDLYDFYRKTERFSCVPLDYMAQQFDISDKMRA 206
Query: 224 ILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWP 283
IL+DWLI+VH KFEL ETL+LT+N++DRFL+ + V R++LQLVG+ ++L+A KYEE+
Sbjct: 207 ILIDWLIEVHDKFELMNETLFLTVNLIDRFLSKQAVARKKLQLVGLVALLLACKYEEVSV 266
Query: 284 PEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMA 343
P V D V +SD+AY+ +L MEKI+L L++ +++PT + FL RF+KA+ D+ + +A
Sbjct: 267 PIVEDLVVISDKAYTRTDVLEMEKIMLSTLQFNMSLPTQYPFLKRFLKAAQSDKKLEILA 326
Query: 344 HFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARL 403
FL EL ++ Y+ + Y PS++AA+AVY A+CT++ WN T + H YSE QL++C R
Sbjct: 327 SFLIELALVDYEMVRYPPSLLAATAVYTAQCTIHGFSEWNSTCEFHCHYSENQLLECCRR 386
Query: 404 LVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQEA 445
+V H G KL V RKYS + G +A A + + ++
Sbjct: 387 MVRLHQKAGTDKLTGVHRKYSSSKFGYIATKYEAAHFLVSDS 428
>At1g76310 putative G2/mitotic-specific cyclin 1 (B-like cyclin)
Length = 418
Score = 259 bits (662), Expect = 2e-69
Identities = 153/394 (38%), Positives = 232/394 (58%), Gaps = 40/394 (10%)
Query: 52 VKPNRPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVT 111
V +RP+TR F AQL AENN + + K E KP
Sbjct: 60 VPVHRPVTRKFAAQL---------AENN-------------------LQIHKEETKKPDL 91
Query: 112 KRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKE 171
+ + +V E +E + + + +E K + + + + + + +E
Sbjct: 92 ISNEALDRIITDVEEGDFNEPMFVQHTEAMLEEIDKMEGI------EMQDSNDIDAEVEE 145
Query: 172 EIVDIDASDVDNELAAVEYIEDIYKFYKMVENESR---PHCYMASQPEINEKMRAILVDW 228
++DID+ D +N L+ VEYI DIY FYK +NE R P YM +Q +INE+MR IL DW
Sbjct: 146 SVMDIDSCDKNNPLSVVEYINDIYCFYK--KNECRSCVPPNYMENQHDINERMRGILFDW 203
Query: 229 LIDVHTKFELSLETLYLTINIVDRFLAV-KTVPRRELQLVGISSMLMAAKYEEIWPPEVN 287
LI+VH KFEL ETLYLTIN++DRFLAV + + R++LQLVG+++ML+A KYEE+ P V+
Sbjct: 204 LIEVHYKFELMEETLYLTINLIDRFLAVHQHIARKKLQLVGVTAMLLACKYEEVSVPVVD 263
Query: 288 DFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLS 347
D + +SD+AY+ +IL MEK++ L++ +PTP+VF+ RF+KA+ D+ + ++ F+
Sbjct: 264 DLILISDKAYTRTEILDMEKLMANTLQFNFCLPTPYVFMRRFLKAAQSDKKLELLSFFMI 323
Query: 348 ELGMMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSF 407
EL ++ Y+ L Y PS +AASA+Y A+ TL W++T + H+ Y+EE L++C+R +V
Sbjct: 324 ELCLVEYEMLQYTPSQLAASAIYTAQSTLKGYEDWSKTSEFHSGYTEEALLECSRKMVGL 383
Query: 408 HCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLM 441
H G GKL V RKY+ + G A + PA L+
Sbjct: 384 HHKAGTGKLTGVHRKYNTSKFGYAARIEPAGFLL 417
>At1g16330 putative mitotic cyclin
Length = 518
Score = 246 bits (629), Expect = 1e-65
Identities = 124/310 (40%), Positives = 200/310 (64%), Gaps = 8/310 (2%)
Query: 135 KEKSVQKKKEDSKKKTLTSVLTARSK--AACGLTNKPKEEIVDIDASDVDNELAAVEYIE 192
K V K + ++K+ TS+L SK G T +P E++ ID D N+L EY++
Sbjct: 211 KVLDVTAKPKSKRRKSFTSLLVNGSKFDEKNGETTEP-EKLPSID--DESNQLEVAEYVD 267
Query: 193 DIYKFYKMVE--NESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIV 250
DIY+FY E N + H Y+++ E++ R IL++WLI+VH KF+L ETLYLT++++
Sbjct: 268 DIYQFYWTAEALNPALGH-YLSAHAEVSPVTRGILINWLIEVHFKFDLMHETLYLTMDLL 326
Query: 251 DRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIIL 310
DR+L+ + + E+QL+G++++L+A+KYE+ W P + D + +S +Y+ EQIL ME+ +L
Sbjct: 327 DRYLSQVPIHKNEMQLIGLTALLLASKYEDYWHPRIKDLISISAESYTREQILGMERSML 386
Query: 311 GRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVY 370
+L++ L PTP+VF+ RF+KA+ ++ + +A +L EL ++ Y+ L Y PS++ ASA+Y
Sbjct: 387 KQLKFRLNAPTPYVFMLRFLKAAQSNKKLEQLAFYLIELCLVEYEALKYKPSLLCASAIY 446
Query: 371 AARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGA 430
ARCTL+ +P W L HT Y+ Q+ DC+ +++ FH G L+V + KY +P+R
Sbjct: 447 VARCTLHMTPVWTSLLNNHTHYNVSQMKDCSDMILRFHKAAKTGNLRVTYEKYINPDRSN 506
Query: 431 VAVLPPAKNL 440
VAVL P L
Sbjct: 507 VAVLKPLDKL 516
>At1g47210 cyclin like protein
Length = 372
Score = 182 bits (462), Expect = 3e-46
Identities = 116/354 (32%), Positives = 194/354 (54%), Gaps = 17/354 (4%)
Query: 100 AVAKREAPKPVT---KRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLT 156
A AKR+A + RV K + E++ +S + ++ +KKE K K
Sbjct: 13 AAAKRKASTAMGIDGDRVNKKRVVLGELLNVS---NVNLLANLNQKKETQKPKRNLKPPP 69
Query: 157 ARSKAACGLTNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESR----PHCYMA 212
A+ + + E DID+ D ++ Y+ DIY++ + +E + + P
Sbjct: 70 AKQIKSAPVAIIDLESKSDIDSRSDDPQMCG-PYVADIYEYLRQLEVKPKQRPLPDYIEK 128
Query: 213 SQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSM 272
Q ++ MR +LVDWL++V +++L ETLYLT++ +DRFL++KTV +++LQLVG+S+M
Sbjct: 129 VQKDVTPSMRGVLVDWLVEVAEEYKLGSETLYLTVSHIDRFLSLKTVNKQKLQLVGVSAM 188
Query: 273 LMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKA 332
L+A+KYEEI PP+V+DF ++D +S + ++ ME IL L++ L PT F+ RF +
Sbjct: 189 LIASKYEEISPPKVDDFCYITDNTFSKQDVVKMEADILLALQFELGRPTINTFMRRFTRV 248
Query: 333 S-----VPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLN-KSPAWNETL 386
+ VP + + +LSEL ++ Y T+ + PS++AASAV+ AR + K WN+ L
Sbjct: 249 AQDDFKVPHLQLEPLCCYLSELSILDYKTVKFVPSLLAASAVFLARFIIRPKQHPWNQML 308
Query: 387 KLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
+ +T Y L C ++ + + G L+ V KY + VA +P + L
Sbjct: 309 EEYTKYKAADLQVCVGIIHDLYLSRRGGALQAVREKYKHHKFQCVATMPVSPEL 362
>At1g44110 mitotic cyclin a2-type, putative
Length = 460
Score = 180 bits (456), Expect = 2e-45
Identities = 103/271 (38%), Positives = 163/271 (60%), Gaps = 9/271 (3%)
Query: 170 KEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHC-YMAS-QPEINEKMRAILVD 227
K +IV+ID+++ D +L A + DIYK + E + RP YM Q ++N MR ILVD
Sbjct: 175 KNQIVNIDSNNGDPQLCAT-FACDIYKHLRASEAKKRPDVDYMERVQKDVNSSMRGILVD 233
Query: 228 WLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVN 287
WLI+V ++ L ETLYLT+N +DR+L+ + R++LQL+G++ M++AAKYEEI P+V
Sbjct: 234 WLIEVSEEYRLVPETLYLTVNYIDRYLSGNVISRQKLQLLGVACMMIAAKYEEICAPQVE 293
Query: 288 DFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKAS-----VPDEGVTNM 342
+F ++D Y +++L ME +L L++ +T PT FL RF++A+ P + M
Sbjct: 294 EFCYITDNTYLKDEVLDMESDVLNYLKFEMTAPTTKCFLRRFVRAAHGVHEAPLMQLECM 353
Query: 343 AHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKS-PAWNETLKLHTDYSEEQLMDCA 401
A++++EL ++ Y L + PS++AASA++ A+ L+ + WN TL+ +T Y +L C
Sbjct: 354 ANYIAELSLLEYTMLSHSPSLVAASAIFLAKYILDPTRRPWNSTLQHYTQYKAMELRGCV 413
Query: 402 RLLVSFHCTVGNGKLKVVFRKYSDPERGAVA 432
+ L T L V KYS + VA
Sbjct: 414 KDLQRLCSTAHGSTLPAVREKYSQHKYKFVA 444
>At5g43080 cyclin A-type
Length = 355
Score = 175 bits (444), Expect = 4e-44
Identities = 113/352 (32%), Positives = 192/352 (54%), Gaps = 31/352 (8%)
Query: 100 AVAKREAPKPVT---KRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLT 156
A KR+A +R+ K + E+ +S ++ +K + QKKK S + ++ T
Sbjct: 14 AATKRKASMEAAIDKERINKKRVVLGELPNLSNIKKSRKATTKQKKKSVS----IPTIET 69
Query: 157 ARSKAACGLTNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHC-YMAS-Q 214
S DID D ++ Y+ I+++ + +E +SRP Y+ Q
Sbjct: 70 LNS---------------DIDTRSDDPQMCG-PYVTSIFEYLRQLEVKSRPLVDYIEKIQ 113
Query: 215 PEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLM 274
++ MR +LVDWL++V +++L +TLYL ++ +DRFL++KTV ++ LQL+G++SML+
Sbjct: 114 KDVTSNMRGVLVDWLVEVAEEYKLLSDTLYLAVSYIDRFLSLKTVNKQRLQLLGVTSMLI 173
Query: 275 AAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASV 334
A+KYEEI PP V+DF ++D Y+ ++I+ ME IL L++ L PT FL RF + +
Sbjct: 174 ASKYEEITPPNVDDFCYITDNTYTKQEIVKMEADILLALQFELGNPTSNTFLRRFTRVAQ 233
Query: 335 PDEGVTN-----MAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLN-KSPAWNETLKL 388
D +++ + +LSEL M+ Y ++ + PS +AASAV+ AR + K WN L+
Sbjct: 234 EDFEMSHLQMEFLCSYLSELSMLDYQSVKFLPSTVAASAVFLARFIIRPKQHPWNVMLEE 293
Query: 389 HTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
+T Y L +C ++ + + G L+ + KY + VA +P + L
Sbjct: 294 YTRYKAGDLKECVAMIHDLYLSRKCGALEAIREKYKQHKFKCVATMPVSPEL 345
>At5g25380 cyclin 3a
Length = 444
Score = 172 bits (435), Expect = 4e-43
Identities = 118/373 (31%), Positives = 192/373 (50%), Gaps = 34/373 (9%)
Query: 70 AQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEISP 129
A++ ++ E N K AC + G+ E K + + ++E S
Sbjct: 86 AESIISTEGNVKVACKRGGKETKQIEEDGLVDVDGEKSK------LAEDLSKIRMVE-SL 138
Query: 130 DEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNELAAVE 189
D K+K V +ED + +T + +IVDID+ D + ++
Sbjct: 139 DASASKQKLVDCAEED--RSDVTDCV----------------QIVDIDSGVQDPQFCSL- 179
Query: 190 YIEDIYKFYKMVENESRPHC-YMAS-QPEINEKMRAILVDWLIDVHTKFELSLETLYLTI 247
Y IY + E E RP YM Q +I+ MR IL+DWL++V +++L +TLYLT+
Sbjct: 180 YAASIYDSINVAELEQRPSTSYMVQVQRDIDPTMRGILIDWLVEVSEEYKLVSDTLYLTV 239
Query: 248 NIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEK 307
N++DRF++ + +++LQL+GI+ ML+A+KYEEI P + +F ++D Y+ ++L ME
Sbjct: 240 NLIDRFMSHNYIEKQKLQLLGITCMLIASKYEEISAPRLEEFCFITDNTYTRLEVLSMEI 299
Query: 308 IILGRLEWTLTVPTPFVFLTRFIKAS-----VPDEGVTNMAHFLSELGMMHYDTLMYCPS 362
+L L + L+VPT FL RFI+A+ VP + +A++ +EL + Y L + PS
Sbjct: 300 KVLNSLHFRLSVPTTKTFLRRFIRAAQASDKVPLIEMEYLANYFAELTLTEYTFLRFLPS 359
Query: 363 MIAASAVYAARCTLNKS-PAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFR 421
+IAASAV+ AR TL++S WN+TL+ +T Y L + + L +
Sbjct: 360 LIAASAVFLARWTLDQSNHPWNQTLQHYTRYETSALKNTVLAMEELQLNTSGSTLIAIHT 419
Query: 422 KYSDPERGAVAVL 434
KY+ + VA L
Sbjct: 420 KYNQQKFKRVATL 432
>At1g80370 putative cyclin
Length = 461
Score = 170 bits (431), Expect = 1e-42
Identities = 105/280 (37%), Positives = 159/280 (56%), Gaps = 11/280 (3%)
Query: 162 ACGLTNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPH--CYMASQPEINE 219
+CG + PK VDID+ D D L ++ Y DIY ++ E + RP +Q ++ E
Sbjct: 172 SCGGASSPK--FVDIDSDDKDPLLCSL-YAPDIYYNLRVAELKRRPFPDFMEKTQRDVTE 228
Query: 220 KMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYE 279
MR ILVDWL++V ++ L +TLYLT+ ++D FL V R+ LQL+GI+ ML+A+KYE
Sbjct: 229 TMRGILVDWLVEVSEEYTLVPDTLYLTVYLIDWFLHGNYVERQRLQLLGITCMLIASKYE 288
Query: 280 EIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKA---SVPD 336
EI P + +F ++D Y+ +Q+L ME +L + + PT FL RF++A S P+
Sbjct: 289 EIHAPRIEEFCFITDNTYTRDQVLEMESQVLKHFSFQIYTPTSKTFLRRFLRAAQVSFPN 348
Query: 337 EGVTN--MAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSP-AWNETLKLHTDYS 393
+ + +A++L+EL +M Y L + PS+IAASAV+ A+ TLN+S WN TL+ +T Y
Sbjct: 349 QSLEMEFLANYLTELTLMDYPFLKFLPSIIAASAVFLAKWTLNQSSHPWNPTLEHYTTYK 408
Query: 394 EEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAV 433
L L L + KY + +VAV
Sbjct: 409 ASDLKASVHALQDLQLNTKGCSLNSIRMKYRQDKFKSVAV 448
>At1g47230 cyclin like protein
Length = 369
Score = 170 bits (430), Expect = 2e-42
Identities = 116/361 (32%), Positives = 189/361 (52%), Gaps = 38/361 (10%)
Query: 100 AVAKREAPK------PVTKR--VTGKPKPVVEVIEISPDEQ--IKKEKSVQKKKEDSKKK 149
A AKR+A PV+K+ V G+ + V+ + E+ +K + SV K KK
Sbjct: 13 AAAKRKASSMALDENPVSKKRVVLGELPNMSNVVAVPNQERETLKAKTSVNTSKRQMKKA 72
Query: 150 TLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESR--- 206
+ E V I++ VD ++ + DI + + +E + +
Sbjct: 73 LMIP-----------------EASVLIESRSVDPQMCE-PFASDICAYLREMEGKPKHRP 114
Query: 207 -PHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQ 265
P Q ++ MRA+LVDWL++V +++L +TLYLTI+ VDRFL+VK + R++LQ
Sbjct: 115 LPDYIEKVQSDLTPHMRAVLVDWLVEVAEEYKLVSDTLYLTISYVDRFLSVKPINRQKLQ 174
Query: 266 LVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVF 325
LVG+S+ML+A+KYEEI PP+V DF ++D ++ ++++ ME IL L++ L PT F
Sbjct: 175 LVGVSAMLIASKYEEIGPPKVEDFCYITDNTFTKQEVVSMEADILLALQFELGSPTIKTF 234
Query: 326 LTRFIKASVPD-----EGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLN-KS 379
L RF + + D + + +LSEL M+ Y + Y PS+++ASAV+ AR + K
Sbjct: 235 LRRFTRVAQEDFKDSQLQIEFLCCYLSELSMLDYTCVKYLPSLLSASAVFLARFIIRPKQ 294
Query: 380 PAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKN 439
WN+ L+ +T Y L C ++ + + L+ V KY + VA +P +
Sbjct: 295 HPWNQMLEEYTKYKAADLQVCVGIIHDLYLSRRGNTLEAVRNKYKQHKYKCVATMPVSPE 354
Query: 440 L 440
L
Sbjct: 355 L 355
>At1g47220 cyclin, putative
Length = 327
Score = 169 bits (427), Expect = 4e-42
Identities = 105/277 (37%), Positives = 160/277 (56%), Gaps = 13/277 (4%)
Query: 175 DIDASDVDNELAAVEYIEDIYKFYKMVENES--RP-HCYMAS-QPEINEKMRAILVDWLI 230
DIDA D ++ + Y+ DIY++ + +E + RP H Y+ Q +I R +LVDWL+
Sbjct: 42 DIDARSDDPQMCGL-YVSDIYEYLRELEVKPKLRPLHDYIEKIQEDITPSKRGVLVDWLV 100
Query: 231 DVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFV 290
+V +FEL ETLYLT++ +DRFL++K V LQLVG+S+M +A+KYEE P+V DF
Sbjct: 101 EVAEEFELVSETLYLTVSYIDRFLSLKMVNEHWLQLVGVSAMFIASKYEEKRRPKVEDFC 160
Query: 291 CLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTN-----MAHF 345
++ Y+ + +L ME+ IL LE+ L PT FL RFI+ + D V N + +
Sbjct: 161 YITANTYTKQDVLKMEEDILLALEFELGRPTTNTFLRRFIRVAQEDFKVPNLQLEPLCCY 220
Query: 346 LSELGMMHYDTLMYCPSMIAASAVYAARCTL--NKSPAWNETLKLHTDYSEEQLMDCARL 403
LSEL M+ Y + + PS++AASAV+ AR + N+ P W++ L+ T Y L C +
Sbjct: 221 LSELSMLDYSCVKFVPSLLAASAVFLARFIILPNQHP-WSQMLEECTKYKAADLQVCVEI 279
Query: 404 LVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
++ + + G K V KY + VA +P + L
Sbjct: 280 MLDLYLSRSEGASKAVREKYKQHKFQYVAAIPVYQEL 316
>At1g15570 putative cyclin (At1g15570)
Length = 450
Score = 167 bits (422), Expect = 1e-41
Identities = 104/297 (35%), Positives = 162/297 (54%), Gaps = 10/297 (3%)
Query: 153 SVLTARSKAACGLTNKPK-EEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRP--HC 209
S TA A G + P + VDID+ D D L + Y +I+ ++ E + RP
Sbjct: 151 SASTAEKSAVIGSSTVPDIPKFVDIDSDDKDPLLCCL-YAPEIHYNLRVSELKRRPLPDF 209
Query: 210 YMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGI 269
Q ++ + MR ILVDWL++V ++ L+ +TLYLT+ ++D FL V R++LQL+GI
Sbjct: 210 MERIQKDVTQSMRGILVDWLVEVSEEYTLASDTLYLTVYLIDWFLHGNYVQRQQLQLLGI 269
Query: 270 SSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRF 329
+ ML+A+KYEEI P + +F ++D Y+ +Q+L ME +L + + PTP FL RF
Sbjct: 270 TCMLIASKYEEISAPRIEEFCFITDNTYTRDQVLEMENQVLKHFSFQIYTPTPKTFLRRF 329
Query: 330 IKAS-----VPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKS-PAWN 383
++A+ P V +A +L+EL ++ Y L + PS++AASAV+ A+ T+++S WN
Sbjct: 330 LRAAQASRLSPSLEVEFLASYLTELTLIDYHFLKFLPSVVAASAVFLAKWTMDQSNHPWN 389
Query: 384 ETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
TL+ +T Y L L L + KY + +VAVL K L
Sbjct: 390 PTLEHYTTYKASDLKASVHALQDLQLNTKGCPLSAIRMKYRQEKYKSVAVLTSPKLL 446
>At1g20590 hypothetical protein
Length = 199
Score = 165 bits (417), Expect = 5e-41
Identities = 75/164 (45%), Positives = 118/164 (71%)
Query: 253 FLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGR 312
FLAV + R++LQLVG++++L+A KYEE+ P V+D + +SD+AYS ++L MEK++
Sbjct: 3 FLAVHQIVRKKLQLVGVTALLLACKYEEVSVPVVDDLILISDKAYSRREVLDMEKLMANT 62
Query: 313 LEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAA 372
L++ ++PTP+VF+ RF+KA+ D+ + ++ F+ EL ++ Y+ L Y PS +AASA+Y A
Sbjct: 63 LQFNFSLPTPYVFMKRFLKAAQSDKKLEILSFFMIELCLVEYEMLEYLPSKLAASAIYTA 122
Query: 373 RCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKL 416
+CTL W++T + HT Y+EEQL+ CAR +V+FH G GKL
Sbjct: 123 QCTLKGFEEWSKTCEFHTGYNEEQLLACARKMVAFHHKAGTGKL 166
>At5g11300 cyclin 3b
Length = 434
Score = 161 bits (408), Expect = 6e-40
Identities = 99/275 (36%), Positives = 159/275 (57%), Gaps = 7/275 (2%)
Query: 172 EIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHC-YMAS-QPEINEKMRAILVDWL 229
++VDID++ D + ++ Y DIY + E + RP YM Q +I+ MR IL+DWL
Sbjct: 155 QVVDIDSNVEDPQCCSL-YAADIYDNIHVAELQQRPLANYMELVQRDIDPDMRKILIDWL 213
Query: 230 IDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDF 289
++V ++L +TLYLT+N++DRFL+ + R+ LQL+G+S ML+A+KYEE+ P V +F
Sbjct: 214 VEVSDDYKLVPDTLYLTVNLIDRFLSNSYIERQRLQLLGVSCMLIASKYEELSAPGVEEF 273
Query: 290 VCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTR---FIKASVPDEGVTNMAHFL 346
++ Y+ ++L ME IL + + L+VPT FL+ I VP + +A++L
Sbjct: 274 CFITANTYTRPEVLSMEIQILNFVHFRLSVPTTKTFLSALFLIIILQVPFIELEYLANYL 333
Query: 347 SELGMMHYDTLMYCPSMIAASAVYAARCTLNKSP-AWNETLKLHTDYSEEQLMDCARLLV 405
+EL ++ Y L + PS+IAASAV+ AR TL+++ WN TL+ +T Y +L + +
Sbjct: 334 AELTLVEYSFLRFLPSLIAASAVFLARWTLDQTDHPWNPTLQHYTRYEVAELKNTVLAME 393
Query: 406 SFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
L KY+ P+ +VA L K +
Sbjct: 394 DLQLNTSGCTLAATREKYNQPKFKSVAKLTSPKRV 428
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,818,525
Number of Sequences: 26719
Number of extensions: 421659
Number of successful extensions: 2047
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 45
Number of HSP's that attempted gapping in prelim test: 1862
Number of HSP's gapped (non-prelim): 134
length of query: 447
length of database: 11,318,596
effective HSP length: 102
effective length of query: 345
effective length of database: 8,593,258
effective search space: 2964674010
effective search space used: 2964674010
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0269a.2


