

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0266.20
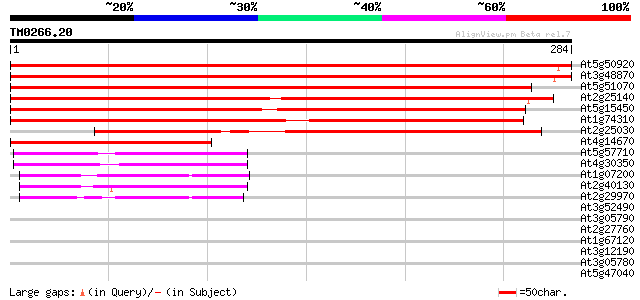
(284 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g50920 ATP-dependent Clp protease, ATP-binding subunit 506 e-144
At3g48870 AtClpC 494 e-140
At5g51070 Erd1 protein precursor (sp|P42762) 334 4e-92
At2g25140 putative ATP-dependent CLPB protein 295 1e-80
At5g15450 clpB heat shock protein-like 288 2e-78
At1g74310 heat shock protein 101 259 1e-69
At2g25030 putative ATP-dependent CLPB protein 167 6e-42
At4g14670 heat shock protein like 145 2e-35
At5g57710 101 kDa heat shock protein; HSP101-like protein 63 2e-10
At4g30350 unknown protein 62 4e-10
At1g07200 unknown protein 54 7e-08
At2g40130 unknown protein 53 2e-07
At2g29970 unknown protein 45 6e-05
At3g52490 putative protein 39 0.004
At3g05790 putative mitochondrial LON ATP-dependent protease 35 0.035
At2g27760 tRNA isopentenyl transferase (IPT2) 35 0.046
At1g67120 hypothetical protein 35 0.060
At3g12190 hypothetical protein 33 0.23
At3g05780 putative mitochondrial LON ATP-dependent protease 32 0.30
At5g47040 Lon protease homolog 1 precursor 32 0.51
>At5g50920 ATP-dependent Clp protease, ATP-binding subunit
Length = 929
Score = 506 bits (1304), Expect = e-144
Identities = 258/288 (89%), Positives = 272/288 (93%), Gaps = 4/288 (1%)
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SFIFSGPTGVGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG
Sbjct: 640 SFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 699
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN
Sbjct: 700 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 759
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
VGSSVIEKGGR+IGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK
Sbjct: 760 VGSSVIEKGGRRIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 819
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
LEVKEIADI+LKEVF+RLK KEI+L VTERF++RVV+EGY+PSYGARPLRRAIMRLLEDS
Sbjct: 820 LEVKEIADILLKEVFERLKKKEIELQVTERFKERVVDEGYNPSYGARPLRRAIMRLLEDS 879
Query: 241 MAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSGAP----ESLPEALP 284
MAEKMLA EIKEGDSVI+D D++G V VLNG SG P E ++LP
Sbjct: 880 MAEKMLAREIKEGDSVIVDVDAEGNVTVLNGGSGTPTTSLEEQEDSLP 927
>At3g48870 AtClpC
Length = 952
Score = 494 bits (1272), Expect = e-140
Identities = 250/290 (86%), Positives = 273/290 (93%), Gaps = 6/290 (2%)
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SFIFSGPTGVGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG
Sbjct: 661 SFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 720
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPYT+VLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN
Sbjct: 721 GQLTEAVRRRPYTLVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 780
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
VGSSVIEKGGR+IGFDLD+DEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK
Sbjct: 781 VGSSVIEKGGRRIGFDLDHDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 840
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
LEVKEIADIMLKEV RL+ KEI+L VTERF++RVV+EG+DPSYGARPLRRAIMRLLEDS
Sbjct: 841 LEVKEIADIMLKEVVARLEVKEIELQVTERFKERVVDEGFDPSYGARPLRRAIMRLLEDS 900
Query: 241 MAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSG------APESLPEALP 284
MAEKML+ +IKEGDSVI+D D++G V+VL+G++G A E++ + +P
Sbjct: 901 MAEKMLSRDIKEGDSVIVDVDAEGSVVVLSGTTGRVGGFAAEEAMEDPIP 950
>At5g51070 Erd1 protein precursor (sp|P42762)
Length = 945
Score = 334 bits (856), Expect = 4e-92
Identities = 161/265 (60%), Positives = 207/265 (77%), Gaps = 1/265 (0%)
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
+ +F GPTGVGK+EL K LA+ YFGSEE+M+RLDMSE+MERHTVSKLIGSPPGYVG+ EG
Sbjct: 659 AMLFCGPTGVGKTELTKALAANYFGSEESMLRLDMSEYMERHTVSKLIGSPPGYVGFEEG 718
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
G LTEA+RRRP+TVVLFDEIEKAHPD+FN++LQ+ EDG LTDS+GR V FKN L+IMTSN
Sbjct: 719 GMLTEAIRRRPFTVVLFDEIEKAHPDIFNILLQLFEDGHLTDSQGRRVSFKNALIIMTSN 778
Query: 121 VGSSVIEKGGR-KIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLT 179
VGS I KG IGF LD DE+ +SY +K+LV EELK YFRPE LNR+DE+++FRQL
Sbjct: 779 VGSLAIAKGRHGSIGFILDDDEEAASYTGMKALVVEELKNYFRPELLNRIDEIVIFRQLE 838
Query: 180 KLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLED 239
K ++ EI ++ML+++ RL + L V+E ++ + ++GYDP+YGARPLRR + ++ED
Sbjct: 839 KAQMMEILNLMLQDLKSRLVALGVGLEVSEPVKELICKQGYDPAYGARPLRRTVTEIVED 898
Query: 240 SMAEKMLAGEIKEGDSVIIDADSDG 264
++E LAG K GD+ + D G
Sbjct: 899 PLSEAFLAGSFKPGDTAFVVLDDTG 923
>At2g25140 putative ATP-dependent CLPB protein
Length = 964
Score = 295 bits (756), Expect = 1e-80
Identities = 146/278 (52%), Positives = 206/278 (73%), Gaps = 8/278 (2%)
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SF+F GPTGVGK+ELAK LA Y F +E A++R+DMSE+ME+H+VS+L+G+PPGYVGY EG
Sbjct: 685 SFMFMGPTGVGKTELAKALAGYLFNTENAIVRVDMSEYMEKHSVSRLVGAPPGYVGYEEG 744
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTE VRRRPY+VVLFDEIEKAHPDVFN++LQ+L+DGR+TDS+GRTV FKN ++IMTSN
Sbjct: 745 GQLTEVVRRRPYSVVLFDEIEKAHPDVFNILLQLLDDGRITDSQGRTVSFKNCVVIMTSN 804
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
+GS I + R + D K++ Y +K V E +Q FRPEF+NR+DE IVF+ L
Sbjct: 805 IGSHHILETLRN-----NEDSKEAVYEIMKRQVVELARQNFRPEFMNRIDEYIVFQPLDS 859
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
E+ +I ++ ++ V + L+ K+I L T+ D + + G+DP+YGARP++R I +++E+
Sbjct: 860 NEISKIVELQMRRVKNSLEQKKIKLQYTKEAVDLLAQLGFDPNYGARPVKRVIQQMVENE 919
Query: 241 MAEKMLAGEIKEGDSVIIDAD---SDGKVIVLNGSSGA 275
+A +L G+ E D+V++D D SD K+++ S A
Sbjct: 920 IAVGILKGDFAEEDTVLVDVDHLASDNKLVIKKLESNA 957
>At5g15450 clpB heat shock protein-like
Length = 968
Score = 288 bits (738), Expect = 2e-78
Identities = 141/261 (54%), Positives = 191/261 (73%), Gaps = 7/261 (2%)
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SF+F GPTGVGK+ELAK LASY F +EEA++R+DMSE+ME+H VS+LIG+PPGYVGY EG
Sbjct: 680 SFMFMGPTGVGKTELAKALASYMFNTEEALVRIDMSEYMEKHAVSRLIGAPPGYVGYEEG 739
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTE VRRRPY+V+LFDEIEKAH DVFN+ LQIL+DGR+TDS+GRTV F NT++IMTSN
Sbjct: 740 GQLTETVRRRPYSVILFDEIEKAHGDVFNVFLQILDDGRVTDSQGRTVSFTNTVIIMTSN 799
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
VGS I + D D + SY IK V + FRPEF+NR+DE IVF+ L +
Sbjct: 800 VGSQFILN-------NTDDDANELSYETIKERVMNAARSIFRPEFMNRVDEYIVFKPLDR 852
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
++ I + L V R+ +++ +++T+ D + GYDP+YGARP++R I + +E+
Sbjct: 853 EQINRIVRLQLARVQKRIADRKMKINITDAAVDLLGSLGYDPNYGARPVKRVIQQNIENE 912
Query: 241 MAEKMLAGEIKEGDSVIIDAD 261
+A+ +L G+ KE D ++ID +
Sbjct: 913 LAKGILRGDFKEEDGILIDTE 933
>At1g74310 heat shock protein 101
Length = 911
Score = 259 bits (661), Expect = 1e-69
Identities = 130/261 (49%), Positives = 188/261 (71%), Gaps = 12/261 (4%)
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SF+F GPTGVGK+ELAK LA F E ++R+DMSE+ME+H+VS+LIG+PPGYVG+ EG
Sbjct: 601 SFLFLGPTGVGKTELAKALAEQLFDDENLLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEG 660
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPY V+LFDE+EKAH VFN +LQ+L+DGRLTD +GRTVDF+N+++IMTSN
Sbjct: 661 GQLTEAVRRRPYCVILFDEVEKAHVAVFNTLLQVLDDGRLTDGQGRTVDFRNSVIIMTSN 720
Query: 121 VGSSVIEKG-GRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLT 179
+G+ + G K+ ++ D V E++++FRPE LNRLDE++VF L+
Sbjct: 721 LGAEHLLAGLTGKVTMEVARD-----------CVMREVRKHFRPELLNRLDEIVVFDPLS 769
Query: 180 KLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLED 239
+++++A + +K+V RL + + L+VT+ D ++ E YDP YGARP+RR + + +
Sbjct: 770 HDQLRKVARLQMKDVAVRLAERGVALAVTDAALDYILAESYDPVYGARPIRRWMEKKVVT 829
Query: 240 SMAEKMLAGEIKEGDSVIIDA 260
+++ ++ EI E +V IDA
Sbjct: 830 ELSKMVVREEIDENSTVYIDA 850
>At2g25030 putative ATP-dependent CLPB protein
Length = 265
Score = 167 bits (423), Expect = 6e-42
Identities = 88/226 (38%), Positives = 142/226 (61%), Gaps = 22/226 (9%)
Query: 44 VSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDS 103
VS+L+G+ PGYVGY +GG+LTE VRRRPY+VV FDEIEK H DVF+++LQ+L+DGR+T+S
Sbjct: 50 VSQLVGASPGYVGYGDGGKLTEVVRRRPYSVVQFDEIEKPHLDVFSILLQLLDDGRITNS 109
Query: 104 KGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRP 163
G + L + +N D K++ Y +K V E ++ FRP
Sbjct: 110 HGSL----HILETIRNNE------------------DIKEAFYEMMKQQVVELARKTFRP 147
Query: 164 EFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPS 223
+F+NR+DE IV + L E+ +I ++ +++V RL+ +I+L T+ D + + G+DP+
Sbjct: 148 KFMNRIDEYIVSQPLNSSEISKIVELQMRQVKKRLEQNKINLEYTKEAVDLLAQLGFDPN 207
Query: 224 YGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVL 269
GARP+++ I +L++ + K+L G+ E +++IDAD +V+
Sbjct: 208 NGARPVKQMIEKLVKKEITLKVLKGDFAEDGTILIDADQPNNKLVI 253
>At4g14670 heat shock protein like
Length = 668
Score = 145 bits (366), Expect = 2e-35
Identities = 70/103 (67%), Positives = 85/103 (81%), Gaps = 1/103 (0%)
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGY-VGYTE 59
SF+F GPTGVGK+ELAK LA F SE ++RLDMSE+ ++ +V+KLIG+PPGY +G+ E
Sbjct: 566 SFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDMSEYNDKFSVNKLIGAPPGYYIGHEE 625
Query: 60 GGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTD 102
GGQLTE VRRRPY VVLFDE+EK H VFN +LQ+LEDGRLTD
Sbjct: 626 GGQLTEPVRRRPYCVVLFDEVEKTHVTVFNTLLQVLEDGRLTD 668
>At5g57710 101 kDa heat shock protein; HSP101-like protein
Length = 990
Score = 62.8 bits (151), Expect = 2e-10
Identities = 35/118 (29%), Positives = 63/118 (52%), Gaps = 8/118 (6%)
Query: 3 IFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQ 62
+FSGP VGK ++ L+S +G+ MI+L + S + G T +
Sbjct: 656 LFSGPDRVGKRKMVSALSSLVYGTNPIMIQLGSRQDAGDGNSS--------FRGKTALDK 707
Query: 63 LTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
+ E V+R P++V+L ++I++A V + Q ++ GR+ DS GR + N + +MT++
Sbjct: 708 IAETVKRSPFSVILLEDIDEADMLVRGSIKQAMDRGRIRDSHGREISLGNVIFVMTAS 765
>At4g30350 unknown protein
Length = 924
Score = 62.0 bits (149), Expect = 4e-10
Identities = 38/118 (32%), Positives = 62/118 (52%), Gaps = 9/118 (7%)
Query: 3 IFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQ 62
+F+GP GKS++A L+ GS+ I L S M+ + G T +
Sbjct: 612 MFTGPDRAGKSKMASALSDLVSGSQPITISLGSSSRMDDGLNIR---------GKTALDR 662
Query: 63 LTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
EAVRR P+ V++ ++I++A + N + +E GR+ DS GR V N ++I+T+N
Sbjct: 663 FAEAVRRNPFAVIVLEDIDEADILLRNNVKIAIERGRICDSYGREVSLGNVIIILTAN 720
>At1g07200 unknown protein
Length = 979
Score = 54.3 bits (129), Expect = 7e-08
Identities = 33/117 (28%), Positives = 60/117 (51%), Gaps = 10/117 (8%)
Query: 6 GPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTE 65
GP VGK ++A TL+ +FG + I +D ++ + G T +T
Sbjct: 665 GPDKVGKKKVAMTLSEVFFGGKVNYICVDFG--------AEHCSLDDKFRGKTVVDYVTG 716
Query: 66 AVRRRPYTVVLFDEIEKAH-PDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV 121
+ R+P++VVL + +EKA PD + + + G++ D GR + KN ++++TS +
Sbjct: 717 ELSRKPHSVVLLENVEKAEFPDQMRLS-EAVSTGKIRDLHGRVISMKNVIVVVTSGI 772
>At2g40130 unknown protein
Length = 907
Score = 52.8 bits (125), Expect = 2e-07
Identities = 35/118 (29%), Positives = 55/118 (45%), Gaps = 9/118 (7%)
Query: 6 GPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGS---PPGYVGYTEGGQ 62
GP VGK ++ LA + SE + +D+ + +G P G T
Sbjct: 577 GPDTVGKRRMSLVLAEIVYQSEHRFMAVDLG------AAEQGMGGCDDPMRLRGKTMVDH 630
Query: 63 LTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
+ E + R P+ VV + IEKA + + + +E G+ DS GR V NT+ +MTS+
Sbjct: 631 IFEVMCRNPFCVVFLENIEKADEKLQMSLSKAIETGKFMDSHGREVGIGNTIFVMTSS 688
>At2g29970 unknown protein
Length = 1002
Score = 44.7 bits (104), Expect = 6e-05
Identities = 32/114 (28%), Positives = 54/114 (47%), Gaps = 11/114 (9%)
Query: 6 GPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTE 65
GP GK ++A LA + G ++ I +D F + ++ + G T +
Sbjct: 679 GPDKAGKKKVALALAEVFCGGQDNFICVD---FKSQDSLDDR------FRGKTVVDYIAG 729
Query: 66 AVRRRPYTVVLFDEIEKAH-PDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMT 118
V RR +VV + +EKA PD + + + G+L DS GR + KN +++ T
Sbjct: 730 EVARRADSVVFIENVEKAEFPDQIRLS-EAMRTGKLRDSHGREISMKNVIVVAT 782
>At3g52490 putative protein
Length = 815
Score = 38.5 bits (88), Expect = 0.004
Identities = 31/121 (25%), Positives = 58/121 (47%), Gaps = 9/121 (7%)
Query: 4 FSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVS------KLIGSPPGYVGY 57
F G K ++A+ LA FGS+++ + + +S F + S K + + Y
Sbjct: 625 FQGLDVDAKEKIARELAKLVFGSQDSFVSICLSSFSSTRSDSAEDLRNKRLRDEQS-LSY 683
Query: 58 TEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIM 117
E + +EAV P V+L ++IE+A + +E GR+ +S G K+ ++I+
Sbjct: 684 IE--RFSEAVSLDPNRVILVEDIEQADYLSQVGFKRAVERGRVCNSSGEEASLKDAIVIL 741
Query: 118 T 118
+
Sbjct: 742 S 742
>At3g05790 putative mitochondrial LON ATP-dependent protease
Length = 942
Score = 35.4 bits (80), Expect = 0.035
Identities = 32/132 (24%), Positives = 61/132 (45%), Gaps = 18/132 (13%)
Query: 5 SGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLT 64
SGPTGVGK+ + +++A + R + + V+++ G Y+G G++
Sbjct: 455 SGPTGVGKTSIGRSIAR---ALDRKFFRFSVGGLSD---VAEIKGHRRTYIG-AMPGKMV 507
Query: 65 EAVRR--RPYTVVLFDEIEKA-----HPDVFNMMLQILEDGR----LTDSKGRTVDFKNT 113
+ ++ +VL DEI+K H D + ML++L+ + L +D
Sbjct: 508 QCLKNVGTENPLVLIDEIDKLGVRGHHGDPASAMLELLDPEQNANFLDHYLDVPIDLSKV 567
Query: 114 LLIMTSNVGSSV 125
L + T+NV ++
Sbjct: 568 LFVCTANVTDTI 579
>At2g27760 tRNA isopentenyl transferase (IPT2)
Length = 466
Score = 35.0 bits (79), Expect = 0.046
Identities = 25/82 (30%), Positives = 37/82 (44%), Gaps = 4/82 (4%)
Query: 3 IFSGPTGVGKSELAKTLASYY----FGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYT 58
+ GPTG GKS+LA LAS++ ++ I + + TV + G P +G
Sbjct: 24 VIMGPTGSGKSKLAVDLASHFPVEIINADAMQIYSGLDVLTNKVTVDEQKGVPHHLLGTV 83
Query: 59 EGGQLTEAVRRRPYTVVLFDEI 80
A R +TV L +EI
Sbjct: 84 SSDMEFTARDFRDFTVPLIEEI 105
>At1g67120 hypothetical protein
Length = 5138
Score = 34.7 bits (78), Expect = 0.060
Identities = 37/135 (27%), Positives = 61/135 (44%), Gaps = 13/135 (9%)
Query: 3 IFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFME-RHTVSKLIGSPPGYVGYTEGG 61
+ GPT GK+ L K LA+ S +R++ E + + + + G + + EG
Sbjct: 963 LLQGPTSSGKTSLVKYLAAI---SGNKFVRINNHEQTDIQEYLGSYMTDSSGKLVFHEGA 1019
Query: 62 QLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGR--LTDSKGRTVD-FKNTLLIMT 118
L +AVR + ++ DE+ A DV + ++L+D R T+ N +L T
Sbjct: 1020 -LVKAVRGGHW--IVLDELNLAPSDVLEALNRLLDDNRELFVPELSETISAHPNFMLFAT 1076
Query: 119 SNVGSSVIEKGGRKI 133
N + GGRKI
Sbjct: 1077 QNPPTLY---GGRKI 1088
Score = 27.3 bits (59), Expect = 9.6
Identities = 24/84 (28%), Positives = 44/84 (51%), Gaps = 8/84 (9%)
Query: 6 GPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTV--SKLIGSPPGYVGYTEGGQL 63
GP+G GKS L + LA S ++ + M + ++ T+ + + PG + + G L
Sbjct: 266 GPSGSGKSALIRKLAD---ESGNHVVFIHMDDQLDGKTLVGTYVCTDQPGEFRW-QPGSL 321
Query: 64 TEAVRRRPYTVVLFDEIEKAHPDV 87
T+A+ + V+ ++I+KA DV
Sbjct: 322 TQAIMNGFW--VVLEDIDKAPSDV 343
>At3g12190 hypothetical protein
Length = 269
Score = 32.7 bits (73), Expect = 0.23
Identities = 22/83 (26%), Positives = 42/83 (50%), Gaps = 4/83 (4%)
Query: 125 VIEKGGRKI----GFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
++EK R+I GF D++EK +R+K + E K+ F + LNR + + R +
Sbjct: 78 ILEKRAREINTADGFRRDFEEKQRKLDRLKREIESEEKKRFLVQKLNRERKFELKRTREQ 137
Query: 181 LEVKEIADIMLKEVFDRLKTKEI 203
+E + D+ L + ++E+
Sbjct: 138 VEALQKNDMKLDVKHSKEMSEEL 160
>At3g05780 putative mitochondrial LON ATP-dependent protease
Length = 924
Score = 32.3 bits (72), Expect = 0.30
Identities = 31/127 (24%), Positives = 60/127 (46%), Gaps = 17/127 (13%)
Query: 5 SGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLT 64
SGP GVGK+ + +++A + R + + V+++ G YVG G++
Sbjct: 446 SGPPGVGKTSIGRSIAR---ALDRKFFRFSVGGLSD---VAEIKGHCQTYVG-AMPGKMV 498
Query: 65 EAVRR--RPYTVVLFDEIEK---AHP-DVFNMMLQILEDGR----LTDSKGRTVDFKNTL 114
+ ++ ++LFDEI+K H D + +L++++ + L T+D L
Sbjct: 499 QCLKSVGTANPLILFDEIDKLGRCHTGDPASALLEVMDPEQNAKFLDHFLNVTIDLSKVL 558
Query: 115 LIMTSNV 121
+ T+NV
Sbjct: 559 FVCTANV 565
>At5g47040 Lon protease homolog 1 precursor
Length = 888
Score = 31.6 bits (70), Expect = 0.51
Identities = 26/86 (30%), Positives = 43/86 (49%), Gaps = 9/86 (10%)
Query: 4 FSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQL 63
F GP GVGK+ LA ++A+ +RL + + + + G Y+G + G+L
Sbjct: 406 FVGPPGVGKTSLASSIAA---ALGRKFVRLSLGGVKDE---ADIRGHRRTYIG-SMPGRL 458
Query: 64 TEAVRRRPY--TVVLFDEIEKAHPDV 87
+ ++R V+L DEI+K DV
Sbjct: 459 IDGLKRVGVCNPVMLLDEIDKTGSDV 484
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,140,680
Number of Sequences: 26719
Number of extensions: 255551
Number of successful extensions: 1083
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 1038
Number of HSP's gapped (non-prelim): 67
length of query: 284
length of database: 11,318,596
effective HSP length: 98
effective length of query: 186
effective length of database: 8,700,134
effective search space: 1618224924
effective search space used: 1618224924
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0266.20


