

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0266.9
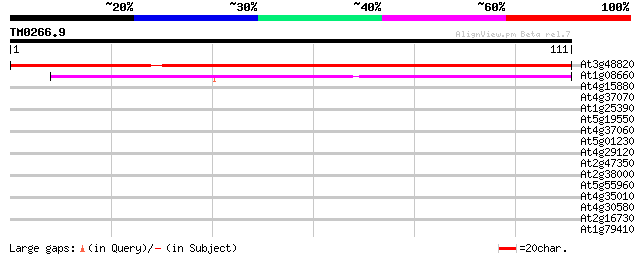
(111 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g48820 unknown protein 149 2e-37
At1g08660 unknown protein 60 2e-10
At4g15880 unknown protein 28 0.57
At4g37070 patatin-like protein 28 0.75
At1g25390 putative protein 27 1.3
At5g19550 aspartate aminotransferase (Asp2) 27 2.2
At4g37060 patatin-like protein 27 2.2
At5g01230 cell division - like protein 26 3.7
At4g29120 unknown protein 26 3.7
At2g47350 unknown protein 25 6.3
At2g38000 unknown protein 25 6.3
At5g55960 unknown protein 25 8.3
At4g35010 beta-galactosidase - like protein 25 8.3
At4g30580 unknown protein 25 8.3
At2g16730 putative beta-galactosidase 25 8.3
At1g79410 unknown protein 25 8.3
>At3g48820 unknown protein
Length = 440
Score = 149 bits (376), Expect = 2e-37
Identities = 71/111 (63%), Positives = 90/111 (80%), Gaps = 2/111 (1%)
Query: 1 MRVLQLGLLLALASGFAAISIYIIGLSDPSVYPTYHLTDEDTQALLSLHNTFQKCVSANG 60
M++L L LLAL +G +A+ IYIIG+S+ +Y + T+ED +AL SL N FQKCVSANG
Sbjct: 1 MKLLHLIFLLALTTGISAVLIYIIGVSN--LYESNRFTNEDLEALQSLQNGFQKCVSANG 58
Query: 61 LGLKAAMSSDYCQTTINFPSDTIPKWKDPKTGELEALSFDFNLCEAVATWE 111
LGL+AAM DYC+ +INFP DT+PKWKDPK+GELE LS++F+LCEAVATWE
Sbjct: 59 LGLQAAMGRDYCKVSINFPKDTVPKWKDPKSGELEGLSYEFDLCEAVATWE 109
>At1g08660 unknown protein
Length = 474
Score = 60.1 bits (144), Expect = 2e-10
Identities = 35/106 (33%), Positives = 50/106 (47%), Gaps = 4/106 (3%)
Query: 9 LLALASGFAAISIYIIGLSDPSVYPTYHLTD---EDTQALLSLHNTFQKCVSANGLGLKA 65
LL L A S+++ + D ED Q L ++ Q+CV+ GLGL A
Sbjct: 12 LLQLLGCVAVFSVFVFTIQSSFFADNNRKLDLQPEDIQILSDFQSSVQQCVANRGLGLSA 71
Query: 66 AMSSDYCQTTINFPSDTIPKWKDPKTGELEALSFDFNLCEAVATWE 111
+ D+C + FP T W + + EAL F +N+CEAV WE
Sbjct: 72 HII-DHCNLILKFPEGTNSTWYNAQFKVFEALEFKYNVCEAVLLWE 116
>At4g15880 unknown protein
Length = 489
Score = 28.5 bits (62), Expect = 0.57
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 3/34 (8%)
Query: 66 AMSSDYCQTTINFPSDTIPKWKDPKTGELEALSF 99
A S+DYC+ NF + K+ D K LEAL F
Sbjct: 88 AKSNDYCEKDANF---FVRKYDDAKRSALEALRF 118
>At4g37070 patatin-like protein
Length = 383
Score = 28.1 bits (61), Expect = 0.75
Identities = 26/92 (28%), Positives = 43/92 (46%), Gaps = 12/92 (13%)
Query: 19 ISIYIIGLSD-PSVYPTYHLTDEDTQALLSLHNTFQKCVSANGLGLKAAMSSDYCQTTIN 77
+S IG S P+ +P ++ ++ED+Q + N V+AN L AM++ Q N
Sbjct: 180 VSDICIGTSAAPTFFPPHYFSNEDSQGNKTEFNLVDGAVTANNPTL-VAMTAVSKQIVKN 238
Query: 78 FPSDTIPKWKDPKTGELEALSFDFNLCEAVAT 109
+P G+L+ L FD L ++ T
Sbjct: 239 ----------NPDMGKLKPLGFDRFLVISIGT 260
>At1g25390 putative protein
Length = 629
Score = 27.3 bits (59), Expect = 1.3
Identities = 21/70 (30%), Positives = 32/70 (45%), Gaps = 7/70 (10%)
Query: 25 GLSDPSVYPTYHLTDEDTQALLSLHNTFQKCVSANGLGLKAAMSSDYCQTTINFPSDTIP 84
G DP + YHLTD+ + S + +S+ K A+ C++ IN S I
Sbjct: 457 GYVDPEYHRCYHLTDKSD--VYSFGVVLVELISS-----KPAVDISRCKSEINLSSLAIN 509
Query: 85 KWKDPKTGEL 94
K ++ T EL
Sbjct: 510 KIQNHATHEL 519
>At5g19550 aspartate aminotransferase (Asp2)
Length = 405
Score = 26.6 bits (57), Expect = 2.2
Identities = 23/84 (27%), Positives = 36/84 (42%), Gaps = 6/84 (7%)
Query: 21 IYIIGLSDPSVYPTYHLTDEDTQALLSLHNTFQKCVSANGLGLKAA--MSSDYCQTTINF 78
I I+G+SD + + D+ A+ T +C+S G A + + Y Q+ I
Sbjct: 70 IPIVGISDFNKLSAKLILGADSPAITESRVTTVQCLSGTGSLRVGAEFLKTHYHQSVIYI 129
Query: 79 PSDTIPKW-KDPKTGELEALSFDF 101
P P W PK L LS ++
Sbjct: 130 PK---PTWGNHPKVFNLAGLSVEY 150
>At4g37060 patatin-like protein
Length = 414
Score = 26.6 bits (57), Expect = 2.2
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 12/92 (13%)
Query: 19 ISIYIIGLSD-PSVYPTYHLTDEDTQALLSLHNTFQKCVSANGLGLKAAMSSDYCQTTIN 77
+S IG S P+ +P Y+ ++ED+Q N V+AN L AM++ Q N
Sbjct: 180 VSDICIGTSAAPTYFPPYYFSNEDSQGKTRHFNLVDGGVTANNPTL-VAMTAVTKQIVNN 238
Query: 78 FPSDTIPKWKDPKTGELEALSFDFNLCEAVAT 109
+P G L L +D L ++ T
Sbjct: 239 ----------NPDMGTLNPLGYDQFLVISIGT 260
>At5g01230 cell division - like protein
Length = 309
Score = 25.8 bits (55), Expect = 3.7
Identities = 13/35 (37%), Positives = 18/35 (51%), Gaps = 4/35 (11%)
Query: 71 YCQTTINFPSDTIPKWKDPKTGELEALSFDFNLCE 105
YCQ + FP+ T K K + +EA F +CE
Sbjct: 177 YCQLKLFFPTVTFAKPKSSRNSSIEA----FAVCE 207
>At4g29120 unknown protein
Length = 334
Score = 25.8 bits (55), Expect = 3.7
Identities = 13/32 (40%), Positives = 16/32 (49%)
Query: 66 AMSSDYCQTTINFPSDTIPKWKDPKTGELEAL 97
A SD T + +PSD DPK+G L L
Sbjct: 91 AEQSDVVFTIVGYPSDVRHVLLDPKSGALSGL 122
>At2g47350 unknown protein
Length = 486
Score = 25.0 bits (53), Expect = 6.3
Identities = 9/17 (52%), Positives = 12/17 (69%)
Query: 74 TTINFPSDTIPKWKDPK 90
TT++FP D +P DPK
Sbjct: 422 TTVSFPIDKVPSLFDPK 438
>At2g38000 unknown protein
Length = 419
Score = 25.0 bits (53), Expect = 6.3
Identities = 10/31 (32%), Positives = 16/31 (51%)
Query: 32 YPTYHLTDEDTQALLSLHNTFQKCVSANGLG 62
+PT + ++TQ L+ T +KC G G
Sbjct: 178 FPTLFVPYKETQVLVPNSETVEKCTGCTGRG 208
>At5g55960 unknown protein
Length = 648
Score = 24.6 bits (52), Expect = 8.3
Identities = 17/89 (19%), Positives = 43/89 (48%), Gaps = 3/89 (3%)
Query: 19 ISIYIIGLSDPSVYPTYHLTDEDTQALLSLHNTFQKCVSANGLGLKAAMSSDYCQTTINF 78
I + I+G ++ +Y + E A+ SL + ++ A +G+K M + ++
Sbjct: 256 IVLMIVGSLTGVIFFSYKIGVEGKDAVYSLKSHVEESNYAEKIGIKQWMDENDVPGMVDM 315
Query: 79 PSDTIPKWKDPKTGELEALSFDFNLCEAV 107
+ K+ + + ++++L+ +N+ E V
Sbjct: 316 YT---TKFYETVSEQIDSLAMQYNMTELV 341
>At4g35010 beta-galactosidase - like protein
Length = 831
Score = 24.6 bits (52), Expect = 8.3
Identities = 12/32 (37%), Positives = 15/32 (46%)
Query: 79 PSDTIPKWKDPKTGELEALSFDFNLCEAVATW 110
P D K+PK G L+ L NLC+ W
Sbjct: 308 PLDEYGLEKEPKYGHLKHLHNALNLCKKPLLW 339
>At4g30580 unknown protein
Length = 356
Score = 24.6 bits (52), Expect = 8.3
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 13/62 (20%)
Query: 12 LASGFAAISIY------IIGLSD------PSVYPTYHLTDEDTQALLSLHNTFQKCVSAN 59
+A +A+ISIY I GL + P+VY + H + D LLSL +F K +S
Sbjct: 166 IAKLWASISIYPFYKINIEGLENLPSSDTPAVYVSNHQSFLDIYTLLSLGKSF-KFISKT 224
Query: 60 GL 61
G+
Sbjct: 225 GI 226
>At2g16730 putative beta-galactosidase
Length = 832
Score = 24.6 bits (52), Expect = 8.3
Identities = 11/32 (34%), Positives = 15/32 (46%)
Query: 79 PSDTIPKWKDPKTGELEALSFDFNLCEAVATW 110
P D ++PK G L+ L NLC+ W
Sbjct: 319 PLDEFGLEREPKYGHLKHLHNALNLCKKALLW 350
>At1g79410 unknown protein
Length = 515
Score = 24.6 bits (52), Expect = 8.3
Identities = 17/61 (27%), Positives = 26/61 (41%), Gaps = 7/61 (11%)
Query: 33 PTYHLTDEDTQALLSLHNTFQKCVSANGLG-------LKAAMSSDYCQTTINFPSDTIPK 85
PT+ DEDT + L+ +K +S G + A++ D Q I +D P
Sbjct: 10 PTHIEEDEDTSSPLTFDKILEKSLSDFGFSQFLQIVLVGLALTFDSQQIFITVFTDAYPT 69
Query: 86 W 86
W
Sbjct: 70 W 70
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,441,696
Number of Sequences: 26719
Number of extensions: 84391
Number of successful extensions: 202
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 191
Number of HSP's gapped (non-prelim): 17
length of query: 111
length of database: 11,318,596
effective HSP length: 87
effective length of query: 24
effective length of database: 8,994,043
effective search space: 215857032
effective search space used: 215857032
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0266.9


